Abstract
Human endogenous retroviruses (HERVs) are a significant component of a wider family of retroelements that constitute part of the human genome. These viruses, perhaps representative of previous exogenous retroviral infection, have been integrated and passed through successive generations within the germ line. The retention of HERVs and isolated elements, such as long-terminal repeats, could have the potential to harm. In this review we describe HERVs within the context of the family of known transposable elements and survey these viruses in terms of superantigens and molecular mimics. It is entirely possible that these mechanisms provide the potential for undesired immune responses.
Keywords: autoimmunity, human endogenous retroviruses, molecular mimicry, superantigens, transposable elements
INTRODUCTION
Viruses are defined as small, infectious molecular parasites and classified on criteria including viral genome (either DNA or RNA), symmetry of the protective protein coat or capsid which may be icosahedral or helical, and the absence (i.e. naked) or presence of a surrounding envelope. At least 30 families of viruses are infectious to humans and other mammals and these microbial pathogens require the manufacture of progeny virions within a host cell for the horizontal transmission and subsequent attachment and disassembly in a permissible target cell for the next infectious cycle [1]. However, over the past 20 years, families of viruses that are transmitted vertically through the germ-line have been detected and classified within the human genome. These viruses, termed human endogenous retroviruses (HERVs), retain some of the hallmarks of exogenous retroviruses, e.g. human immunodeficiency virus (HIV), human T cell lymphotrophic virus (HTLV) such as genomic structure, i.e. group-associated antigen (gag), polymerase (pol) and envelope (env) genes sandwiched between long terminal repeat (LTR) regions [2,3]. So far as we know, HERVs are not infectious but many have been subjected to repeated amplification and transposition events giving rise to multi-copy proviruses within the DNA of all cells. Thus HERVs reflect past exogenous retroviral infection in which proviral DNA has been integrated, passed on and retained/trapped within the genome. It is estimated that HERVs constitute about 4·8% of the human genome, which is significant when bearing in mind that only 3% of human DNA appears to make up the 25 000 + genes essential for protein manufacture [4–6]. What makes these viruses (coined ‘fossil viruses’) so intriguing ? In a previous paper we demystified HERVs [7] with some emphasis on the beneficial potential of these viruses. After all, HERVs have been within our genome for a considerable period of time (30 million years) and some transposition events may have modified/modulated gene expression to confer a selective advantage to the host. This may include retroviral receptor blockade, interference by antisense RNA, immunosuppressive peptides and the fusogenic properties of HERV envelope gene products that may play a role in placenta formation [7–9]. However, it should also be appreciated that there is a potential for harm [10], for the latter transposition and the opportunity for mutation has been cited as a possible role for HERVs in certain types of cancer [11,12]. In this paper, we consider transposable elements including HERVs and their potential in disease states mediated possibly by superantigens and molecular mimics.
HERVS − WITHIN THE CONTEXT OF TRANSPOSABLE ELEMENTS
Repetitive DNA in the human genome
Overall, the human genome comprises just over 3 billion base pairs (bp) of DNA, of which around half this mass, or length, is unique-sequence or low-copy DNA. A small percentage of this DNA represents coding sequences for proteins, the remaining unique/low-copy DNA is made up of introns or spacer DNA. Repetitive DNA in humans includes genes that encode ribosomal components, i.e. the 28S, 18S and 5·8S subunits, arranged in tandem arrays repeated 150–200 times. Similarly, highly repetitive DNA comprises very short sequences that may be repeated up to a million times per genome, e.g. minisatellite DNA consists of units 10–100 bp long repeated in arrays of 0·5–40 kb [5]. The variable nature of the repeat patterns of some of these sequences between individuals was the original basis of genetic fingerprinting [13]. Most of the intermediate repetitive class of DNA sequences is made up of distinct families of transposable elements (approaching 46·4% of the total genome), as shown in Table 1[14–18]. In this context, HERVs represent only one member of a group of repetitive DNA elements [19].
Table 1.
Relative contributions of the main classes and common examples of interspersed repeat DNA in the human genome (derived from 5 and 6)
| Repeat sequence family | Fraction of genome (%) | Typical unit (encoded proteins) | Typical length kb | Ref. (Accession nos†) |
|---|---|---|---|---|
| LINEs | 21 | P□□A/T | 6–8* | [14–16] |
| Long interspersed elements(e.g. LINE1) | (17·4) | (reverse transcriptase and endonuclease) | (M22333) | |
| SINEs | 13·6 | PA/T | 0·1–0·3 | [17,18] |
| Short interspersed elements (e.g. Alu) | (10·7) | (non-protein coding) | (L35531) | |
| LTRs | 8·6 | ⇒□□□⇒ | 1·5–11 | [3,7,9,19,69] |
| Long-terminal repeats(e.g. HERV class I +II + III, MaLR III) | (4·8, 3·8) | (reverse transcriptase, protease, RNAse H and integrase) | (AY208136, M14123, AF020092, U07856) | |
| DNA transposons | 3·0 | ⇒□□⇐ | 0·08–3·0 | [17] |
| (e.g. MER-1 Charlie) | (1·4) | (transposonase) | (L13659) | |
| Unclassified | 0·15 | |||
| Total | Circa 46·4% |
Further details of human repeat sequences can be found at http://www.girinst.org/Repbase_Update.html. Note HERV are contained within the repeat sequence LTRs.
Reverse transcription having primed at the 3′ end often fails to proceed to the 5′ end. so many LINEs are shorter than 1kb. SINEs share a common 3′ sequence for reverse transcription so active SINEs can exploit a LINE reverse transcriptase. SINEs and LINEs may also show short flanking repeat sequences (e.g. 5′TTAAAA/3′AATTTT) which act as signal sequences for integration [17–20]. Code; ⇒: Repeat sequence, Ρ: RNA polymerase promoter (LINE RNA pol II, SINE RNA pol III), A/T: polyA/polyT sequences, □□: open reading frame (ORF).
Mobile genetic elements
All the families shown in Table 1 are distinguished by their present (‘active’) or past mobility within the genome. SINEs (short interspersed elements), LINEs (long interspersed elements) and LTRs, including HERV elements, rely upon the action of reverse transcriptase (RT) on RNA copies for transposition (‘copy and paste’) [20]. There are three distinct SINE families (Alu, MIR and MIR3) in humans and these genetic elements, lacking an RT, must utilize this enzyme activity from a LINE that recognizes a common sequence at the 3′ end [21]. While a range of LTRs are found in eukaryotes, only four main classes (HERV I, II, III and MalR) are found in humans comprised of many different families [5,6,22]. Most LTRs have lost much of their original sequences through homologous recombination between their flanking repeat sequences, and so exist as ‘solitary’ elements [5]. At least seven major classes of DNA transposons are present in humans, with some reflecting ancient eukaryotic elements such as the ‘mariner’ sequence found in both mammals and insects [full details of these families are found in Repbase (Table 1)][23].
Transposonase can act on deleted or mutated versions of an inactive element and so can use these as substrates when the enzyme returns to the nucleus from the cytosol following translation. Hence, inactive elements may accumulate rapidly in a genome. In contrast, LINE proteins (such as RT) tend to associate with intact RNA molecules free of deletions or mutations from which they were translated. Thus LINEs are less prone to a loss of function because of the intimacy of their association with complete and functional RNA substrates. The interactions of repetitive DNA elements may blur their origins. For example, a hybrid HERV has been described which appears to be a product of a recent retrotransposition event that has also acquired inverted repeats characteristic of DNA transposons [24]. Hence, while some transposable elements may become inactive over time, others can retain mobility within and perhaps between genomes.
LTRs and DNA transposons are distributed evenly between AT- and GC-rich DNA, but Alu elements are preferentially retained (although not necessarily targeted) in GC-rich regions. Generally there is a strong correlation between the density of actively transcribed genes, reflecting GC content and the density of Alu elements. Some chromosomes, such as 19, have an unusually high number of such elements. Conversely, chromosome Y shows a low density of Alu elements relative to its GC content. This may reflect an accumulation of pseudogenes (non-functional sequences that closely resemble operating genes) on this chromosome [5]. In contrast, LINEs accumulate in AT rich regions [21]. The latter contain fewer genes than GC-rich DNA with insertions here presumably extracting a lower mutational penalty. However, as SINEs such as Alu depend upon LINE transposition, their enriched presence in GC-rich DNA is intriguing. Many SINEs are expressed under conditions of stress, their RNA products then binding to a specific protein kinase (PKR), so enhancing protein translation repressed previously by PKR. Consequently, there may be a selective pressure to retain SINEs such as Alu within gene-rich areas of open chromatin where they may be transcribed rapidly to stimulate protein translation [15]. The distribution of integrated DNA sequences derived from HERV elements is clearly not random. For example, one group of sequences derived as pseudogenes from Class I HERVs mediated by LINE retrotransposition shows a bias for chromosomes 3, 4, X, and especially Y. In the latter case this probably reflects a common phenomenon of the limited frequency of recombination, in the absence of a homologous partner chromosome, allowing the survival of repetitive DNA including HERVs. The presence of these incomplete HERV sequences in AT-rich regions perhaps reflects the selection pressure on their guiding LINEs. There is no obvious insertion motif for retroviruses in general although, like LINEs, they are influenced by topological features of the host DNA such as nucleosome structure and chromatin condensation [25].
Evolutionary fate and disease
It has been suggested that 47 human genes were clearly derived from retroelements, virtually all of them from DNA transposons [5,26]. LINE1 retrotransposition of RNA transcribed from host genes has also contributed to the development of several genes, while several hundred genes have transcription termination signals derived from LTRs. Potentially retroviruses could evolve from LTR elements (which contain gag and pol genes encoding replication and transposition functions) by acquiring genes for infectivity such as those encoding envelope proteins. Conversely, the loss of such genes by exogenous retroviruses could produce retrotransposons marooned in their host cell [17]. It is probable that half the human genome can be derived from the insertion of repetitive DNA elements with many translated elements found in proteins. Often such elements were not inserted into the original open reading frames of genes but became part of the genes by alternative splicing of introns that shorten or extend the coding region [27]. Reverse transcriptase from LINEs may act on mRNA from other genes, resulting in the transposition of a copy to a fresh site. As introns will normally have been processed from the mRNA template the new copy will represent exon sequences only and produce a pseudogene [28]. Overall insertions of transposable elements into genes in the host genome may have important mutagenic consequences. For example, Alu insertions can cause some cases of breast cancer, Huntingdon's disease, agammaglobulinaemia and haemophilia. Similarly, L1 insertions are associated with some cases of muscular dystrophy and haemophilia. Furthermore, more than 40 examples of Alu mispairing and crossing-over that lead to deletions have been reported, covering a variety of diseases [29]. It is estimated that perhaps as many as one in 600 human mutations are caused by retrotransposition events, with one in 1100 and one in 1500 caused by Alu and LINE1, respectively [28]. Consequently, retroelements such as HERVs are potentially significant agents of mutation.
HERVS: CLASSIFICATION
The detection and classification of HERVs has been reviewed extensively [3,7,9,22], together with the availability of a useful database (http://herv.img.cas.cz). A raft of criteria have been employed, although it is now generally accepted that HERVs may be ascribed principally to Class I or Class II HERVs that are related to gammaretrovirus sequences and betaretrovirus sequences, respectively, based on their genetic similarity in the pol region (Table 2). A limitation in terms of research on endogenous retroviruses in general has been the relatively few molecular probes and antibody reagents to detect HERV products, although this issue is being addressed. Currently, monoclonal antibodies have been developed [30,31] together with recombinant phage antibodies to HERV-K10 [32]. With the molecular approaches, e.g. polymerase chain reaction, specific primers are available [33] for detecting specific HERVs compared with primers recognizing consensus retroviral regions (e.g. the reverse transcriptase segment) with a short intervening ‘fingerprint’ region [34].
Table 2.
Details of several Class I and Class II human endogenous retroviruses: full details provided in references [3,7,9]
| Family | Family members | No. of copies /previous classification | mRNA expression | Protein expression | Genome structure | Primer binding site | Chromosomal location | Comment | |
|---|---|---|---|---|---|---|---|---|---|
| HERV-H/RTVL-HClass I | RTVL-H2; RGH2 | Multicopy type C; 800–1000 copies plus ∼ 1000 solitary LTRs | LTR-gag-pol-env-LTR mRNA expression in lung squamous cell carcinoma, teratocarcinoma cell line, multiple sclerosis derived B-lymphoblastoid cell lines | 62 kDa protein detected for env gene | Severely truncated provirus | tRNAHis | Copies in all chromosomes with concentration on chromosomes 1p and 7q | Complementation and full expression may occur under circumstances yet to be described | |
| HERV-EClass I | HERV-E clone 4–1 | Multicopy type C retrovirus; 50–100 copies | Several LTR and en- containing mRNAs are expressed in normal placenta, breast and colon carcinoma cells | 38 kDa env protein, no particle formation | Full length genome = 8·8-kilobase Truncate genome = 6-kilobase (lack env sequence) | tRNAGlu | Altered tissue-specific gene expression in salivary gland for the amylase gene complex due to insertion of HERV-E LTR into its promoter region | ||
| ERV-9Class I | Type C retroviruses | Detected ERV-9 RNA transcripts of 8, 2, and 1·5 kb in undifferen- tiated NT2/D1cells, teratocarcinoma cells | Severely truncated provirus | tRNAArg | Selective expression of zinc finger protein in cell lineages associated with an ERV9 promoter | ||||
| HERV-RClass I | ERV3 | Single copy type C retrovirus | High expression in placenta and cell line U937, low expression in thymus, breast, lung, pancreas | 65 KDa env protein | Full length = 9·9 kilobase | tRNAArg | Chromosome 7 | mRNA expression mediated by steroid hormones | |
| HRES-1Class I | Single copy element per haploid genome | Detected for LTR, gag regions | Encodes a 28 kDa gag protein in H9 human T cells | Mapped to a common fragile site: chromosome 1 at 1q42 | Antibodies to HRES-1 specific synthetic peptides were found in patients with MS, SLE, Sjogren syndrome | ||||
| HERV-W(MSRV)Class I | Multicopy type C retrovirus | LTR-gag-pol-env-LTR mRNA expression in placenta and testis | env gene codes for Syncytin protein (65 kDa env) | Detected in MS, human genome contain at least 70 100, and 30 HERV-W-related gag, pro, and env regions, respectively | |||||
| HERV-KClass II | HERV-K10 | Multicopy mosaic- type endogenous proviruses; 50 copies per haploid genome | LTR-gag-pol-env-LTR, rev mRNA expression; high in teratocarcinoma cell lines, testicular tumours; expression of HERV-K env gene in human breast cancer cells; low in placenta and normal tissues; expressed in peripheral blood lymphocytes | gag, cORF, protease, integrase, polymerase, env; rev, retroviral particle formation | Complete nucleotide sequence of the HERV-K10 clone = 8·8-kilobase | tRNALys | Induced HERV-K mRNA expression in human breast cancer cells by addition of oestradiol and progesterone | ||
| HERV-LClass II | Multicopy type | pol, LTR-gag-pol mRNA expression; tissues: rheumatoid arthritis, synovial fluid cells, placenta, breast cancer | Unknown | tRNALeu |
HERVS: POTENTIAL MODULATORS
Because retroelements appear to show rapid mutation and deletion they could be viewed as accidentally integrated viral elements with no function. On the other hand, there is growing evidence that such integration events may have important ramifications for the host genome [35]. DNA sequences from HERVs and LINEs may be highly expressed in tissues with embryonic features, such as the germ line or tumours. For example, HERV-K envelope transcripts are expressed in most human breast cancers although not in normal control breast tissues − such expression patterns then providing novel markers of the disease [11]. Overall, a diverse range of disease states could arise from loss of function mutations that result from the integration of DNA sequences into the host genome.
Intriguingly, integration events may also deliver disease phenotypes by gain of function provided by alterations in target gene expression conferred by incoming retroelement control sequences [36]. Landry et al. [37] describe the transcription of the opitz syndrome gene mid1, particularly in placental and embryonic kidney tissues, from a promoter element that originated from an LTR of a HERV-E element. In some cases the particular consensus sequences responsible for enhancing transcription of adjacent human genes have been identified. For example, ERV-9 LTRs contain GATA, CCAAT and CCACC motifs which can bind transcription factors expressed in placental and haematopoietic tissues. Enhanced RNA synthesis could be demonstrated in such embryonic tissues from a site just downstream of a TATA box motif, mediated presumably by RNA polymerase II [38]. Such alterations in gene transcription by retroelements may be widespread. One group [39] was able to identify using computer-based searches (i.e. ‘in silico’ genetics) of the draft human genome that 25% of human promoters contained sequences derived from transposable elements. Intriguingly, many transcription factor binding sites could be identified in these sequences plus scaffold or matrix attachment regions and locus control regions that regulate multiple genes. Disease states caused by the integration of transposable elements into genes are drawn to our attention by the dramatic phenotype of the patient, e.g. haemophilia. However, more subtle alterations in gene transcription caused by such events may be much more significant and profound, providing advantages for the host organism by extending the control of gene expression [40]. For example, the phenomenon of X chromosome inactivation found during mammalian female embryogenesis may rely upon the enrichment of LINEs found on this chromosome relative to autosomes. The initial signal for inactivation of the majority of genes on one of the X chromosomes originates and then spreads from a particular locus specific to this chromosome. The resulting physical changes in the chromatin of the various X chromosome domains is postulated to be amplified by regions of high L1 content − regions escaping inactivation being deficient in these modulating L1 sequences [41]. The abundance of retroelements including HERVs within the genome and the effect on protein expression of genes linked to LTRs (e.g. cytochrome C1 and salivary amylase) [7] makes them intriguing agents for altering host gene/protein expression.
HERVS: POTENTIAL SUPERANTIGENS
A superantigen (SAG) bypasses the normal route of MHC-peptide–T cell receptor (TCR) interaction in binding to a given T cell Vβ chain and MHC Class II protein [42]. As a consequence, a large number of T cells (5–20%, hence the term ‘superantigen’) bearing the particular Vβ segment are expanded [43]. SAGs produced by bacteria or virus modulate the immune system through the release of cytokines with disruption of lymphocyte homeostasis, the polarization of CD4 Th1/Th2 phenotypes plus changes in antibody production [44,45]. Over time, associations of T cell receptor usage with particular diseases have been cited, e.g. Vβ14, Vβ8 (and Vβ13), Vβ7 (and Vβ13) with rheumatoid arthritis, chronic cutaneous lupus erythematosus and insulin-dependent diabetes mellitus, respectively [46–48]. Furthermore, inferences of enhanced frequencies of T cells expressing particular Vβ segments have been attributed to certain viruses such as Epstein–Barr virus (EBV) (Vβ13), cytomegalovirus (Vβ12) and rabies virus (Vβ8) [49]. Could the chance of selective expansions of T cells by viruses and in set disease states be coincidental, or is there a missing link ?
It is plausible that an endogenous virus could be of significance because immune modulation is evident by the mouse mammary tumour virus (MMTV) SAG encoded within the ORF of the 3′-LTR of the integrated viral genome [50]. There is some evidence to suggest that some common human viruses might encode superantigens. For example, it was demonstrated that EBV infection of B cells induced the activation of human T cells expressing the Vβ13 element of the TCR [51]. Unfortunately, exhaustive analysis of the EBV genome failed to identify any EBV superantigen. However, a recent study by Sutkowski et al. [52] has revealed a possible connection between EBV infection and superantigen activity, which might account for the suggested aetiological link between EBV and certain autoimmune diseases [53,54]. Mapping studies had demonstrated that HERV-K18 has three alleles, found in the first intron of CD48 in reverse orientation, one allele of which is truncated and corresponds to IDDMK1,222 (now HERV-K18·1). The envelope genes of each of the alleles encode a superantigen with identical specificity for Vβ7+ and Vβ13+ T cells. Sutkowski et al. speculated that EBV transactivated the CD48-associated HERV-K18 superantigen-encoding gene, thereby explaining the association between EBV infection and Vβ-specific T cell activation [51]. RNAse-protection assays showed that the expression of HERV-K18 was indeed up-regulated in EBV-infected lymphoblastoid cell lines and following EBV infection of EBV-negative Burkitt's lymphoma lines. More recently, transcription of HERV-K has been shown to be induced following infection with a relative of EBV, namely the herpes simplex type-1 virus (HSV-1), an effect that was mediated specifically by the HSV-1 immediate early protein, ICP0, and required the AP-1 binding site present on the HERV-K LTR [55]. Furthermore, LTR-directed transcription of the HERV-W is also induced by the HSV-1 infection, an effect mediated partly by the action of the HSV-1 immediate early protein 1 (IE1) [56]. These studies suggest an indirect connection between viral infection and the host immune response, whereby viral up-regulation of the expression of host superantigen-encoding genes can affect dramatically the nature of the immune response, possibly towards a response that is more autoimmune in nature. Stauffer et al. [57] have also established a link between viral infections and interferon (IFN)-α. From this work expression of HERV-K18 SAGs was inducible by IFN-α and sufficient to stimulate Vβ7 T cells. Thus there remains the possibility that diverse viral infections (i.e. not simply herpes viruses) might elicit SAG-like responses through HERV intermediaries.
In terms of EBV it is not clear at this time what the superantigen response might contribute to its life cycle. One possibility is that it might contribute to the T cell help necessary for the generation of long lived EBV-infected memory B cells and so contribute to EBV persistence in the normal host. In addition, HERV-mediated stimulation of T cells might also contribute to EBV-induced B-lymphomagenesis. The idea that common viruses can activate endogenous superantigens is already accepted in some animal systems. For example, in mice there is strong evidence that the BM5 murine leukaemia virus complex activates endogenous mouse mammary tumour virus (MMTV) genes [58]. However, the extension of this principle to humans is relatively new [49]. While herpesviruses share genomic similarities there are differences that reflect development along distinct evolutionary pathways. For example, the Kaposi's sarcoma herpesvirus (KSHV) genome encodes a number of cellular homologues not present in EBV [59]. These include, among others, viral IL-6 (vIL-6) and viral cyclin D (v-cyclin D) genes. However, the EBV-latent membrane protein-1 (LMP1) is able to induce the expression of both cellular IL-6 and cellular cyclin D genes [60,61]. Therefore, both viruses have evolved different means (expression of viral homologues in the case of KSHV versus induction of cellular genes by EBV) to produce the same functional end-points. It therefore appears that EBV, in the same way as described above, is able to take advantage of the retroviral superantigen strategy without actually acquiring its own superantigen-encoding genes. It remains to be established whether KSHV can induce superantigen expression and whether the mechanism of induction is similar to that of EBV. It should also not be overlooked that EBV is in its own right an oncogenic virus and the possibility exists that EBV induction of HERVs might play an important role in tumour development that may not necessarily be linked to the induction of superantigen expression.
HERVS: POTENTIAL MIMICS
The premise for molecular mimicry is that an external pathogenic agent possesses similarity to human protein(s) such that an ensuing immune response is unable to distinguish between the host and inciting component. As a result collateral damage occurs. Hence, the concept of molecular mimicry has been accepted as a plausible mechanism in autoimmune diseases [62,63]. In regard to exogenous agents, cross-reactivity with the p24 gag protein of human immunodeficiency virus type I (HIV-I) was demonstrated from sera of non-HIV-infected patients with primary Sjögren's syndrome (SS) [64]. Furthermore, antibodies to p24 Gag of HIV-I were found in patients with systemic lupus erythematosus (SLE). In these patients, immunological cross-reactivity between the p24 Gag protein and the small ribonucleoprotein (snRNP) Sm was demonstrated [65]. Subsequently the presence of antibodies to retroviral products has been highlighted in rheumatoid arthritis [66,67], polymyositis and SLE patients, but no evidence was found of exogenous viral infection (HIV and HTLV) using polymerase chain reaction (PCR). Other studies investigating patients with multiple sclerosis, SLE and SS have also shown higher levels of antibodies to synthetic peptides that represent the major epitopes of HTLV-I p19 and p24 Gag proteins, and its endogenous counterpart HTLV-related endogenous sequence type I (HRES-I), than sera of normal donors [68,69]. Currently, endogenous viruses such as HERV-K have been cloned and sequenced from patients with rheumatoid diseases [34]. In addition, the use of specific oligonucleotide primers in reverse transcription-polymerase chain reaction (RT-PCR) has helped to identify HERV-K10 from rheumatoid arthritis (RA) synovial fluid cells [33], confirming earlier studies of endogenous retroviruses implicated in RA [70]. Overall, the consensus from these findings suggests HERVs as important mediators of autoimmune diseases.
Latterly, immunoblotting using recombinant proteins of HERV-K10/IDDMK1,222 showed that 32–47% of patients with autoimmune rheumatic disease were positive compared to 29% of controls [71]. Importantly, an immunodominant region (GKTCPKEIPKGSKNT: single amino acid code) was identified by four patients through epitope mapping and highlighted the potential for an antigen-driven immune response. Evidently the need for epitope mapping of additional HERVs should not be underestimated. This is illustrated by an unrelated investigation of mapping antibodies to IgG that highlighted immunodominant regions (APIEKTISKAKGQPR and KPREE) that were also recognized by rheumatoid factors [72]. It is speculated that exogenous/endogenous agents could help drive this immune response.
Overall, the potential for mimicry (and mayhem ?) is unquestionable, but research has been biased largely towards proteins of exogenous retroviruses, synthetically derived viral peptides and recombinant proteins to ascertain the presence of antiretroviral/HERV antibodies to the exclusion of T cell studies. However for molecular mimicry to be a mechanism for HERVs, there remains an underlying assumption that HERVs with intact open reading frames produce full-length or truncated retroviral proteins. In essence this is substantiated, as retrovirus-like particles (VLP) have been observed by electron microscopy in autoimmune disease tissues, e.g. synovial tissue [73]. VLPs derived from an HERV-K provirus have also been identified from a human teratocarcinoma-derived cell line (GH) and a hormone-stimulated human breast carcinoma-derived T47D cell line [30,74–76]. To date, HERV-K, ERV-3 and HERV-W are among the major HERVs with the capacity to code for viral products and potential molecular mimics.
CONCLUSIONS
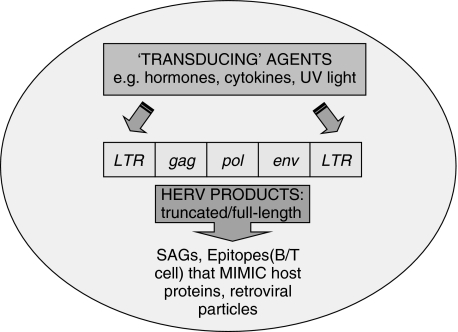
HERVs constitute but one group of transposable elements or retroelements within the human genome. Having been part of our molecular evolution, retroelements and isolated LTRs may be of benefit to the host in promoting plasticity and the regulation of gene expression, i.e. through promoters and cis-regulatory sequences. Intriguingly, polymorphism in LTRs linked to HLA genes may explain the variation in immune responses between individuals of similar phenotype. However, there is also an opportunity to harm because retroelements may provide the potential for mutation, modulation and the undesired expression of full-length or truncated products. Indeed LTRs may act as ‘biological transducers’, as HERV expression may be altered through hormonal and environmental signals (Fig. 1). Consequently, the potential of environmental agents on the host could be of particular significance to HERV research. It is now well established that herpes viruses such as cytomegalovirus (CMV) [34] and EBV transactivate HERV-K. Clearly, herpes viruses may be seen as helper viruses and a mechanism of ‘switching on’ HERV production. However, in the case of EBV and superantigens discussed in this article, who is helping whom, or is this case of viral hijacking ? (HIV has also been suggested to utilize the pol region of HERVs [77].) Interestingly, it has also been shown that HERV-W envelope glycoprotein can be used to form pseudotypes with HIV-1 virions [78]. Hence, there is a possibility that complementation with virion proteins encoded by different HERVs and/or other viruses could produce infectious viruses. Ultimately superantigens can pose a ‘danger’ to the immune system, with undesired outcomes for the host. Similarly, viral products that mimic host proteins may also have the potential to harm, perhaps as a result of collateral damage following immune activation and damage through epitope spreading. However, underlying the mechanism of molecular mimicry is a requirement for HERV peptides and protein products. What drives and influences their manufacture (including other viruses and the cellular microenvironment) is an important area of investigation as it is also apparent that certain biological agents over-ride premature stop codons: a case in point for some HERVs with (multiple) termination signals [7]. A final comment is that while HERV expression may be a potential ‘danger’ to humans, their exposure and replication (i.e. outside the germline) would be somewhat detrimental to the virus in question. Overall, while a number of articles [11,34,52,57,71] highlight some steps in determining the role of HERVs, it will be crucial in the next few years to establish firmly clear associations of HERVs that may contribute to the pathology of disease states and/or aetiology [79].
Fig. 1.
Schematic view of HERV activation via long-terminal repeats that may augment the production of superantigens and peptide mimics that could lead to autoimmune diseases. The ‘transducing’ elements within HERVs enable environmental agents to modulate HERV expression.
Acknowledgments
The authors thank the Multiple Sclerosis Society, the James Beattie Charitable Trust, the Graduate School, University of Wolverhampton and the South Staffordshire Medical Foundation for recent support.
REFERENCES
- 1.Knipe DM, Howley PM. Fields virology. 4. Philadelphia USA: Lippincott, Williams & Wilkins; 2001. [Google Scholar]
- 2.Griffiths DJ. Endogenous rretroviruses in the human genome sequence. Genome Biol. 2001;2:1017.1–1017.5. doi: 10.1186/gb-2001-2-6-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–84. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennisi E. Gene counters struggle to get the right answer. Science. 2003;301:1040–1. doi: 10.1126/science.301.5636.1040. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Nelson PN, Carnegie PR, Martin J, et al. Demystified. Human endogenous retroviruses. J Clin Pathol Mol Pathol. 2003;56:11–8. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsson E, Andersson G. Beneficial role of human endogenous retroviruses: facts and hypotheses. Scand J Immunol. 1998;48:329–38. doi: 10.1046/j.1365-3083.1998.00428.x. [DOI] [PubMed] [Google Scholar]
- 9.Blond JL, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–9. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perl A. Role of endogenous retroviruses in autoimmune diseases. Rheum Dis Clin North Am. 2003;29:123–43. doi: 10.1016/s0889-857x(02)00098-4. [DOI] [PubMed] [Google Scholar]
- 11.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–35. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 12.Sauter M, Schommer S, Kremmer E, et al. Human endogenous retrovirus K10: expression of gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–21. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffries AJ, Wilson V, Thein SL. Individual specific ‘fingerprints’ of human DNA. Nature. 1985;316:76–9. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 14.Smit AF, Toth G, Riggs AD, Jurka J. Ancestral mammalian wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–17. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 15.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol. 2002;14:343–50. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 16.Ovchinnikov I, Rubia A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci USA. 2002;99:10522–7. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smit AFA. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–8. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 18.Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci USA. 2002;99:327–32. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–65. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 20.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–7. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid CW. Does SINE evolution preclude Alu function ? Nucl Acids Res. 1998;26:4541–50. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 23.Robertson HM, Lampe DJ. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol Biol Evol. 1995;12:850–62. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 24.Hughes JF, Coffin JM. A novel endogenous retrovirus related element in the human genome resembles a DNA transposon: evidence for an evolutionary link ? Genomics. 2002;80:453–5. [PubMed] [Google Scholar]
- 25.Pavlicek A, Paces J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retrovirus generated by LINEs; their integration, stability and distribution. Genome Res. 2002;12:391–9. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetika. 1999;107:209–38. [PubMed] [Google Scholar]
- 27.Li WH, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–9. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 28.Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Ann Rev Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 29.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–93. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 30.Bieda K, Hoffman Boller K. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol. 2001;82:591–6. doi: 10.1099/0022-1317-82-3-591. [DOI] [PubMed] [Google Scholar]
- 31.Nelson PN, Astley JS, Warren P. Crocker J, Murray PG. Molecular biology in cellular pathology. Chichester: John Wiley & Sons Ltd; 2003. Monoclonal antibodies. the generation and application of ‘tools of the trade’ within biomedical science; pp. 329–49. ch.16: [Google Scholar]
- 32.Smith K, Sampath S, Richards Warren P, Nelson PN, Greenman J. Selection and characterisation of phage antibodies to HERV-K. Immunology. 2003;110:S79. [Google Scholar]
- 33.Ejtehadi HD, Ali HA, Serhan E, Bowman S, Nelson PN. The potential role of human endogenous retrovirus K10 in pathogenesis of rheumatoid arthritis. Immunology. 2002;107:S109. doi: 10.1136/ard.2004.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson PN, Lever AML, Smith S, et al. Molecular investigations implicate human endogenous retroviruses as mediators of anti-retroviral antibodies in autoimmune rheumatic diseases. Immun Invest. 1999;28:277–89. doi: 10.3109/08820139909060862. [DOI] [PubMed] [Google Scholar]
- 35.Taruscio D, Floridia G, Zoraqi GK, Mantovani A, Falbo V. Organisation and integration sites in the human genome of endogenous retroviral sequences belonging to the HERV-E families. Mamm Genome. 2002;13:216–22. doi: 10.1007/s00335-001-2118-7. [DOI] [PubMed] [Google Scholar]
- 36.Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA. 1997;94:7704–11. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landry JR, Rouhi A, Medstrand P, Mager DL. The opitz syndrome gene mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol. 2002;19:1934–42. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 38.Ling J, Pi W, Bollag R, et al. The solitary long terminal repeats of ERV-9 edogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–23. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King Jordan I, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 40.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–85. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000;97:6634–9. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundberg EJ, Li Y, Mariuzza RA. So many ways of getting in the way: diversity in the molecular architecture of superantigen-dependent T-cell signaling complexes. Curr Opin Immunol. 2002;14:36–44. doi: 10.1016/s0952-7915(01)00296-5. [DOI] [PubMed] [Google Scholar]
- 43.Schafer R, Sheil JM. Superantigens and their role in infectious disease. Adv Pediatric Infect Dis. 1995;10:369–90. [PubMed] [Google Scholar]
- 44.Cameron SB, Nawijn MC, Kum WW, Savelkoul HF, Chow AW. Regulation of helper T cell responses to staphylococcal superantigens. Eur Cytokine Netw. 2002;12:210–22. [PubMed] [Google Scholar]
- 45.Rago C, Tocce K, Ficarro S, Masters G, Riggs J. Superantigen disruption of CD8+ T and B lymphocyte homeostasis. Immunobiology. 2000:202. doi: 10.1016/S0171-2985(00)80107-2. [DOI] [PubMed] [Google Scholar]
- 46.Paliard X, West SG, Lafferty JA, et al. Evidence for the effects of a superantigen in rheumatoid arthritis. Science. 1991;253:325–9. doi: 10.1126/science.1857971. [DOI] [PubMed] [Google Scholar]
- 47.Furukawa F, Iwasaki-Inuzuka K, Takigawa M, Matisushita K. Selective expansion of T cells expressing V beta 8 and V beta 13 in skin lesions of patients with chronic cutaneous lupus erythematosus. J Dermatol. 1999;23:670–6. doi: 10.1111/j.1346-8138.1996.tb02679.x. [DOI] [PubMed] [Google Scholar]
- 48.Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–113. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 49.Woodland DL. Immunity and retroviral superantigens in humans. Trends Immunol. 2002;23:57–8. doi: 10.1016/s1471-4906(01)02146-9. [DOI] [PubMed] [Google Scholar]
- 50.Choi Y, Kappler JW, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of mouse mammary tumour virus. Nature. 1991;350:203–7. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 51.Sutkowski N, Palkama T, Ciurli C, Sekaly RP, Thorley-Lawson DA, Huber BT. An Epstein–Barr-virus-associated superantigen. J Exp Med. 1996;184:971–80. doi: 10.1084/jem.184.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT. Epstein–Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity. 2001;15:579–90. doi: 10.1016/s1074-7613(01)00210-2. [DOI] [PubMed] [Google Scholar]
- 53.Incaprera M, Rindi L, Bazzichi A, Garzelli C. Potential role of the Epstein–Barr virus in systemic lupus erythematosus autoimmunity. Clin Exp Rheumatol. 1998;16:289–94. [PubMed] [Google Scholar]
- 54.Levin LI, Munger KL, Rubertone MV, et al. Multiple sclerosis and Epstein–Barr virus. JAMA. 2003;289:1533–6. doi: 10.1001/jama.289.12.1533. [DOI] [PubMed] [Google Scholar]
- 55.Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 2002;86:93–100. doi: 10.1016/s0168-1702(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 56.Lee WJ, Kwun HJ, Kim HS, Jang KL. Activation of the human endogenous retrovirus W long terminal repeat by herpes simplex virus type 1 immediate early protein 1. Mol Cells. 2003;15:75–80. [PubMed] [Google Scholar]
- 57.Stauffer Y, Marguerat S, Meylan F, et al. Interferon-alpha-induced endogenous superantigen: a model linking environment and autoimmunity. Immunity. 2001;15:591–601. doi: 10.1016/s1074-7613(01)00212-6. [DOI] [PubMed] [Google Scholar]
- 58.Hugin AW, Tang Y, Vacchio MS, et al. Viral superantigens. FL, USA: CRC Press Inc.; 1995. Is a superantigen involved in the pathogenesis of MAIDS ? pp. 207–18. [Google Scholar]
- 59.Sharp TV, Boshoff C. Kaposi's sarcoma-associated herpesvirus: from cell biology to pathogenesis. IUBMB Life. 2000;49:97–104. doi: 10.1080/15216540050022395. [DOI] [PubMed] [Google Scholar]
- 60.Eliopoulos AG, Gallagher NJ, Blake SM, Dawson CW, Young LS. Activation of the p38 mitogen-activated protein kinase pathway by Epstein–Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem. 1999;274:16085–96. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 61.Arvanitakis L, Yaseen N, Sharma S. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol. 1995;155:1047–56. [PubMed] [Google Scholar]
- 62.Levin MC, Lee SM, Kalume F, et al. Autoimmunity due to molecular mimicry as a cause of neurological disease. Nat Med. 2002;8:509–13. doi: 10.1038/nm0502-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oldstone M. Molecular mimicry and immune mediated disease. FASEB J. 1998;12:1255–65. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Talal N, Dauphinee MJ, Dany M, Alexander SS, Hart DJ, Garry RF. Detection of serum antibodies to retroviral proteins in patients with primary Sjögren's syndrome (autoimmune exocrinopathy) Arth Rheum. 1990;33:774–81. doi: 10.1002/art.1780330603. [DOI] [PubMed] [Google Scholar]
- 65.Talal N, Garry RF, Schur PH, et al. A conserved idiotype and antibodies to retroviral proteins in systemic lupus erythematosus. J Clin Invest. 1990;85:1866–71. doi: 10.1172/JCI114647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson PN, Lever AML, Bruckner FE, Isenberg DA, Kessaris N, Hay FC. Polymerase chain reaction fails to incriminate exogenous retroviruses HTLV-I and HIV-1 in rheumatological diseases although a minority of sera cross-reacts with retroviral antigens. Ann Rheum Dis. 1994;53:749–54. doi: 10.1136/ard.53.11.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson PN, Bowman SJ, Hay FC, Lanchbury JS, Panayi GS, Lever AML. Absence of exogenous retrovirus in Felty's syndrome. Br J Rheumatol. 1995;34:185–7. doi: 10.1093/rheumatology/34.2.185. [DOI] [PubMed] [Google Scholar]
- 68.Brookes SM, Pandolfino YA, Mitchell TJ, et al. The immune response to and expression of cross-reactive retroviral Gag sequences in autoimmune disease. Br J Rheumatol. 1992;31:735–42. doi: 10.1093/rheumatology/31.11.735. [DOI] [PubMed] [Google Scholar]
- 69.Banki K, Hurley ME, Ablonczy E, et al. Human T-cell lymphotropic virus (HTLV)-related endogenous sequence, HRES-1, encodes a 28-kDa protein: a possible autoantigen for HTLV-I gag-reactive autoantibodies. Proc Natl Acad Sci USA. 1992;89:1939–43. doi: 10.1073/pnas.89.5.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakagawa K, Brusic V, McColl G, Harrison LC. Direct evidence for the expression of multiple endogenous retroviruses in the synovial compartment in rheumatoid arthritis. Arthritis Rheum. 1997;40:627–38. doi: 10.1002/art.1780400407. [DOI] [PubMed] [Google Scholar]
- 71.Herve CA, Lugli EB, Brand A, Griffiths DJ, Venables PJW. Autoantibodies to human endogenous retrovirus-K are frequently detected in health and disease and react with multiple epitopes. Clin Exp Immunol. 2002;128:75–82. doi: 10.1046/j.1365-2249.2002.01735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson PN, Westwood OMR, Soltys A, et al. Characterisation of epitopes of pan-IgG/anti-G3m(u) and anti-Fc monoclonal antibodies. Immunol Lett. 2003;88:77–83. doi: 10.1016/s0165-2478(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 73.Stransky G, Vernon J, Aicher WK, Moreland LW, Gay RE, Gay S. Virus-like particles in synovial fluids from patients with rheumatoid arthritis. Br J Rheumatol. 1993;32:1044–8. doi: 10.1093/rheumatology/32.12.1044. [DOI] [PubMed] [Google Scholar]
- 74.Boller K, Janssen O, Schuldes H, Tonjes RR, Kurth R. Charactersation of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 1997;71:4581–8. doi: 10.1128/jvi.71.6.4581-4588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seifarth W. Retrovirus-like particles released from human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–16. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patience C, Simpson GR, Colletta AA, Welch HM, Weiss RA, Boyd MT. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–7. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Towler EM, Gulnik SV, Bhat TN, et al. Functional characerization of the protease of human endogenous retrovirus K10: can it complement HIV-1 protease ? Biochemistry. 1998;37:17137–44. doi: 10.1021/bi9818927. [DOI] [PubMed] [Google Scholar]
- 78.An DS, Xie YM, Chen IS. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J Virol. 2001;75:3488–9. doi: 10.1128/JVI.75.7.3488-3489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stoye JP. The pathogenic potential of endogenous retroviruses: a sceptical view [Letter] Trends Microbiol. 1999;7:430. doi: 10.1016/s0966-842x(99)01616-9. [DOI] [PubMed] [Google Scholar]