Abstract
Streptococcus pneumoniae is a leading cause of otitis media, sinusitis, pneumonia, bacteraemia and meningitis worldwide. The drawbacks associated with the limited number of various capsular polysaccharides that can be included in the polysaccharide-based vaccines focuses much attention on pneumococcal proteins as vaccine candidates. We extracted an enriched cell wall fraction from S. pneumoniae WU2. Approximately 150 soluble proteins could be identified by 2D gel electrophoresis. The proteins were screened by 2D-Western blotting using sera that were obtained longitudinally from children attending day-care centres at 18, 30 and 42 months of age and sera from healthy adult volunteers. The proteins were further identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Seventeen proteins were antigenic in children and adults, of which 13 showed an increasing antibody response with age in all eight children analysed. Two immunogenic proteins, fructose–bisphosphate aldolase (FBA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and a control protein with known low immunogenicity, heat shock protein 70 (DnaK), were expressed in Escherichia coli, purified and used to immunize mice. Mouse antibodies elicited to the recombinant (r) FBA and rGAPDH were cross-reactive with several genetically unrelated strains of different serotypes and conferred protection to respiratory challenge with virulent pneumococci. In addition, the FBA used in this study (NP_345117) does not have a human ortholog and warrants further investigation as a candidate for a pneumococcal vaccine. In conclusion, the immunoproteomics based approach utilized in the present study appears to be a suitable tool for identification of novel S. pneumoniae vaccine candidates.
Keywords: immunoproteomics, proteomics, Streptococcus pneumonie, vaccine
INTRODUCTION
Streptococcus pneumoniae is a leading cause of bacteraemia, meningitis and pneumonia worldwide [1], with infants and the elderly being most vulnerable. An epidemiological study conducted in the United States showed that the incidence of invasive pneumococcal disease is highest in children 0–2 years of age (145 per 100 000 cases) but diminishes thereafter to 25 cases per 100 000 at 3–4 years of age, and in the population aged between 5 and 54 the incidence is only around five cases per 100 000. There is then a gradual resurgence in the incidence of disease in the elderly with 15, 21 and 75 cases per 100 000 in the 55–64-, 65–74- and >75-year age groups, respectively [2]. The significant levels of morbidity and mortality and the persistent emergence of antibiotic resistant strains of S. pneumoniae have heightened the need for the development of more effective means of prevention.
The immunologically variant capsular polysaccharides of S. pneumoniae are used widely for the typing of clinical isolates. There are more than 90 capsular serotypes and their prevalence among human isolates varies with age, disease type and, to some extent, geographical origin [3]. A 23-valent capsular polysaccharide-based vaccine is licensed for use in adults [4,5], but it does not elicit an efficient antibody response or protection in children below 2 years of age and in immunocompromised patients [6,7]. To overcome this lack of responsiveness to the T cell independent polysaccharide antigens in young children the conjugate pneumococcal vaccines were developed. These vaccines consist of seven to 11 most prevelant in pneumococcal infection, S. pneumoniae capsular polysaccharides linked covalently to a protein carrier to stimulate T cell responses to the vaccine. These vaccines are highly effective in preventing invasive pneumococcal disease in infants [8], but there are some drawbacks associated with the complexity of the manufacturing process that increase costs and the limited number of various capsular polysaccharides that can be included in the vaccine. Vaccination with conjugate pneumococcal vaccines has been shown recently to result in a shift in serotype distribution towards those pneumococcal capsular polysaccharides that are not present in the vaccine [9–11]. In addition, geographical variations in the prevalence of clinically important serotypes of S. pneumoniae have been described [12]. These concerns, combined with increasing antibiotic resistance [13], are driving research efforts to develop a ‘universal’ pneumococcal vaccine that is immunogenic in all age groups and broadly cross-protective against all serotypes.
Antibodies to S. pneumoniae protein antigens develop in humans during the asymptomatic carriage in adults [14] and invasive disease [15]. Importantly, infants below 2 years of age who are at most risk from pneumococcal infections are responding efficiently to protein vaccination [16]. We have shown previously, using sera longitudinally collected from healthy children, that there is an age-dependent enhancement of the antibody response to S. pneumoniae surface protein antigens [17]. This enhancement of antibody responses against specific pneumococcal surface proteins with increasing age is implicated in the development of natural immunity and thus might be useful in identifying candidate antigens for a vaccine [18].
In this study, we have used proteomics and immunoproteomics to investigate the age-dependent antigenicity of children's sera to S. pneumoniae cell wall-associated proteins and identify potential vaccine antigens. Two such proteins were found to elicit cross-strain protective immunity in mice.
MATERIALS AND METHODS
Reagents
Unless otherwise stated all chemicals and biochemicals of highest purity available were purchased from Sigma-Aldrich Corp. (St Louis, MO, USA).
Bacterial strains, growth conditions and growth medium
The S. pneumoniae strains used in this study were: strain WU2 (capsular serotype 3) obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA); strain 14DW (serotype 14) − its unencapsulated mutant 14·8 and WU2 unencapsulated mutant 3·8 are a kind donation of Dr D. Watson (Dallas, TX, USA); and strains 14R (serotype 14), 6BR (serotype 6B) and 9VR (serotype 9 V) from the Collection of Pediatric Infectious Disease Unit, Soroka University Medical Center (Beer Sheva, Israel). Except for the strains 14R and 9V all the other strains of S. pneumoniae used in this study are genetically unrelated (as assessed by Pulse Field Gel Electrophoresis; data not shown). Pneumococci were grown to mid-logarithmic growth phase as determined by OD in Todd–Hewitt broth (Difco Laboratories, Detroit, MI, USA) supplemented by yeast extract (Difco Laboratories), and confirmed by colony-forming units (CFU) counts on blood agar plates at 37°C under microaerobic conditions. The following Escherichia coli strains were used in this study: DH5α UltraMAX (Invitrogen Corp, Carlsbad, CA, USA) and BL21(DE3)pLysS (Promega Corp, Madison, WI, USA). E. coli were grown in Luria Broth (LB) medium. When required, ampicillin was added to the E. coli culture medium at a final concentration of 100 µg/ml.
Isolation of S. pneumoniae cell wall proteins
The proteins were isolated by the method of Siegel et al. [19]. Briefly, bacterial cells were harvested by centrifugation at 4700 g for 15 min, washed with phosphate buffered saline (PBS) and incubated with mutanolysin (200 U/ml mutanolysin in 20% sucrose, 2·5 mm MgCl2, 5 mm Tris-Cl, pH 7·4) for 1 h at 37°C. Soluble proteins released from bacteria treated with cell wall degrading enzymes were collected in the supernatant after centrifugation and stored at −70°C. Less than 10% of cytoplasmic proteins are reported to be found in the supernatant due to leakage using this procedure [19], but for comparison, pneumococcal cytoplasmic proteins were also isolated and separated on 2D gels. Bacteria were sonicated and the supernatant, containing soluble cytoplasmic proteins, was collected following centrifugation at 4700 g for 15 min.
Protein gel electrophoresis and staining
One-dimensional sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the Hoefer mini VE vertical electrophoresis system (Amersham Biosciences, San Francisco, CA, USA). Protein samples were suspended in Laemmli sample buffer (62·5 mm Tris-HCl, pH 6·8, 2% SDS, 25% glycerol, 0·01% bromophenol blue, Bio-Rad Laboratories, Inc., Hercules, CA, USA) and boiled for 5 min prior to electrophoresis. Two-dimensional SDS-PAGE involved separation of proteins by isoelectric point and by molecular weight, respectively. Isoelectric focusing (pI 4–6·5) was carried out in a Hoefer SE 220 Mighty Small Tube Gel Adaptor (Amersham Biosciences, San Francisco, CA, USA) according to the manufacturer's instructions. The proteins were then separated in the second dimension using 10% polyacrylamide gels in a Hoefer mini VE vertical electrophoresis system (Amersham Biosciences, San Francisco, CA, USA). The proteins were stained in the gels using Coomassie brilliant blue [20].
Human sera
Sera were collected longitudinally from eight healthy children attending day-care centres at 18, 30 and 42 months of age. Starting at 12 months of age, nasopharyngeal swabs were taken from the children on a bimonthly schedule over the 2·5 years of the study. Pneumococcal isolates were characterized by inhibition with optochin and a positive slide agglutination test (Phadebact, Pharmacia Diagnostics, Uppsala, Sweden). Serogrouping and serotyping of S. pneumoniae was performed by means of the Quellung reaction using antisera from Statens Serum Institute of Copenhagen, Denmark [21]. In addition, sera were collected from eight healthy adults. Experiments with human sera were approved by Soroka University Medical Center Ethics Committee, Beer Sheva, Israel. Informed written consent was obtained from parents of the children and from the adults participating in this study.
Western blot analysis
Protein mixtures were separated by one-dimensional (25 µg protein) and two-dimensional (40 µg protein) SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad Laboratories, Inc., Carlsbad, CA, USA), as described elsewhere [20]. The immunological detection of immobilized proteins was performed as described previously [20]. The results were analysed using MultiImageII light cabinet (DE-500) Alpha Innotech Corporation, San Leandro, California, USA.
Identification of proteins
Proteins spots of interest were excised from the gel using a pipette-tip. The gel plugs were washed three times for 20 min in 100 µl solution A (freshly prepared 80% 50 mm ammonium bicarbonate/20% acetonitril) and then air-dried for 10–15 min. Dithiothreitol (DTT; 100 µl of 10 mm DTT in 50 mm ammonium bicarbonate) was then added and incubated at 65°C for 30 min to reduce the disulphide bonds after which time the liquid was removed. Iodacetamide (100 µl of 100 mm iodoacetamide in 50 mm ammonium bicarbonate) was added and incubated for 30 min at room temperature in the dark to alkylate the sulphur-groups. Trypsin (5 µl containing 50 ng trypsin in 10 mm ammonium bicarbonate) was then added and incubated at 37°C for 2–4 h to digest the proteins. Trypsin was inactivated by adding 5% formic acid for 20 min, after which time the plugs were frozen rapidly in liquid nitrogen and stored at −80°C. For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis, the spots were analysed at the John Innes Centre Protein Sequencing Facility using a Reflex III (Bruker). Peptide peak lists were searched against the database of S. pneumoniae TIGR4 (http://www.matrixscience.com/).
Cloning, expressing and purification of recombinant proteins
Fructose–bisphosphate aldolase (FBA; SP0605 S. pneumoniae TIGR4), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; SP2012 S. pneumoniae TIGR4) and heat shock protein 70 (DnaK; SP0517 S. pneumoniae TIGR4) genes were amplified from S. pneumoniae strain WU2 genomic DNA by polymerase chain reaction (PCR) with primers FBA forward (5′-TCGGATC CATGGCAATCGTTTCAGCAGA-3′) and FBA reverse (5′-TCGAGCTCTGCTTTACCTTCTGAACCGA-3′), GAPDH forward (5′-TCGGATCCATGATTTTCATAAGGAGGA-3′) and GAPDH reverse (5′-TCGAGCTCTTTAGCAATTTTTGC GAAG-3′), and DnaK forward (5′-TCGGATCCAAGAGTATC AAAAAGAAAAA-3′) and DnaK reverse (5′-TCGAGCTCTT CATCCAATACACTCAT-3′). The forward and reverse primers contain BamHI and SacI recognition sequences, respectively, and all primers contain 5′-TC spacers. The primers flank the entire open reading frames. The amplified and BamHI-SacI (Takara Bio Inc, Shiga, Japan) digested DNA-fragments were cloned into the pHAT expression vector (BD Biosciences Clontech, Palo Alto, CA, USA) and transformed in DH5α UltraMAX ultracompetent E. coli cells. Ampicillin-resistant transformants were cultured and plasmid DNA was analysed by PCR. The pHAT-GAPDH, pHAT-FBA and pHAT-DnaK vectors were purified from DH5α UltraMAX cells using Qiagen High Speed Plasmid Maxi Kit (Qiagen GMBH, Hilden, Germany) and transformed in E. coli host expression strain BL21(DE3)pLysS. The identity of the inserts was confirmed by sequencing. Bacteria were grown overnight and expression of the recombinant his-tagged proteins was induced by the addition of 1 mm IPTG to BL21(DE3)pLysS+GAPDH, BL21(DE3)pLysS+FBA and BL21(DE3)pLysS+DnaK cells for 5 h. The cells were harvested by centrifugation and lysed in lysis buffer (8 m urea, 0·1 m NaH2PO4, 0·01 m Tris-Cl pH 8·0). The histidine-tagged recombinant proteins were purified using a Ni-Nta column (Qiagen GMBH, Hilden, Germany), following binding for 1 h at room temperature. The column was then washed with wash buffer (8 m urea, 0·1 m NaH2PO4, 0·01 m Tris-Cl pH 6·3), and the recombinant proteins were recovered from the column using elution buffer (8 m urea, 0·1 m NaH2PO4, 0·01 m Tris-Cl, pH 5·9). Isolation of the proteins was confirmed by Western blot analysis using anti-HAT antibodies (BD Biosciences Clontech, Palo Alto, CA, USA) and by MALDI-TOF mass spectrometry sequencing. The recombinant protein preparations were stored in elution buffer at −20°C.
Immunization and infection of mice
Six-week-old (n = 164) BALB/c female mice (Harlan Laboratories, Israel) were housed in sterile conditions under 12-h light/dark cycles and fed Purina chow and tap water ad libitum. Animal experimental protocols were reviewed and approved by the Institutional Animal Care and Use Ethic Committee of the Ben-Gurion University of the Negev, Beer Sheva, Israel. Mice were immunized intraperitoneally (i.p.) with 25 µg of rFBA (n = 21), rGAPDH (n = 23) and rDNAk (n = 20), and 75 µl of Inject Alum adjuvant (Pierce Biotechnology, Inc., Rockford, IL, USA) on days 0 (primary immunization) and 21 (booster). Control mice (n = 20) were sham-immunized with adjuvant. The experiments were performed at least on three different occasions and the results were pooled. Blood samples were collected from mice 1 week prior to immunization, 1 week after booster immunization. The sera were pooled for immunological assays.
For respiratory challenge with S. pneumoniae strain WU2 mice were anaesthetized with pentobarbital sodium (0·6 mg/kg) and inoculated intranasally (i.n.) with 2–5 × 108 bacteria (in 25 µl PBS). This inoculum size was used as it was found to be the lowest that causes 100% mortality in our mouse model system within 96 h. Survival was monitored daily. Kaplan–Maier statistical analysis for survival was performed. An additional 10 mice were immunized with FBA and 10 mice with GAPDH and challenged with S. pneumoniae strain 9VR with 3 × 108 CFU.
Bioinformatic analysis
The FBA protein sequence (Swiss-Prot Acc. NP_345117) was compared to the following GenBank databases: nr protein (using BLASTP), nr nucleotide, est, htgs, gss, dbSTS and wgs (using TBLASTN). In addition, both the protein and nucleotide sequences (AJ005697) of FBA were compared to the entire human genome sequence at NCBI and at UCSC. None of the searches revealed a human orthologue. GAPDH (NP_346439) showed 46% identity to the human GAPDH protein (AAH23632).
RESULTS
Cell wall extract of S. pneumoniae contains immunogenic proteins
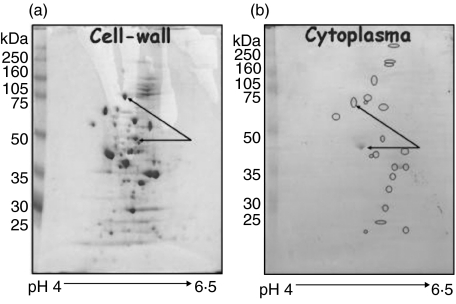
Cell wall proteins were extracted from S. pneumoniae strain WU2 by mutanolysin treatment and then collecting soluble cell wall proteins were collected by centrifugation. Approximately 150 proteins were isolated by the cell wall extraction procedure, as shown by two-dimensional gel electrophoresis (Fig. 1a). When an equivalent amount of protein (40 µg) from total cytoplasmic protein extract was separated by 2D gel electrophoresis and stained with Coomassie brilliant blue the spots were not as easily visible due to the increased complexity of the cytoplasmic protein mixture. The distribution of the major protein spots in the cytoplasmic extract (circled) was also different to that of the cell-wall extract (Fig. 1b).
Fig. 1.
2D PAGE of S. pneumoniae cell wall proteins. (a)Forty µg of enriched cell wall proteins were extracted by mutanolysin treatment of S. pneumoniae, separated by 2D PAGE and stained with Coomassie brilliant blue. More than 150 proteins residing in the pI range 4–6·5 could be visualized. (b) Forty µg of cytoplasmic protein extract prepared by sonication of mutanolysin treated bacteria. The increased complexity of the cytoplasmic protein mixuture is evident from the decreased spot intensity. The distribution of the major protein spots in the cytoplasmic extract (circled) was also different to that of the cell-wall extract. Extensive differences in protein concentration exemplified for two proteins (arrows)
Immunogenicity of the S. pneumoniae cell wall proteins was assessed by 2D Western blot analysis with human sera from children and adults. The sera were collected from healthy adult volunteers and longitudinally from healthy children at 18, 30 and 42 months of age who were attending day-care centres. The children were shown to be carriers of multiple strains of pneumococcus starting at 12 months of age. Over the course of the study, child 1 (DCC study no. 1014) carried S. pneumoniae serotypes 6A, 19F and 23F; child 2 (DCC study no. 1020) carried S. pneumoniae serotypes 11, 15, 23A, and 23F; child 3 (DCC study no. 1021) carried S. pneumoniae serotypes 6A, 11, 15, 19F and 23A; child 4 (DCC study no. 1032) carried S. pneumoniae serotypes 6A, 12, 15 and 23F; child 5 (DCC study no. 1007) carried S. pneumoniae serotypes 6A, 15, 19 A, 19F, 23A and 23F; child 6 (DCC study no. 1029) carried S. pneumoniae serotypes 19F and 23A; child 7 (DCC study no. 1001) carried S. pneumoniae serotypes 6A, 6B, 15, 19F and 23F; and child 8 (DCC study no. 1048) carried S. pneumoniae serotypes 6A, 6B, 14, 15, 19A and 23F.
The proteins were identified using MALDI-TOF mass spectrometry. Seventeen proteins were antigenic using sera from children and adults. In Table 1 we present 13 proteins that showed an increasing antibody response with age in all the eight children analysed (Fig. 2). Antigenicity of other proteins did not change over the course of the study (Table 1). Eight proteins are involved in glycolysis, i.e. l-lactate dehydrogenase, UDP-glucose 4-epimerase, enolase, fructose–bisphosphate aldolase (FBA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucose-6-phosphate isomerase, phosphoglycerate kinase and 6-phosphoglutamate dehydrogenase. Two proteins involved in protein synthesis were identified, i.e. glutamyl-tRNA amidotransferase and glutamyl-tRNA synthetase. One chaperon a protein was also identified, i.e. DnaK/HSP70. The low immunogeneicity of DnaK found in the current study is in agreement with previously published data [22]. A group of proteins belonging to the other physiological pathways were found among them: NADP glutamate dehydrogenase, aminopeptidase C, carbamoyl-phosphate synthase, aspartate carbamoyltransferase and pyruvate oxidase. In addition, an open reading frame (NP358083) without known function was identified.
Table 1.
Identification of Pnc surface proteins with age-dependent immunogenicity
| Immunoreactivity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spot | By MALDI-TOF analysis Homology | SP number | Acc.number | Mascot* | MW | PI | Age 1·5 | Age 2·5 | Age 3·5 | Adult |
| Proteins with low immunogenicity | ||||||||||
| 1* | DNA K | SP0517 | NP_345035 | 173 | 64·8 | 4·6 | * | * | ||
| 23 | NADP-specific glutamate dehydrogenase | SP1306 | NP_345769 | 186 | 49 | 5·3 | * | * | * | |
| Proteins with increased immunogenicity | ||||||||||
| 7 | Glutamyl-tRNA Amidotransferase subunit A | SP0437 | NP_344959 | 83 | 52 | 4·9 | * | ** | ** | *** |
| 13 | l-lactate dehydrogenase | SP1220 | NP_345686 | 134 | 35·9 | 5·2 | * | ** | * | ** |
| 14 | Glyceraldehyde 3-phosphate dehydrogenase | SP2012 | NP_346439 | 350 | 37·1 | 5·7 | * | ** | *** | *** |
| 15 | Fructose–biphosphate aldolase | SP0605 | NP_345117 | 106 | 31·5 | 5 | ** | *** | *** | *** |
| 16 | UDP-glucose 4-epimerase | SP1607 | NP_346051 | 116 | 37·5 | 4·8 | ** | * | * | ** |
| 22 | Glutamyl-tRNA synthetase | SP2069 | NP_346492 | 194 | 56 | 4·9 | * | ** | ** | |
| 27 | Phosphoglycerate kinase | SP0499 | NP_345017 | 109 | 41·9 | 4·9 | * | ** | ** | ** |
| 29 | Glucose-6-phosphate isomerase | SP2070 | NP_346493 | 96 | 51·3 | 5·2 | * | * | ** | |
| 30 | 6-phosphogluconate dehydrogenase | SP0375 | NP_344902 | 58 | 53·7 | 4·9 | ** | ** | ||
| 31 | Aminopeptidase C | SP0281 | NP_344819 | 120 | 33·7 | 4·8 | ** | ** | ||
| x | Hypothetical protein | SPr0489 | NP_358083 | 15 | 5·2 | * | ** | |||
| 33 | Carbamoyl-phosphate synthase | SP1275 | NP_345739 | 230 | 116·5 | 4·8 | * | ** | *** | |
| 65 | Aspartate carbamoyltransferase | SP1277 | NP_345741 | 44 | 34·7 | 5·1 | * | * | ** | ** |
| Proteins with high immunogenicity | ||||||||||
| 18 | Pyruvate oxidase | SP0730 | NP_345231 | 168 | 65·3 | 5·1 | *** | *** | *** | *** |
| 25 | Enolase (2-phosphoglycerate dehydratase) | SP1128 | NP_345598 | 215 | 47·1 | 4·7 | ** | ** | ** | ** |
The extent of surface protein recognition by the sera was determined by the optical density as measured by the imager used in our study (αInnotech). The Sp or the SPr are the numbers designated to genes at the TIGR Microbial Resource (http://www.tigr.org/togr_scrips/CMR2/CMRHomePage.spl).
Low;
intermediate;
high.
Fig. 2.
Age-dependent enhancement of antibody response to S. pneumoniae surface proteins. S. pneumoniae surface proteins were separated by 2D gel electrophoresis, transferred to nitrocellulolose membrane and probed with pooled (a), 1·5, (b) 2·5 and (c) 3·5-year-old children’s and (d) adult sera. The results demonstrate enhancement of antibody response and widening of the antibody repertoire to S. pneumoniae cell wall proteins along with maturation of the host immune system. The proteins arrowed above were identified by MALDI-TOF mass spectrometry using the protein mass fingerprint technique and the Mascot search tool (Matrix Science, http://www.matrixscience.com). DnaK: heat shock protein 70; FBA: fructose–bisphosphate aldolase; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Immune responses to recombinant fructose biphosphate aldolase (FBA) glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and heat shock protein 70 (DnaK) in mice
Three proteins were selected for further studies: GAPDH and FBA represent proteins that were increasingly antigenic in children over time, whereas DnaK was selected as a control protein with relatively low immunogenicity and was weakly antigenic in all sera tested. The fba, gapdh, and hsp70 (dnaK) genes from S. pneumoniae strain WU2 were cloned and expressed in E. coli as histidine-tagged proteins and purified for immunization of mice. Expression of the recombinant proteins of the correct size was confirmed by polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (Fig. 3a) and with Western blot analysis using anti-HAT antibodies (Fig. 3b).
Fig. 3.
Cloning and expression of recombinant S. pneumoniae fructose–bisphosphate aldolase and glyceraldehyde-3-phosphate dehydrogenase and DnaK-heat shock protein 70. Bacteria were transformed and the proteins as purified as described in the Methods section. (a) Following lysis of the bacteria, recombinant Hat-tagged proteins were purified on Ni-Nta columns, resolved on PAGE and stained with Coomassie brilliant blue. The representative gel demonstrates the presence of single protein bands of expected molecular weight in the eluates. (b) The recombinant FBA and GAPDH were separated on PAGE, transferred onto nitrocellulose membranes and probed with anti-HAT antibodies. (c) Total cell wall (CW) proteins of S. pneumoniae strains 9VR, 14R, 6B, 14·8, 3·8 and WU2 were separated on PAGE, transferred onto nitrocellulose membranes and probed sera from mice immunized with the respective recombinant proteins. Sera of the mice immunized with rFBA and rGAPDH recognized respective native proteins in all preparations, as judged by generation of the band of expected molecular weight. Preimmune sera were used as controls.
Six-week-old female BALB/c mice were immunized intraperitoneally with recombinant FBA (rFBA), GAPDH (rGAPDH) or DnaK (rDnaK) mixed with adjuvant or with adjuvant alone as a control. Sera from mice immunized with rFBA, rGAPDH, rDnaK but not preimmune sera recognized the purified recombinant form of these antigens as shown by Western blot analysis (Fig. 3c). Immune sera to rFBA, rGAPDH and rDnaK were also shown to detect a protein band of the expected size in Western blots containing cell wall protein extracts from the homogeneous strain WU2 (Fig. 3c). Antiserum raised against rFBA and rGAPDH detected the respective proteins extracted from Pnc strains 6BR, 9VR, 14R and 14DW (Fig. 3c), which were confirmed to also be genetically diverse using pulse field gel electrophoresis [23]. Positive staining was observed using flow cytometry analysis with sera from immunized mice and no staining could be seen with preimmune sera. This further confirmed the surface localization of these proteins (see the inserts in Fig. 4a and c).
Fig. 4.
Survival of FBA or GAPDH immunized mice following S. pneumoniae challenge. Mice immunized with the respective protein were intranasally challenged with 1–2 × 108 CFU of S. pneumoniae strain WU2. Survival was monitored daily and compared to that of control sham-vaccinated mice (n = 20). (a) Mice immunization with rFBA protein (n = 21, *P < 0·05). Insert: flow cytometry of WU2 bacteria stained with mouse anti-FBA antibodies (geometric mean 9·26) in comparison to control bacteria stained with a secondary antibody only (geometric mean 2·64) (b) Mice, immunized with rGAPDH (n = 23) proteins (n = 23, *P < 0·05). Insert: flow cytometry of WU2 bacteria stained with mouse anti-GAPDH antibodies (geometric mean 10·96) in comparison to control bacteria stained with a secondary antibody only (geometric mean 2·64) (c) Mice, immunized with rDnaK (n = 20) protein (n = 20). (d) Mice immunized with rFBA (n = 10) and rGAPDH (n = 10) were intranasally challenged with 3 × 108 CFU of S. pneumoniae strain 9VR. Survival was monitored daily and compared to that of control, sham-vaccinated mice (n = 20). *P < 0·05.
Immunization with rFBA and rGAPDH elicits protection against respiratory challenge with S. pneumoniae
BALB/c mice were immunized with rFBA, rGAPDH, rDnaK or adjuvant alone as a control. One week after the booster immunization mice were challenged. Intranasal inoculation with 5 × 108 CFU of S. pneumoniae strain WU2 caused lethal infection in 100% of non-vaccinated mice (data not shown). All control and rDnaK immunized mice succumbed to lethal infection within 120 h post-challenge, whereas 36% of the mice vaccinated with either rFBA and rGAPDH survived 21 days post-challenge (Fig. 4a,b,c, P < 0·05 compared to control).
Interestingly, immunization of the mice with FBA or GAPDH protected the mice from a challenge with a genetically unrelated strain (strain 9VR) to the same extent as challenge with strain WU2 (Fig. 4d).
Bioinformatics analysis
It should be noted that while GAPDH had 46% identity to human GAPDH, sequence database searches with FBA revealed no human orthologues.
DISCUSSION
Pneumococcal surface proteins have a wide range of functions, including adherence to host tissues, binding to specific immune system components, protein processing, nutrient acquisition and uptake of DNA from the environment [24]. Immunization with several pneumococcal surface proteins involved in virulence such as PspA [25–27], PsaA [28,29] autolysin [30,31] and neuraminidase [32] has been shown to confer protection against pneumococcal infection in animal models.
Recently, Wizemann et al. [33] proposed a whole genome-based approach for identification of novel vaccine proteins for protection against pneumococcal infection. This approach is based on identifying surface proteins from the genome sequence by searching for sequence motifs commonly related to their secretion or surface binding. From the 130 identified open reading frames with secretion motifs or similarity to predicted virulence factors, 108 proteins were used for immunization of mice, and six proteins conferred protection against disseminated pneumococcal infection.
To identify novel protective antigens, we screened an enriched pneumococcal cell wall protein extract using 2D gel electrophoresis and Western blotting with human sera from children of different age groups for antigenic proteins. In addition, reactivity of sera from healthy adults was assessed. Our hypothesis was that longitudinal study of protein antigenicity in day-care children who are frequently exposed to S. pneumoniae would identify protein antigens involved in the development of natural immunity to S. pneumoniae. In a complex mixture of around 150 cell wall extracted proteins, 70 were identified using mass spectrometry and 30 were antigenic.
We found that the antigenic proteins from the enriched cell wall extract fell into three groups. The first group comprised proteins with low antigenicity. The second group consists of antigens for which the antigenicity seemed to increase with age of children attending day-care centres, while the third group of proteins was highly antigenic with all sera tested. The existence of serum antibodies to a certain bacterial protein does not necessarily indicate their capacity to elicit protective immune response against the bacteria. However, the increase in the antibody response to bacterial proteins which coincides with the diminution in morbidity described in children [2] encouraged us to test these proteins for their ability to elicit protection against S. pneumoniae.
This second group of proteins included known virulence factors for S. pneumoniae, such as enolase [34] and pyruvate oxidase [35]. Other proteins belonging to this group, such as dipeptidyl aminopeptidase, were described previously as virulence factors for other protozoa, such as Leishmania spp. [36] and leucil aminopeptidase Porphyromonas gingivalis[37]. Some of the proteins identified were reported previously to be associated with the cell surface of Streptococcus agalactiae, among them phosphoglycerate kinase, glucose 6 phosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase [38]. Recently, similar observations were published regarding S. pyogenes[39].
The detection of several metabolic cytoplasmic enzymes on the cell surface of S. pneumoniae is puzzling because none of those proteins contain signal peptides, LPXTG motif or choline binding repeats. However, anchorless adhesins and invasins have been described previously, such as PavA and enolase α on the surface S. pneumoniae and S. pyogenes[40]. Furthermore, the importance of the surface-displayed enzymes to bacteria virulence was demonstrated previously by Zysk et al. [41], who showed that antisera raised against such surface proteins (ornithine carbamoyltransferase and phosphoglycerate kinase) can protect passively against infection in a neonatal murine model of S. agalactiae infection. Therefore, these enzymes might belong to a family of dual-function proteins and act as a virulence factor in addition to being ‘housekeeping’ proteins. Pneumococcal α-enolase functions as an enolase and as a plasmin-binding protein [34]. Staphylococcal GAPDH has also been assigned a function as a virulence factor − it serves as a transferrin-binding protein [42,43] and a plasminogen-binding protein [44]. Interestingly, the GAPDH and FBA proteins, effective in eliciting protection against S. pneumoniae upon vaccination, have been demonstrated to elicit protection against Schistosoma mansonii[45] and Onchocerca volvulus[46], respectively. Database searches revealed no human orthologue to the FBA used in the current study.
Surprisingly, none of the choline binding proteins and none of the putative cell wall anchored proteins (sortase substrates) were identified among the 18 immunogenic cell wall proteins. We have described previously the immunological detection of PspA in S. pneumoniae cytoplasmic membrane protein preparation from Pnc strain 14DW [17]. Therefore, we checked for the presence of PspA in the enriched cell wall extracts and membrane protein preparation from WU2 strain by 2D Western blot analysis using anti-PspA antibodies. PspA was detected by 2D-Western blot analysis mainly in the membrane preparation but was not visible on a Coomassie brilliant blue-stained 2D-protein gel, indicating that the abundance of this protein in our preparations is relatively low.
We conclude that the immunogenic enzymes with an age-dependent increase in antigenicity of S. pneumoniae found in enriched cell wall extract may represent a novel class of vaccine candidates, under the stipulation that they either will not share homologies to the human enzymes or proven not to elicit autoimmune diseases. These proteins should also be conserved among different S. pneumoniae strains as demonstrated in this study. GAPDH and FBA enzymes were shown to elicit protective level immune responses in mice and afford significant protection against respiratory challenge with virulent S. pneumoniae. Given that these enzymes play a crucial role in glycolysis they are likely to be conserved and present in all strains of S. pneumoniae. Taken together, these findings show that the immunoproteomics-based approach implemented in the present study has the ability to identify novel targets for S. pneumoniae vaccine and FBA is a promising candidate antigen that warrants further studies.
Acknowledgments
This work was supported by grants from the Israeli MOH numbers 4476 and 5540, BGNegev Biotechnology, BGU seed money no. 80904101 to YMN and European Bacterial Proeomic Thematic Network QLK2-CT-2000–01536 to JW and KO. Special thanks to Ms Marilou Shagan for her excellent technical help, and to Dr Angel Progador for his help with the flow cytometry analysis.
REFERENCES
- 1.Siber GP. Pneumococcal disease: prospects for a new generation of vaccines. Science. 1994;265:1385–7. doi: 10.1126/science.8073278. [DOI] [PubMed] [Google Scholar]
- 2.Zangwill KM, Vadheim CM, Vannier AM, Hemenway LS, Greenberg DP, Ward JL. Epidemiology of invasive pneumococcal disease in southern California: implications for the design and conduct of a pneumococcal conjugate vaccine efficacy trial. J Infect Dis. 1996;174:752–9. doi: 10.1093/infdis/174.4.752. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Preventing pneumococcal disease among infants and young children: recommendations of the advisory Committee on Immunization Practices (ACIP) MMWR. 2000;49:888–92. (no. RR-9). [PubMed] [Google Scholar]
- 4.Hutchison BG, Oxman AD, Shannon HS, Lloyd S, Altmayer CA, Thomas K. Clinical effectiveness of pneumococcal vaccine. Meta-analysis. Can Fam Physician. 1999;45:2381–93. [PMC free article] [PubMed] [Google Scholar]
- 5.Wuorimaa T, Käyhty H. Current state of pneumococcal vaccines. Scand J Immunol. 2000;56:111–29. doi: 10.1046/j.1365-3083.2002.01124.x. [DOI] [PubMed] [Google Scholar]
- 6.Breiman RF, Keller DW, Phelan MA, et al. Evaluation of effectiveness of the 23-valent pneumococcal capsular polysaccharide vaccine for HIV-infected patients. Arch Intern Med. 2000;160:2633–8. doi: 10.1001/archinte.160.17.2633. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization (WHO) Pneumococcal vaccines. Wkly Epidemiol Rec. 1999;74:177–83. . WHO position paper. [PubMed] [Google Scholar]
- 8.Whitney CG, Farley MM, Hadler J, et al. Active bacterial core surveillance of the emerging infections program network. Decline in invasive pneumococcal disease after the introduction of protein–polysaccharide conjugate vaccine. N Engl J Med. 2003;348:1737–46. doi: 10.1056/NEJMoa022823. [DOI] [PubMed] [Google Scholar]
- 9.Butler JC. Epidemiology of pneumococcal serotypes and conjugate vaccine formulations. Microb Drug Resist. 1997;3:125–9. doi: 10.1089/mdr.1997.3.125. [DOI] [PubMed] [Google Scholar]
- 10.Dagan R, Givon-Lavi N, Zamir O, et al. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J Infect Dis. 2002;185:927–36. doi: 10.1086/339525. [DOI] [PubMed] [Google Scholar]
- 11.Goldblatt D. Conjugate vaccines. Clin Exp Immunol. 2000;119:1–3. doi: 10.1046/j.1365-2249.2000.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hausdorff WP, Siber G, Paradiso PR. Geographical differences in invasive pneumococcal disease rates and serotype frequency in young children. Lancet. 2001;357:950–2. doi: 10.1016/S0140-6736(00)04222-7. [DOI] [PubMed] [Google Scholar]
- 13.Hakenbeck R. Beta-lactam-resistant Streptococcus pneumoniae: epidemiology and evolutionary mechanism. Chemotherapy. 1999;45:83–94. doi: 10.1159/000007170. [DOI] [PubMed] [Google Scholar]
- 14.McCool T, Cate T, Moy G, Weiser J. The immune response to pneumococcal proteins during experimental human carriage. J Exp Med. 2002;195:359–65. doi: 10.1084/jem.20011576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zysk G, Bethe G, Nau R, et al. Immune response to capsular polysaccharide and surface proteins of Streptococcus pneumoniae in patients with invasive pneumococcal disease. J Infect Dis. 2003;187:330–3. doi: 10.1086/367701. [DOI] [PubMed] [Google Scholar]
- 16.Virolainen A, Jero J, Chattopadhyay P, Karrma P, Eskola J, Leinonen M. Comparison of serum antibodies to pneumolysin with those of pneumococcal capsular polysaccharides in children with acute otitis media. Pediatr Infect Dis. 1996;15:128–33. doi: 10.1097/00006454-199602000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Lifshitz S, Dagan R, Shani-Sekler M, et al. Age-dependent preference in human antibody responses to Streptococcus pneumoniae polypeptide antigens. Clin Exp Immunol. 2002;127:344–53. doi: 10.1046/j.1365-2249.2002.01745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapola S, Jantti V, Eerola M, Makela PH, Kayhty H, Kilpi T. Anti-PsaA and the risk of pneumococcal AOM and carriage. Vaccine. 2003;21:3608–13. doi: 10.1016/s0264-410x(03)00409-2. [DOI] [PubMed] [Google Scholar]
- 19.Siegel JL, Hurst SF, Liberman ES, Coleman SE, Bleiweis AS. Mutanolysin-induced sphroplasts of Streptococcus mutans are true protoplasts. Infect Immun. 1981;31:808–15. doi: 10.1128/iai.31.2.808-815.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. Canada: Analysis of protein. Wiley & Sons; 2003. 10.2.1–10.8.18. [Google Scholar]
- 21.Austrian R. The Quellung reaction, a neglected microbiologic technique. Mount Sinai J Med. 1976;43:669–709. [PubMed] [Google Scholar]
- 22.Kolberg J, Hoiby EA, Aase A, et al. Streptococcus pneumoniae heat shock protein 70 does not induce human antibody responses during infection. FEMS Immunol Med Microbiol. 2000;29:289–94. doi: 10.1111/j.1574-695X.2000.tb01536.x. [DOI] [PubMed] [Google Scholar]
- 23.Mizrachi Nebenzahl Y, Porat N, Lifshitz S, et al. Virulence of Streptococcus pneumoniae may be determined independently of capsular polysaccharides. FEMS Microbiol Let. 2004;233:147–52. doi: 10.1016/j.femsle.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 24.McCullers JA, Tuomanen EI. Molecular pathogenesis of pneumococcal pneumonia. Front Biosci. 2000;6:D877–89. doi: 10.2741/mccullers. [DOI] [PubMed] [Google Scholar]
- 25.Briles DE, Tart RC, Wu HY, Ralph BA, Russell MW, McDaniel LS. Systemic and mucosal protective immunity to pneumococcal surface protein A. Ann NY Acad Sci. 1996;797:118–26. doi: 10.1111/j.1749-6632.1996.tb52954.x. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel LS, Sheffield JS, Delucchi P, Briles DE. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–8. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talkington DF, Brown BG, Tharpe JA, Koenig A, Russell H. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA) Microb Pathog. 1996;21:17–22. doi: 10.1006/mpat.1996.0038. [DOI] [PubMed] [Google Scholar]
- 28.Briles DE, Ades E, Paton JC, et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun. 2000;68:796–800. doi: 10.1128/iai.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briles DE, Hollingshead S, Brooks-Walter A, et al. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine. 2000;18:1707–11. doi: 10.1016/s0264-410x(99)00511-3. [DOI] [PubMed] [Google Scholar]
- 30.Berry AM, Lock RA, Hansman D, Paton JC. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–30. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lock RA, Hansman D, Paton JC. Comparative efficacy of autolysin and pneumolysin as immunogens protecting mice against infection by Streptococcus pneumoniae. Microb Pathog. 1992;12:137–43. doi: 10.1016/0882-4010(92)90116-6. [DOI] [PubMed] [Google Scholar]
- 32.Lock RA, Paton JC, Hansman D. Purification and immunological characterization of neuraminidase produced by Streptococcus pneumoniae. Microb Pathog. 1988;4:33–43. doi: 10.1016/0882-4010(88)90046-0. [DOI] [PubMed] [Google Scholar]
- 33.Wizemann T, Heinrichs J, Adamou J, et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect Immun. 2001;69:1593–8. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. α-Enolase of Streptococcus pneumoniae is a plasmin (ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40:1273–87. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 35.Spellerberg B, Cundell DR, Sandros J, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19:803–13. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 36.Morty RE, Morehead J. Cloning and characterization of leucil aminopeptidase from three pathogenic Leishmania species. J Biol Chem. 2002;277:26057–65. doi: 10.1074/jbc.M202779200. [DOI] [PubMed] [Google Scholar]
- 37.Yagishita H, Kumagai Y, Konishi K, Takahashi Y, Aoba T, Yoshikawa M. Histopathological studies on virulence of dipeptidyl aminopeptidase IV (DPPIV) of Porphyromonas gingivalis in a mouse abscess model: use of a DPPIV-deficient mutant. Infect Immun. 2001;69:7159–61. doi: 10.1128/IAI.69.11.7159-7161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes MJ, Moore JC, Lane JD, et al. Identification of major outer surface proteins of Streptococcus agalactiae. Infect Immun. 2002;70:1254–9. doi: 10.1128/IAI.70.3.1254-1259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Costa SS, Romer TG, Boyle MD. Analysis of expression of a cytosolic enzyme on the surface of Streptococcus pyogenes. Biochem Biophys Res Commun. 2000;278:826–32. doi: 10.1006/bbrc.2000.3884. [DOI] [PubMed] [Google Scholar]
- 40.Chhatwal GS. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 2002 May;10:205–8. doi: 10.1016/s0966-842x(02)02351-x. [DOI] [PubMed] [Google Scholar]
- 41.Zysk G, Bongaerts RJ, ten Thoren E, Bethe G, Hakenbeck R, Heinz HP. Detection of 23 immunogenic pneumococcal proteins using convalescent-phase serum. Infect Immun. 2000;68:3740–3. doi: 10.1128/iai.68.6.3740-3743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–26. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modun B, Williams P. The staphylococcal transferring-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67:1086–92. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergmann S, Rohds M, Hammerschmidt S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun. 2004;72:2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Argiro L, Kohlstadt S, Henri S, et al. Identification of a candidate vaccine peptide on the 37 kDa Schistosoma mansoni GAPDH. Vaccine. 2000;18:2039–48. doi: 10.1016/s0264-410x(99)00521-6. [DOI] [PubMed] [Google Scholar]
- 46.McCarthy JS, Wieseman M, Tropea J, et al. Onchocerca volvulus glycolytic enzyme fructose−1,6-biphosphate aldolase as a target for a protective immune response in humans. Infect Immun. 2002;70:851–8. doi: 10.1128/IAI.70.2.851-858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]