Abstract
To provide a basis for beginning to explore the CD94/NKG2 family of molecules in rhesus monkeys, we sought to characterize the expression of these inhibitory and activating cell signalling molecules in peripheral blood mononuclear cells (PBMCs) from healthy rhesus monkeys. We developed and employed a semiquantitative polymerase chain reaction (PCR)-based assay to evaluate mRNA expression levels of nine NKG2 molecules in PBMCs from the monkeys. In addition to quantitating NKG2A, NKG2B, NKG2C2, NKG2C and NKG2D expression, mRNA expression of transmembrane-deleted forms of these molecules was also evaluated. Significant variability in NKG2 mRNA expression in the PBMCs was detected, with 15 unique NKG2 expression level profiles detected in a study of 15 monkeys. We also found that the ratio of the expressed levels of mRNA of the four NKG2 splice variants, NKG2A, NKG2B, NKG2AΔtm, and NKG2BΔtm, was variable between the monkeys as well as in an individual monkey over a period of 1·5 years. These findings indicate the dynamic nature of NKG2 mRNA expression in the rhesus monkey.
Keywords: CD94/NKG2, natural killer cells, non-human primate, semi-quantitative PCR
INTRODUCTION
The function of a lymphocyte is often dramatically altered by its level of expression of selected cell-surface signalling molecules. For example, the ability of a T lymphocyte to proliferate in response to interleukin (IL)-2 is modulated by the expression of the high affinity chain of the IL-2 receptor on that cell's surface [1]. Whether a T cell will respond to an interaction with a B7 molecule by activation or anergy is controlled by the cell's relative surface expression of CD28 and CTLA-4 [2]. Moreover, infection by pathogens can alter the cell surface expression of signalling molecules, and these changes can alter the very nature of the host immune cell–pathogen interaction. Immune activation can cause an up-regulation of CCR5 expression on a T cell that in turn increases the susceptibility of that cell to HIV infection [3–5]. Furthermore, adenovirus interferes with MHC class II up-regulation [6]. These types of observations suggest that an understanding of the biology of lymphocyte signalling molecules requires an understanding of the expression levels of these molecules in vivo.
Surface-expressed cell signalling molecules play a central role in the regulation of natural killer (NK) cells in vivo. Many viruses down-regulate major histocompatibility complex (MHC) class I expression to evade the host cytotoxic T lymphocyte (CTL) response, and NK cells are specifically activated by their recognition of these changes in MHC class I expression [7]. The expression by NK cells of the CD94/NKG2 family of signalling molecules is involved in the recognition of these events. The CD94/NKG2 molecules are a heterodimeric family composed of an invariant CD94 molecule bound covalently to various NKG2 molecules (NKG2A, NKG2B, NKG2C and NKG2E) [8–12]. They interact with the non-classical MHC molecule HLA-E, which presents a peptide derived from an MHC class I signal sequence [13,14]. CD94/NKG2A is able to inhibit NK and T cell cytolytic activity. This inhibition of lytic activity is mediated by two immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic domain of NKG2A [15]. In contrast, NKG2C lacks ITIMs, but contains a charged lysine in its transmembrane domain that is essential for interaction with the immunoreceptor tyrosine-based activation motif (ITAM)-bearing DAP12 [15,16]. Through this interaction, the CD94/NKG2C heterodimer can activate NK cell cytolytic activity. NKG2D, a homodimer that also functions to activate NK cell lysis, binds to the non-classical MHC molecule MICA [17,18].
We recently characterized the CD94/NKG2 family in peripheral blood mononuclear cells (PBMCs) of the rhesus monkey and found its NKG2A, NKG2B, NKG2C and NKG2D molecules to be highly homologous to their human counterparts [19,20]. A novel molecule, NKG2C2, was also identified that is identical to rhesus NKG2C except for a 15-amino acid insertion in the stalk region of the extracellular domain. Kravitz et al. [21] have identified a novel rhesus monkey molecule, termed NKG2C62, that contains a 15-amino acid insertion in the stalk of the extracellular domain identical to that seen in rhesus monkey NKG2C2. The 15-amino acid insertion in both NKG2C2 and NKG2C62 appears to be the result of an exon duplication [22]. As reported previously in humans and mice, alternatively spliced forms of the CD94/NKG2 molecules that lack the transmembrane domain were also identified in the rhesus monkey and termed CD94Δtm, NKG2AΔtm, NKG2BΔtm, NKG2CΔtm and NKG2DΔtm [19].
To provide a basis for beginning to explore the role of the CD94/NKG2 family of molecules in both health and disease, we sought to characterize the expression of these inhibitory and activating cell signalling molecules in PBMCs of healthy rhesus monkeys. Because a complete panel of monoclonal antibodies that distinguishes the NKG2 family members does not exist, mRNA expression studies were needed to assess the expression of these molecules. In fact, a recent study demonstrated that NKG2 expression in human lymphocytes appears to be regulated at least at the transcriptional level as PBMCs of 52 evaluated individuals possessed the genes encoding NKG2A, NKG2C, NKG2D and NKG2E, yet mRNA expression was not detected for all molecules in each individual [23]. We therefore developed a semiquantitative polymerase chain reaction (PCR)-based assay that allowed us to evaluate mRNA expression of nine NKG2 molecules in rhesus monkey PBMCs.
MATERIALS AND METHODS
Animals and viruses
EDTA-anticoagulated blood samples were obtained from normal rhesus monkeys (Macaca mulatta). Normal monkeys are defined as not having been infected with a pathogen and not having any known disease. These animals were maintained in accordance with the guidelines of the Committee on Animals for the Harvard Medical School and the Guide for Care and Use of Laboratory Animals.
RNA isolation and cDNA generation
EDTA-anticoagulated whole blood was subjected to Ficoll diatrizoate density gradient centrifugation to isolate leucocytes (Ficopaque; Pharmacia Chemical Co., Piscataway, NJ, USA). Total RNA was extracted from 1 × 107 cells using the RNeasy mini kit (Qiagen, Inc., Chatsworth, CA, USA). DNase treatment was performed during the RNA extraction with RNase-free DNase I (Qiagen, Inc.) to avoid DNA contamination. To generate cDNA, 0·5 µg of RNA was reverse transcribed using the Reverse Transcription System (Promega, Madison, WI, USA).
Primers
PCR primers were designed to recognize either GAPDH or specific NKG2 family members. The forward primer A.fam.F1 (5′-GATGGATAACCAAGGAGTAATCTAC-3′) and the reverse primer A.fam.R1 (5′-CTTACAATGATATATTTTTGA AGATCCAC-3′) were used to amplify NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm. The forward primer C.fam.F4 (5′-ATTGATCAAATATATGACTGCCAAG-3′) and the reverse primer C.fam.R1 (5′-GTAGCATTGCACAGTTATGTTCAGC-3′) were used to amplify NKG2C2, NKG2C and NKG2CΔtm. The forward primer D.fam.F1 (5′-GATGGGGTGGATTCGTGGTC-3′) and the reverse primer D.fam.R1 (5′-GGAGCCATCTTC CCACTGC-3′) were used to amplify NKG2D and NKG2DΔtm. The forward primer GAP.F1 (5′-AGGCTGGGGCTCATTTGC-3′) and the reverse primer GAP.R1 (5′-GTGCTCAGTGTAGC CCAGGATC-3′) were used to amplify GAPDH.
Semi-quantitative PCR assay
All PCR samples were first incubated at 94°C for 10 min to activate the polymerase AmpliTaq Gold (Perkin Elmer, Foster City, CA, USA). PCR reactions using the A primer pair were performed for 30 cycles (94°C for 30 sec, 64°C for 45 sec, 72°C for 60 sec). PCR reactions using the C primer pair were performed for 30 cycles (94°C for 30 sec, 66°C for 45 sec, 72°C for 60 sec). PCR reactions using the D primer pair were performed for 30 cycles (94°C for 30 sec, 69°C for 45 sec, 72°C for 60 sec). GAPDH was amplified using specific primers for 25 cycles (94°C for 30 sec, 66°C for 45 sec, 72°C for 60 sec). At least three serial twofold dilutions of cDNA were used as template for each series of PCR reactions.
PCR products were separated on a 5% polyacrylamide gel. The gels were incubated in a 1 : 10 000 dilution of SYBR-Gold (Molecular Probes, Eugene, OR, USA) in TE for 15 min in the dark. PCR products were then visualized by fluorescent detection using the Storm Instrument (Molecular Dynamics, Sunnyvale, CA, USA). Bands were quantified and assigned an area value using ImageQuant software (Molecular Dynamics). Each area value was divided by the appropriate dilution to obtain a 1× value for each sample. Two 1× area values within the linear range for each sample were averaged. Samples were considered within the linear range if the area values differed by less than 15%. If values for a particular sample were not in the linear range, additional cDNA dilutions were PCR amplified. To obtain a normalized value for each sample, the average 1× NKG2 area value was divided by the average 1× GAPDH area value.
Statistical analysis
Statistical data in normal rhesus monkeys was performed with the two-sided Wilcoxon rank sum test.
Transient transfections
293T cells were transfected by the calcium phosphate method using the following constructs: pNKG2A, pNKG2A′, pNKG2B, pNKG2C2, pNKG2C2′, pNKG2C, pNKG2C′, pNKG2C′′, pNKG2C′′′, pNKG2D or pCD94. Cells were harvested with 5 mm EDTA 48 h post-transfection.
Flow cytometric analysis and cell sorting
The antihuman NKG2A (Z199) monoclonal antibody (Coulter, Miami, FL, USA) used for this study was conjugated to phycoerythrin (PE). The conjugated antibody was used to stain either transfected cells or 100 µl of fresh whole blood from each monkey. The monoclonal antibodies used for the cell sorting studies were conjugated to FITC, PE-texas red (ECD) or allophycocyanin (APC). The following MoAbs were used: anti-CD4-ECD and anti-CD8α-FITC (Becton Dickinson, Franklin Lakes, NJ, USA). The MoAb FN18, which recognizes rhesus monkey CD3, a gift from Dr D. M. Neville Jr (National Institutes of Health, Bethesda, MD, USA), was directly coupled to APC. The conjugated antibodies were used to stain 40–60 × 106 PBMC from each monkey. A Coulter EPICS Elite ESP was used to sort stained lymphocytes into CD3+ CD8+ CD4− and CD3− CD8+ CD4− cell populations.
RESULTS
Development of a semiquantitative PCR-based assay to evaluate NKG2 mRNA expression in rhesus monkey lymphocytes
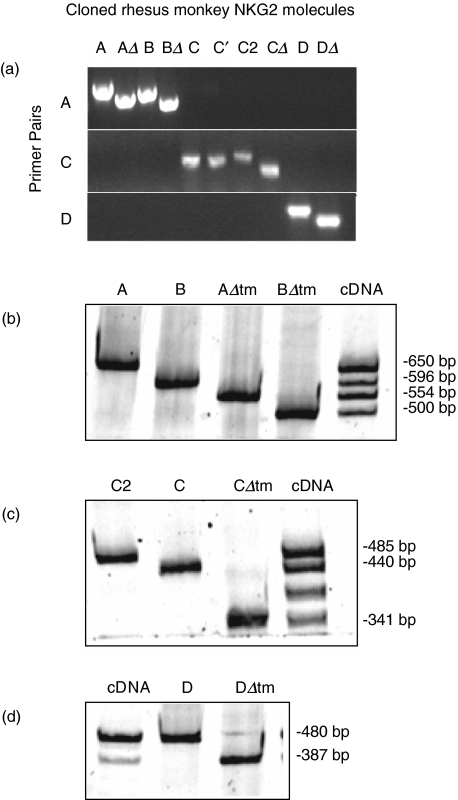
To quantify the magnitude of NKG2 expression in lymphocytes of the rhesus monkey, we developed a PCR-based assay to evaluate the mRNA expression of nine NKG2 molecules. We designed PCR primer pairs that would specifically amplify each member of the rhesus monkey NKG2 family of molecules. Initially, cloned cDNAs corresponding to nine members of the rhesus monkey NKG2 family were amplified with three specific primer pairs (Fig. 1a). In the cases where an individual primer pair amplified more than one cloned cDNA, these molecules could be distinguished by size (Fig. 1). For instance, NKG2B, NKG2AΔtm and NKG2BΔtm are each alternatively spliced forms of NKG2A. Because the 5′ and 3′ ends of these four molecules are identical, the A primer pair simultaneously amplified NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm. As shown in Fig. 1b, NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm amplified from plasmids containing these cloned NKG2s, as well as from lymphocyte-derived cDNA, were detected readily and could be distinguished by size (Fig. 1b). NKG2CΔtm could similarly be distinguished from NKG2C and NKG2C2 (Fig. 1c). Rhesus monkey NKG2D and NKG2DΔtm were also detected using this approach (Fig. 1d). Although we did not analyse mRNA expression of rhesus monkey NKG2FE, NKG2F2 or NKG2Ce [19], the primer pairs used in this study did not amplify these additional molecules (data not shown). Thus, we were able to demonstrate that each NKG2 cDNA could be detected specifically by PCR using specific primer pairs followed by size separation by PAGE. This specific detection of each NKG2 molecule facilitated the analysis of mRNA expression of each NKG2 family member.
Fig. 1.
PCR amplification of rhesus monkey NKG2 family member cDNA is family specific and NKG2 splice variants can be distinguished on the basis of differing size. (a) The indicated primer pairs were used to amplify cloned molecules and the PCR products were run on a 1% agarose gel. C and C′ represent allelic variants of NKG2C; all other cloned cDNAs represent distinct NKG2 family members. (b) The A-specific primer pair was used to amplify cloned NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm cDNAs, as well as cDNA derived from primary rhesus monkey lymphocytes. PCR products were separated by PAGE and the four NKG2 molecules could be distinguished by size. (c) Cloned NKG2C2, NKG2C and NKG2CΔtm cDNA and rhesus monkey lymphocyte-derived cDNA were amplified with the C primer pair and separated by PAGE. The PCR product intermediate in size between NKG2C and NKG2CΔtm has not been identified, but this does not hinder the analysis of NKG2C2, NKG2C or NKG2CΔtm mRNA expression. (d) The d-specific primer pair was used to amplify cloned NKG2D and NKG2DΔtm cDNA, as well as rhesus monkey lymphocyte-derived cDNA, and the PCR products were separated by PAGE.
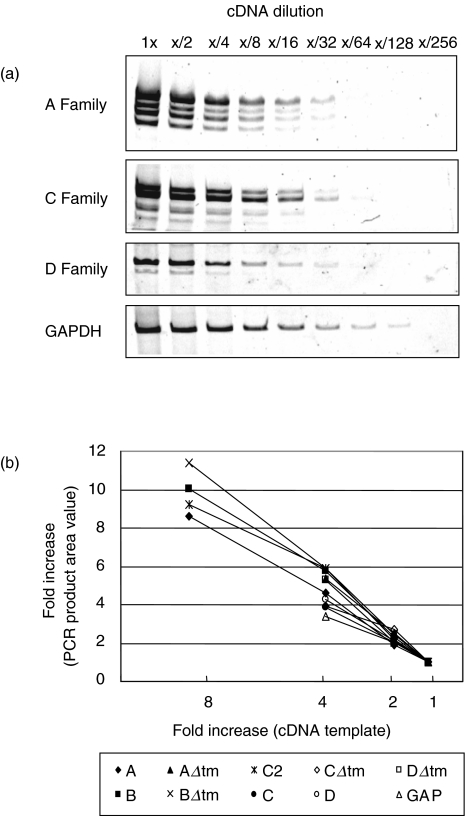
To validate that this PCR amplification of rhesus monkey NKG2 mRNA could be applied to quantify the expression of these molecules, we sought to ensure that a linear range of amplification could be achieved with each primer pair. To this end, we amplified serial twofold dilutions of cDNA using each of the specific A, C and D family primer pairs as well as the GAPDH primer pair. PCR products were separated by polyacrylamide gel electrophoresis (PAGE), stained with SYBR-Gold, and visualized with the Storm instrument (Fig. 2a). ImageQuant software was then used to quantify the PCR products. As demonstrated in Fig. 2b, PCR amplification was linear over at least a twofold difference in starting template for each of the molecules. More specifically, a twofold increase in template cDNA resulted in a twofold increase in the amount of PCR product. These results indicate that PCR amplification under the conditions employed in these studies was linear for each of the 10 examined molecules.
Fig. 2.
PCR amplification of lymphocyte-derived cDNA with NKG2 family-specific primers (a, c and d) and GAPDH primers is sensitive and linear. (a) PCR reactions with the indicated primer pairs were performed to amplify serial twofold dilutions of rhesus monkey lymphocyte-derived cDNA. PCR products were separated by PAGE and visualized with SYBR-Gold DNA stain. (b) Quantification of the PCR products shown in (a). As the template is increased, the amount of PCR product is proportionally increased. The PCR amplification is linear over at least a twofold difference of template.
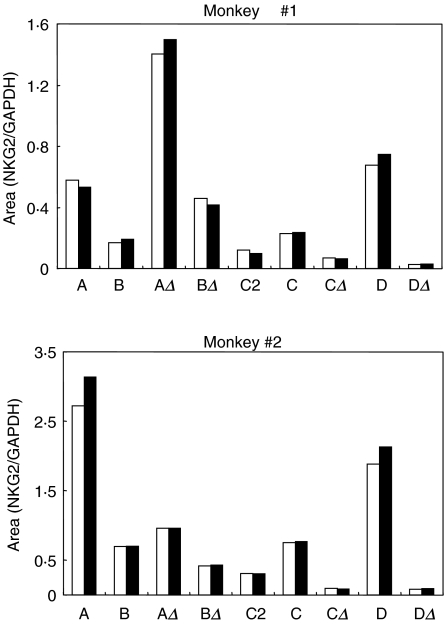
To assess the reproducibility of this assay, we quantified NKG2 expression in replicate cDNA samples from two rhesus monkeys. Serial twofold dilutions of cDNA from each monkey were amplified with the NKG2A, NKG2C and NKG2D specific primers as well as the GAPDH primers. PCR products were separated by PAGE and quantified as described above. Values for NKG2 expression were obtained by normalization of the obtained values to those for GAPDH expression. As demonstrated in Fig. 3, area values for NKG2 expression obtained using this assay were reproducible in duplicate samples from each monkey.
Fig. 3.
Quantification of PCR products amplified with NKG2-specific primer pairs is reproducible. The NKG2-specific and GAPDH-specific primer pairs were used to amplify serial twofold dilutions of cDNA derived from PBMCs of two rhesus monkeys. PCR products were run on a 5% polyacrylamide gel, stained with SYBR-Gold and detected and quantified using imagequant software. NKG2 expression was normalized by GAPDH expression in each sample. Black and white bars indicate duplicate samples.
NKG2 mRNA expression in lymphocytes of normal rhesus monkeys
We utilized the PCR-based assay to evaluate NKG2 mRNA expression by lymphocytes of 15 normal rhesus monkeys. Total RNA was isolated from peripheral blood lymphocytes from the monkeys. cDNA was generated and at least two serial twofold dilutions of cDNA were PCR amplified using the A, C and D family-specific and GAPDH primer pairs.
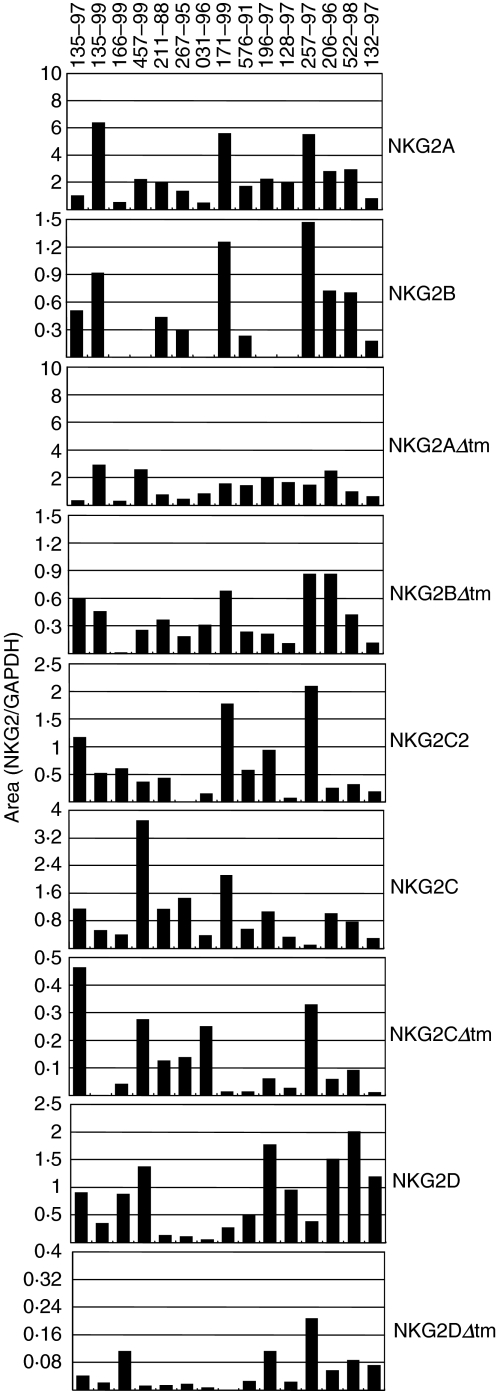
Interestingly, significant variability in NKG2 mRNA expression was observed in the lymphocytes of the 15 rhesus monkeys (Fig. 4). A more than 40-fold difference in NKG2C mRNA expression, more than 38-fold difference in NKG2D mRNA expression and more than 90-fold difference in NKG2BΔtm mRNA expression was detected in these lymphocyte populations.
Fig. 4.
mRNA expression of specific NKG2 molecules is extremely variable in PBMCs of normal rhesus monkeys. The expression of NKG2A, NKG2B, NKG2AΔtm, NKG2BΔtm, NKG2C2, NKG2C, NKG2CΔtm, NKG2D and NKG2DΔtm mRNA in PBMCs of 15 normal rhesus monkeys was evaluated by semiquantitative PCR. Each sample was analysed in duplicate and the resulting values were averaged. The mRNA expression of one NKG2 molecule (either NKG2B, NKG2C2, NKG2CΔtm or NKG2DΔtm) was undetectable in the lymphocytes of eight of the 15 monkeys analysed. All values are shown; for samples without a visible bar, no mRNA expression for that molecule was detected.
Lymphocytes from eight of the 15 evaluated monkeys expressed undetectable levels of one of the NKG2 molecules. NKG2B expression was undetectable in lymphocytes of five of the monkeys. Because NKG2B is a splice variant of NKG2A and the lymphocytes of these monkeys expressed NKG2A as well as the two other NKG2A splice variants, the absence of NKG2B expression was not due to the absence of the genomic copy of this molecule in these cell populations. Lymphocytes of three monkeys did not express NKG2C2, NKG2CΔtm or NKG2DΔtm mRNA. This absence of NKG2CΔtm and NKG2DΔtm mRNA expression was not due to a deletion of the genomic copies of these molecules in these cell populations, as the full-length counterpart of each of these NKG2 molecules was expressed by the same cells. None of the evaluated normal monkeys had undetectable expression of more than one NKG2 molecule.
To determine if this observed variability simply reflected variability in the relative sizes of the NK cell and CD8+ T lymphocyte populations in the PBMC samples, we determined the relative magnitude of NK cell (CD8+ CD3−) and CD8+ T lymphocyte (CD8+ CD3+) populations by flow cytometry in PBMC samples from four of these monkeys. No correlation was apparent between NKG2 mRNA expression profiles and the percentage of NK or CD8+ T cells in the sampled cell populations (data not shown). Thus, the observed variability in the mRNA expression of nine NKG2 molecules in PBMCs of normal rhesus monkeys was not simply a reflection of changes in the relative representation of NK and CD8+ T cells in these cell populations.
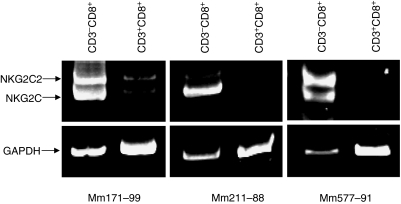
Because NKG2 molecules are expressed on NK cells as well as a subset of T cells in rhesus monkeys, we sought to determine which cell subpopulation expressed NKG2 mRNA. Flow cytometric sorting was used to isolate NK cells (CD8+ CD3− CD4−) and CD8+ T cells (CD8+ CD3+ CD4−) from the PBMCs of three normal rhesus monkeys. We then evaluated the mRNA expression of the splice variants NKG2C2 and NKG2C by these two cell populations. As shown in Fig. 5, NKG2C2 and NKG2C expression was detected predominately in CD8+ CD3− cells. Therefore, the variability in expression of the activating NKG2 molecule reflects the normal biology of NK cells.
Fig. 5.
NKG2C and NKG2C2 expression is observed predominately in CD8+ CD3− lymphocytes in PBMCs of rhesus monkeys. Flow cytometry was used to sort CD8+ CD3+ and CD8+ CD3− lymphocyte populations from PBMCs of three normal rhesus monkeys. PCR was then utilized to analyse NKG2C, NKG2C2 and GAPDH expression in each lymphocyte population.
Variable patterns of NKG2A splice variant mRNA expression in rhesus monkeys
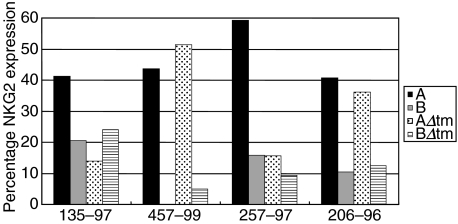
We also sought to assess the variability in the relative mRNA expression of the four NKG2A splice variants in PBMCs of the normal monkeys. To determine the percentage expression of each of the NKG2A splice variants, the values for NKG2A, NKG2B, NKG2AΔtm or NKG2BΔtm were divided by the sum of the values of these four molecules and multiplied by 100. There was no consistent pattern in the relative expression of the four NKG2A splice variants in PBMCs of these four normal rhesus monkeys (Fig. 6).
Fig. 6.
Variable patterns of NKG2A splice variant mRNA expression are seen in PBMCs of normal rhesus monkeys. The percentage of NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm mRNA expression was compared in PBMCs of four normal rhesus monkeys. NKG2 percentages were obtained by dividing the area value for each individual NKG2 molecule by the sum of the NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm area values. The relative expression level of each of the four NKG2A splice variant mRNAs is variable when individual rhesus monkeys are compared.
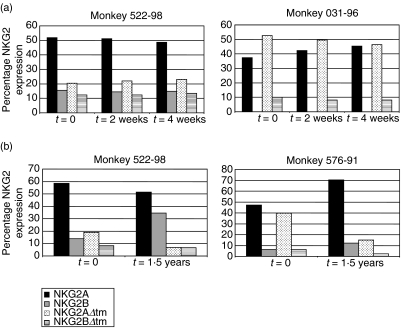
We then sought to determine if the relative expression of the NKG2A splice variants was constant or variable over time in a single monkey. mRNA expression was analysed from PBMC samples obtained from two rhesus monkeys at three time-points, each 2 weeks apart. As shown in Fig. 7a, the relative expression of the NKG2A splice variants was constant during that time period. However, when mRNA expression was examined from samples obtained from the same animals over the course of 1·5 years, the relative expression of the NKG2A splice variants was highly variable (Fig. 7b). In PBMCs from monkey 576–91, the relative expression of NKG2AΔtm mRNA was decreased dramatically at the later time-point. In contrast, the relative expression of NKG2B mRNA in PBMCs from monkey 522–98 was increased in the sample obtained at this later time-point. Thus, while the relative expression of the NKG2A splice variants was consistent over a short period of time, it changed dramatically over a longer interval of time.
Fig. 7.
(a) The relative expression of NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm in PBMCs is constant over a 1-month period of time. NKG2 percentages were obtained by dividing the area value for each individual NKG2 molecule by the sum of the NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm area values. NKG2A splice variant mRNA expression was assessed at 2-week intervals for a total of three time-points in two normal monkeys. (b) Patterns of NKG2A splice variant mRNA expression in PBMCs of individual monkeys are variable over a period of 1·5 years. The relative expression of NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm mRNA in PBMCs was assessed in two rhesus monkeys over a period of 1·5 years. NKG2 percentages were obtained by dividing the area value for each individual NKG2 molecule by the sum of the NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm area values. All values are shown; for samples without a visible bar, no mRNA expression for that molecule was detected.
Variability in NKG2 protein expression
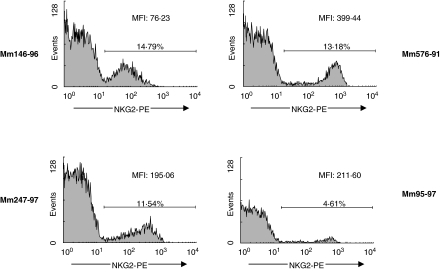
We next sought to determine if the variability seen in NKG2 mRNA expression was also apparent in NKG2 protein expression. Transient transfection experiments demonstrated that the only available antihuman NKG2A monoclonal antibody that cross-reacts with homologous rhesus monkey molecules recognizes NKG2A, NKG2B, NKG2C2 and NKG2C of the monkey (data not shown). Therefore, we used this antibody as a tool to assess the expression of these NKG2 family members on the surface of rhesus monkey lymphocytes. Whole blood of four normal rhesus monkeys was stained with this fluorochrome-labelled monoclonal antibody and gated lymphocytes were assessed by flow cytometry for NKG2 expression (Fig. 8). The percentage of PBMCs expressing NKG2 ranged from 5·70% to 15·03% in the evaluated monkeys. Furthermore, the mean fluorescence intensity of the stained cell populations varied dramatically, even between monkeys with similar percentages of NKG2-expressing cells. Thus, the variability we saw in mRNA expression by PBMCs of various monkeys was also apparent in the cell surface expression of these proteins.
Fig. 8.
NKG2 protein expression on the surface of rhesus monkey lymphocytes is variable. Whole blood from four normal rhesus monkeys was stained with the antihuman NKG2A monoclonal antibody Z199. PBMCs were gated on the lymphocyte population and NKG2 expression was evaluated. Percentage of NKG2 expressing cells and MFI values are indicated.
DISCUSSION
The rhesus monkey CD94/NKG2 molecules are remarkably similar to their human homologues, displaying a high degree of sequence homology. Moreover, rhesus monkey NKG2A and NKG2B encode two ITIMs in their cytoplasmic domains, and rhesus monkey NKG2C and NKG2D both lack ITIMs and contain a charged amino acid in their transmembrane domains. These features suggest that the CD94/NKG2 family of molecules is functional in the rhesus monkey. Importantly, the similarities between these monkey and human genes suggest that the rhesus monkey should be a valuable model for studying the in vivo biology of the NKG2 molecules.
Because cells of most individuals express the entire complement of NKG2 genes, variability in the expression of these molecules appears to stem from a differential regulation of the expression of these genes. Because monoclonal antibodies that distinguish the various cell-surface NKG2 proteins are not available for studies in the rhesus monkey, we developed a PCR-based assay to evaluate semiquantitatively mRNA expression of nine rhesus monkey NKG2 molecules. In addition to facilitating an evaluation of the expression of NKG2A, NKG2B, NKG2C2, NKG2C and NKG2D, this PCR-based assay allowed us to assess expression of the transmembrane deleted splice variants, molecules that could not be recognized specifically by monoclonal antibodies.
The present studies demonstrate a significant variability in NKG2 mRNA expression levels in rhesus monkeys. Variability in NKG2 mRNA expression was apparent in comparing PBMCs from 15 different rhesus monkeys as well as in studies of individual monkeys over a period of 1·5 years. This variability in NKG2 expression was confirmed at the protein level using an antihuman NKG2A monoclonal antibody that recognizes all NKG2A, NKG2B, NKG2C2 and NKG2C alleles in the rhesus monkey. Additional studies in our laboratory have demonstrated that this variability in NKG2 expression is due to the different abilities of NKG2 molecules to pair with CD94 (manuscript in preparation). In addition to the variability in expression of one NKG2 molecule in a comparison of 15 monkeys, the repertoire of NKG2 expression levels in each monkey was also extremely variable. In fact, 15 unique NKG2 expression repertoires were observed in an analysis of the 15 monkeys.
The expression levels of the NKG2A splice variants were highly variable in the evaluated monkeys. The relative expression of these splice variants was fairly consistent over a 4-week time period in normal monkeys, yet changed dramatically over a period of 1·5 years. No work has yet been conducted to evaluate the function of the transmembrane-deleted NKG2 molecules. While it is possible that these molecules are non-functional by-products of an alternative-splicing pathway, the very fact that their expression levels are so variable suggests that they may be of functional importance. In an analysis of NKG2A, NKG2B, NKG2AΔtm and NKG2BΔtm in PBMCs of the 15 monkeys evaluated, no consistent expression level profile was observed. While we can only speculate as to the significance of this variability, it does suggest that the NK cell receptor repertoire is dynamic in the rhesus monkey.
Acknowledgments
We would particularly like to thank Jason LaBonte for help with data analysis and manuscript preparation. We also thank David Margolin, Karen Hershberger and Igor Koralnik for helpful suggestions. Finally, we thank Joern Schmitz and Michelle Lifton for assistance with flow cytometric analysis. This work was supported by NIH grant AI20729.
REFERENCES
- 1.Harel-Bellan A, Bertoglio J, Quillet A, et al. Interleukin 2 (IL-2) up-regulates its own receptor on a subset of human unprimed peripheral blood lymphocytes and triggers their proliferation. J Immunol. 1986;136:2463–9. [PubMed] [Google Scholar]
- 2.Slavik JM, Hutchcroft JE, Bierer BE. CD28/CTLA-4 and CD80/CD86 families: signaling and function. Immunol Res. 1999;19:1–24. doi: 10.1007/BF02786473. [DOI] [PubMed] [Google Scholar]
- 3.Choe H, Farzan M, Sun Y, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 4.Hariharan D, Douglas SD, Lee B, Lai JP, Campbell DE, Ho WZ. Interferon-gamma upregulates CCR5 expression in cord and adult blood moconuclear phagocytes. Blood. 1999;93:1137–44. [PubMed] [Google Scholar]
- 5.Clerici M, Butto S, Lukwiya M, et al. Immune activation in Africa is environmentally-driven and is associated with upregulation of CCR5. Italian–Ugandan AIDS Project. AIDS. 2000;14:2083–92. doi: 10.1097/00002030-200009290-00003. [DOI] [PubMed] [Google Scholar]
- 6.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–18. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen GB, Gandhi RT, Davis DM, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–71. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 8.Houchins JP, Yabe T, McSherry C, Bach FH. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J Exp Med. 1991;173:1017–20. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol. 1996;157:4741–5. [PubMed] [Google Scholar]
- 10.Berg SF, Dissen E, Westgaard IH, Fossum S. Two genes in the rat homologous to human NKG2. Eur J Immunol. 1998;28:444–50. doi: 10.1002/(SICI)1521-4141(199802)28:02<444::AID-IMMU444>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Cantoni C, Biassoni R, Pende D, et al. The activating form of CD94 receptor complex: CD94 covalently associates with the Kp39 protein that represents the product of the NKG2-C gene. Eur J Immunol. 1998;28:327–38. doi: 10.1002/(SICI)1521-4141(199801)28:01<327::AID-IMMU327>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Lohwasser S, Hande P, Mager DL, Takei F. Cloning of murine NKG2A, B and C: second family of C-type lectin receptors on murine NK cells. Eur J Immunol. 1999;29:755–61. doi: 10.1002/(SICI)1521-4141(199903)29:03<755::AID-IMMU755>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Braud VM, Allan DS, O'Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 14.Lee N, Llano M, Carretero M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houchins JP, Lanier LL, Niemi EC, Phillips JH, Ryan JC. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J Immunol. 1997;158:3603–9. [PubMed] [Google Scholar]
- 16.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 17.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Song Y, Bakker AB, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–2. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 19.LaBonte ML, Levy DB, Letvin NL. Characterization of rhesus monkey CD94/NKG2 family members and identification of novel transmembrane-deleted forms of NKG2-A, B, C, and D. Immunogenetics. 2000;51:496–9. doi: 10.1007/s002510050650. [DOI] [PubMed] [Google Scholar]
- 20.LaBonte ML, Hershberger ML, Korber B, Letvin NL. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol Rev. 2001;183:25–40. doi: 10.1034/j.1600-065x.2001.1830103.x. [DOI] [PubMed] [Google Scholar]
- 21.Kravitz RH, Grendell RL, Slukvin II, Golos TG. Selective expression of NKG2-A and NKG2-C mRNAs and novel alternative splicing of 5′ exons in rhesus monkey decidua. Immunogenetics. 2001;53:69–73. doi: 10.1007/s002510000289. [DOI] [PubMed] [Google Scholar]
- 22.Glienke J, Sobanov Y, Brostjan C, et al. The genomic organization of NKG2C, E, F, and D receptor genes in the human natural killer gene complex. Immunogenetics. 1998;48:163–73. doi: 10.1007/s002510050420. [DOI] [PubMed] [Google Scholar]
- 23.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]