Asthma is a chronic inflammatory disease of the small airways, being clinically characterized by reversible airway obstruction and bronchial hyperreactivity [1]. These clinical features result from a chronic inflammation of the airways, caused by a migration of leucocytes and an increase of inflammatory mediators in the bronchial wall [2]. This pathological reaction in asthma is thought to arise from a complex interaction between genes and the environment.
Although allergens and infection appear to be environmental modifiers of asthma [3,4], it is now estimated that at least a dozen polymorphic genes regulate asthma and control the chronic inflammatory response and production of immunoglobulin E (IgE), cytokines and chemokines. By genetic-linkage analysis on 460 pairs of siblings from asthmatic families in the USA and UK, Van Eerdewegh et al. [5] identified a locus on the short arm of chromosome 20 which was linked to asthma and bronchial hyperreactivity. They identified the ADAM-33 gene as significantly associated with asthma. ADAMs are a subfamily of metalloproteinases expressed on the cell surface, and have proteolytic functions such as shedding tumour necrosis factor receptors, and other cell-surface cytokines, adhesion molecules, and growth factors and receptors that are involved in inflammation, cell proliferation, and cell death. ADAM-33 is expressed by lung fibroblasts and bronchial smooth muscle cells, and is suspected to be associated with small-airway remodeling in patients with asthma.
Regarding immunological aspects of asthma genes, the human homologue of Tim1 has been identified as an asthma susceptibility gene [6]. Tim1 lies at chromosome 5q33.2, a region that has been repeatedly linked to asthma, and codes for the cellular receptor for hepatitis A virus [6]. The Tim1 gene product is expressed on T cells and appears to regulate the production of interleukin-4 (IL-4) in T cells by affecting CD4+ T cell differentiation, the development of TH2 cells and development of airway hyperreactivity. As observed in the Tim1 hypothesis, the inappropriate TH2 response causes pulmonary inflammation, airway eosinophilia, and airway hyperreactivity to a variety of specific and nonspecific stimuli that result in the clinical symptoms of asthma.
In the TH1-TH2 paradigm, TH1 cell responses are thought to protect against asthma, by dampening the activity of TH2 responses [7–9]. However, other investigators have shown that TH1 cells may exacerbate asthma, as human asthma is associated with the production of IFN-γ which appears to contribute to pathogenesis of asthma [10,11]. Allergen-specific TH1 cells, when adoptively transferred into naïve recipients, migrate to the lungs but fail to counterbalance TH2 cell-induced airway hyperreactivity. Instead, allergen-specific TH1 cells cause severe airway inflammation [12]. Moreover, Dahl et al. [13] showed virus-induced TH1-dependent enhancement of allergic pulmonary inflammation via TH1-polarized dendritic cells (DCs). Thus, although TH2 cells play an important role in the pathogenesis of asthma, the binary TH1-TH2 paradigm cannot explain all the immunological processes that occur in asthma. These processes in asthma may be much more complex than is hypothesized by a TH1-TH2 paradigm. In this regard, Kruschinski et al. [14], in this issue of Clinical and Experimental Immunology, show a critical role for CD26+ T cells, which are TH1 cells, in asthma, by examining CD26-deficient and CD26-reduced rats in ovalubmin (OVA)-induced asthma models. They show in their article that the decrease in T cell recruitment to the airway observed in CD26low and CD26-deficient rats is associated with significantly reduced OVA-specific IgE-titres. They suggest a role for CD26+ T cells in the pathogenesis of asthma by means of T cell migration and T cell-dependent IgE production in the airway.
CD26/dipeptidyl peptidase IV (DPPIV) is a 110-kDa cell surface glycoprotein that belongs to the serine protease family [15]. It is expressed on a variety of tissues including T lymphocytes, endothelial and epithelial cells. It is composed of a short cytoplasmic domain of 6 amino acids, a transmembrane region of 22 amino acids, and an extracellular domain of 738 amino acids, with DPPIV activity which selectively removes the N-terminal dipeptide from peptides with proline or alanine in the second position [16]. Possible substrates of DPPIV include several critical cytokines and chemokines. Activity of CCL5 (RANTES, regulated on activation, normal T cell expressed and secreted) is altered by the enzymatic cleavage of DPPIV, as CD26-processed CCL5(3–68) has a more than 10 times lower chemotactic potency for monocytes and eosinophils. CCL5(3–68) also has impaired binding and signalling properties through CCR1 and CCR3, but remains fully active on CCR5, leading to TH1 polarization [17,18]. Other important chemokines that appear to be substrates of the enzymatic activity of DPPIV include CCL11 (eotaxin), CCL22 (macrophage-derived chemokine), CXCL10 and CXCL11 (interferon inducible chemokines), and other chemokines [19,20]. Besides its ability to regulate the effect of biological factors through its enzymatic activity, CD26/DPPIV has an essential role in human T cell physiology.
Originally characterized as a T-cell differentiation antigen, CD26 is preferentially expressed on a specific population of T lymphocytes, the subset of CD4+ memory T cells, and is up-regulated after T cell activation [21,22]. As well as its enhanced expression on activated T cells, various lines of evidence have converged to demonstrate that CD26 is functionally associated with T cell signal transduction processes, which are capable of transmitting signals relating to T cell activation [22,23]. In addition CD26 serves as a functional collagen receptor with a role in T cell activation, as well as having a potential role in thymic ontogeny [24]. The enzymatic activity of CD26 appears to be very important in enhancing cellular responses to external stimuli. For example, Jurkat cells transfected with wild type CD26 consistently demonstrated greater activation than parental CD26 negative Jurkat or cells transfected with CD26 mutated at the DPPIV enzymatic site [25].
CD26 expression is tightly regulated on human T lymphocytes, with its density being markedly elevated following T cell activation [21,26]. At the resting state in the peripheral blood, CD26 is preferentially expressed on the helper/memory T cell population [21]. High CD26 cell surface expression is correlated with the production of TH1-like cytokines by T-cell clones, and CD26 expression is induced by stimuli that favour the development of the TH 1 response [27,28]. CD26 is able to conduct IL-2-dependent comitogenic signals in conjunction with activation through the CD3/T cell receptor complex or the CD2 pathway of mature human T lymphocytes when crosslinked with solid-phase immobilized antibodies [22].
Meanwhile, recombinant soluble CD26/DPPIV molecule up-regulates expression of CD86 on antigen presenting cells (APC), leading to greater APC–T cell interaction and enhanced T cell proliferation, with important implications for immunoregulation [29]. More recently, T cell proliferation via CD26 driven by recall antigens such as tetanus toxoid is mediated by means of caveolin-1 on APC, leading to up-regulation of the costimulatory molecule CD86 [30]. Thus, CD26+ T cells play a critical role in inflammation responding to recall antigen.
Of clinical relevance, patients with autoimmune diseases such as Graves’ disease and rheumatoid arthritis have been found to increase numbers of CD26+ T cells in inflamed tissues such as thyroid and synovial fluids [26,31]. In addition, enhancement of CD26 expression in these autoimmune diseases may correlate with disease severity [32,33]. Moreover, we have shown that T cells migrating through endothelial cell monolayers in vitro express high levels of CD26 [34], and the fact that chemokines play a key role in T cell migration supports the notion that CD26/DPPIV may interact with chemokines [18–20]. These findings imply that CD26+ T cells play a role in the inflammation process and subsequent tissue damage not only in autoimmune diseases, but also in asthma.
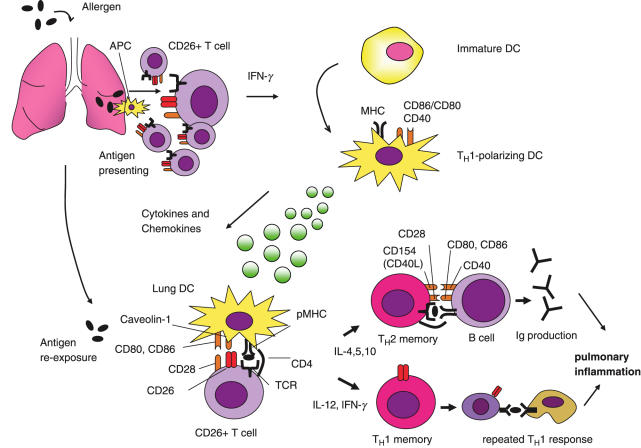
In the context that CD26+ T cells preferentially play an important role in allergic pulmonary inflammation, it is hypothesized that allergens such as OVA incite a robust TH1-type cytokine response, such as IFN-γ, in the lung, promoting the development of durable TH1-polarizing DCs, partly via a CD26–caveolin-1 interaction, subsequently leading to enhancement of CD28-CD86 costimulation (Fig. 1). These DCs support subsequent immunity in a TH2-dependent process of allergen-induced pulmonary inflammation, and so enhance both TH1 and TH2 immune cytokines and IgE production.
Fig. 1.
TH1-dependent enhancement of allergic pulmonary inflammation. Initially, antigens such as sensitizing-allergens stimulate CD26+ T cells via antigen presenting cells (APC), and subsequently generate CD26+ TH1 response to produce IFN-γ and stable TH1-polarizing DCs. Later, these DCs are capable of augmenting both TH1 and TH2 controlled immune responses in allergen-induced pulmonary inflammation, partly via interaction of CD26 on T cells and caveolin-1 on APC loaded with sensitized allergens. Ig, immunoglobulin; MHC, major histocompatibility complex; pMHC, peptide-loaded major histocompatibility complex.
Interestingly, a recent report on single-nucleotide polymorphism in asthmatic patients revealed that polymorphism of DPP-10, a CD26/DPPIV-like peptidase, was observed [35]. Further elucidation on DPPs and asthma will open an avenue for our understanding the pathophysiology of asthma.
Although the precise effects rely on the exact timing, intensity and dose of cytokines and antigens as well as how these interactions alter T helper regulatory cells and cytokines, CD26+ T cells play an important role in asthma and targeting CD26/DPPIV may contribute to the elucidation of the pathophysiology and therapeutic means to treat asthma, and other inflammatory disorders.
References
- 1.Sheffer AL. Guidelines for the diagnosis and management of asthma. National Heart, Lung, and Blood Institute. National Asthma Education Program. Expert Panel Report. J Allergy Clin Immunol. 1991;88:425–34. [PubMed] [Google Scholar]
- 2.Bittleman DB, Casale TB. Allergic models and cytokines. Am J Respir Crit Care Med. 1994;150:S72–6. doi: 10.1164/ajrccm/150.5_Pt_2.S72. [DOI] [PubMed] [Google Scholar]
- 3.Ball TM, Castro-Rodriguez JA, Griffith KA, Holberg CJ, Martinez FD, Wright AL. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343:538–43. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- 4.Riedler J, Braun-Fahrlander C, Eder W, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–33. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 5.Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 6.McIntire JJ, Umetsu SE, Akbari O, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–16. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 7.Coffman RL, Seymour BW, Lebman DA, et al. The role of helper T cell products in mouse B cell differentiation and isotype regulation. Immunol Rev. 1988;102:5–28. doi: 10.1111/j.1600-065x.1988.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 8.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 9.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 10.Corrigan CJ, Kay AB. CD4 T-lymphocyte activation in acute severe asthma. Relationship to disease severity and atopic status. Am Rev Respir Dis. 1990;141:970–7. doi: 10.1164/ajrccm/141.4_Pt_1.. [DOI] [PubMed] [Google Scholar]
- 11.Cembrzynska-Nowak M, Szklarz E, Inglot AD, Teodorczyk-Injeyan JA. Elevated release of tumor necrosis factor-alpha and interferon-gamma by bronchoalveolar leukocytes from patients with bronchial asthma. Am Rev Respir Dis. 1993;147:291–5. doi: 10.1164/ajrccm/147.2.291. [DOI] [PubMed] [Google Scholar]
- 12.Hansen G, Berry G, DeKruyff RH, Umetsu DT. Allergen-specific Th1 cells fail to counterbalance Th2 cell-induced airway hyperreactivity but cause severe airway inflammation. J Clin Invest. 1993;103:175–83. doi: 10.1172/JCI5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl ME, Dabbagh K, Liggitt D, Kim S, Lewis DB. Viral-induced T helper type 1 responses enhance allergic disease by effects on lung dendritic cells. Nat Immunol. 2004;5:337–43. doi: 10.1038/ni1041. [DOI] [PubMed] [Google Scholar]
- 14.Kruschinski C, Skripuletz T, Bedoui S, Tschering T, Pabst R, Nassenstein C, Braun A, von Horsten S. CD26 (dipeptidyl-peptidase IV)-dependent recruitment of T cells in a rat asthma model. Clin Exp Immunol. 2004;139:17–24. doi: 10.1111/j.1365-2249.2005.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morimoto C, Schlossman SF. The structure and function of CD26 in the T-cell immune response. Immunol Rev. 1998;161:55–70. doi: 10.1111/j.1600-065x.1998.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T, Camerini D, Seed B, Torimoto Y, Dang NH, Kameoka J, Dahlberg HN, Schlossman SF, Morimoto C. Cloning and functional expression of the T cell activation antigen CD26. J Immunol. 1992;149:481–6. [PubMed] [Google Scholar]
- 17.Schrum S, Probst P, Fleischer B, Zipfel PF. Synthesis of the CC-chemokines MIP-1alpha, MIP-1beta, and RANTES is associated with a type 1 immune response. J Immunol. 1996;157:3598–604. [PubMed] [Google Scholar]
- 18.Oravecz T, Pall M, Roderiquez G, Gorrell MD, et al. Regulation of the receptor specificity and function of the chemokine RANTES (regulated on activation, normal T cell expressed and secreted) by dipeptidyl peptidase IV (CD26)-mediated cleavage. J Exp Med. 1997;186:1865–72. doi: 10.1084/jem.186.11.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsuki T, Hosono O, Kobayashi H, Munakata Y, Souta A, Shioda T, Morimoto C. Negative regulation of the anti-human immunodeficiency virus and chemotactic activity of human stromal cell-derived factor α by CD26/dipeptidyl peptidase IV. FEBS Lett. 1998;431:236–40. doi: 10.1016/s0014-5793(98)00763-7. [DOI] [PubMed] [Google Scholar]
- 20.Proost P, Struyf S, Schols D, et al. Truncation of macrophage-derived chemokine by CD26/ dipeptidyl-peptidase IV beyond its predicted cleavage site affects chemotactic activity and CC chemokine receptor 4 interaction. J Biol Chem. 1999;274:3988–93. doi: 10.1074/jbc.274.7.3988. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto C, Torimoto Y, Levinson G, Rudd CE, Schrieber M, Dang NH, Letvin NL, Schlossman SF. 1F7, a novel cell surface molecule, involved in helper function of CD4 cells. J Immunol. 1989;142:3430–9. [PubMed] [Google Scholar]
- 22.Dang NH, Torimoto Y, Deusch K, Schlossman SF, Morimoto C. Comitogenic effect of solid-phase immobilized anti-1F7 on human CD4 T cell activation via CD3 and CD2 pathways. J Immunol. 1990;144:4092–100. [PubMed] [Google Scholar]
- 23.Ishii T, Ohnuma K, Murakami A, Takasawa N, Kobayashi S, Dang NH, Schlossman SF, Morimoto C. CD26-mediated signaling for T cell activation occurs in lipid rafts through its association with CD45RO. Proc Natl Acad Sci USA. 2001;98:12138–43. doi: 10.1073/pnas.211439098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang NH, Torimoto Y, Schlossman SF, Morimoto C. Human CD4 helper T cell activation. functional involvement of two distinct collagen receptors, 1F7 and VLA integrin family. J Exp Med. 1990;172:649–52. doi: 10.1084/jem.172.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, Kameoka J, Yaron A, Schlossman SF, Morimoto C. The costimulatory activity of the CD26 antigen requires dipeptidyl peptidase IV enzymatic activity. Proc Natl Acad Sci USA. 1993;90:4586–90. doi: 10.1073/pnas.90.10.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eguchi K, Ueki Y, Shimomura C, et al. Increment in the Ta1+ cells in the peripheral blood and thyroid tissue of patients with Graves’ disease. J Immunol. 1989;142:4233–40. [PubMed] [Google Scholar]
- 27.Reinhold D, Bank U, Buhling F, Lendeckel U, Faust J, Neubert K, Ansorge S. Inhibitors of dipeptidyl peptidase IV induce secretion of transforming growth factor-β1 in PWM-stimulated PBMC and T cells. Immunology. 1997;91:354–60. doi: 10.1046/j.1365-2567.1997.d01-2258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willheim M, Ebner C, Baier K, et al. Cell surface characterization of T lymphocytes and allergen-specific T cell clones: correlation of CD26 expression with T (H1) subsets. J Allergy Clin Immunol. 1997;100:348–55. doi: 10.1016/s0091-6749(97)70248-3. [DOI] [PubMed] [Google Scholar]
- 29.Ohnuma K, Munakata Y, Ishii T, et al. Soluble CD26/dipeptidyl peptidase IV induces T cell proliferation through CD86 up-regulation on APCs. J Immunol. 2001;167:6745–55. doi: 10.4049/jimmunol.167.12.6745. [DOI] [PubMed] [Google Scholar]
- 30.Ohnuma K, Yamochi T, Uchiyama M, et al. CD26 up-regulates expression of CD86 on antigen-presenting cells by means of caveolin-1. Proc Natl Acad Sci USA. 2004;101:14186–91. doi: 10.1073/pnas.0405266101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizokami A, Eguchi K, Kawakami A, et al. Increased population of high fluorescence 1F7 (CD26) antigen on T cells in synovial fluid of patients with rheumatoid arthritis. J Rheumatol. 1996;23:2022–6. [PubMed] [Google Scholar]
- 32.Muscat C, Bertotto A, Agea E, Bistoni O, et al. Expression and functional role of 1F7 (CD26) antigen on peripheral blood and synovial fluid T cells in rheumatoid arthritis patients. Clin Exp Immunol. 1994;98:252–6. doi: 10.1111/j.1365-2249.1994.tb06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerli R, Muscat C, Bertotto A, Bistoni O, et al. CD26 surface molecule involvement in T cell activationand lymphokine synthesis in rheumatoid and other inflammatory synovitis. Clin Immunol Immunopathol. 1996;80:31–7. doi: 10.1006/clin.1996.0091. [DOI] [PubMed] [Google Scholar]
- 34.Masuyama J, Berman JS, Cruikshank WW, Morimoto C, Center DM. Evidence for recent as well as long-term activation of T cells migrating through endothelial cell monolayers in vitro. J Immunol. 1992;148:1367–74. [PubMed] [Google Scholar]
- 35.Allen M, Heinzmann A, Noguchi E, et al. Positional cloning of a novel influencing asthma from chromosome 2q14. Nat Genet. 2003;35:258–63. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]