Abstract
Eosinophils are principal effector cells of inflammation in allergic asthma, characterized by their accumulation and infiltration at inflammatory sites mediated by the chemokine eotaxin and their interaction with adhesion molecules expressed on bronchial epithelial cells. We investigated the modulation of nuclear factor-κB (NF-κB) and the mitogen-activated protein kinase (MAPK) pathway on the in vitro release of chemokines including regulated upon activation normal T cell expressed and secreted (RANTES), monokine induced by interferon-γ (MIG), monocyte chemoattractant protein-1 (MCP-1), interleukin (IL)-8, and interferon-inducible protein-10 (IP-10) upon the interaction of human bronchial epithelial BEAS-2B cells and eosinophils. Gene expression of chemokines was evaluated by RT-PCR and the induction amount of chemokines quantified by cytometric bead array. NF-κB and p38 MAPK activities were assessed by electrophoretic mobility shift assay and Western blot, respectively. The interaction of eosinophils and BEAS-2B cells was found to up-regulate the gene expression of the chemokines IL-8, MCP-1, MIG, RANTES and IP-10 expression in BEAS-2B cells, and to significantly elevate the release of the aforementioned chemokines except RANTES in a coculture of BEAS-2B cells and eosinophils. IκB-α phosphorylation inhibitor, BAY 11–7082, and p38 MAPK inhibitor, SB 203580 could decrease the release of IL-8, IP-10 and MCP-1 in the coculture. Together, the above results show that the induction of the release of chemokines in a coculture of epithelial cells and eosinophils are regulated by p38 MAPK and NF-κB activities of BEAS-2B cells, at least partly, through intercellular contact. Our findings therefore shed light on the future development of more effective agents for allergic and inflammatory diseases.
Keywords: eosinophils, epithelial cells, chemokines, p38, NF-κB
Introduction
The inflammatory and airway remodeling processes in allergic asthma are the consequence of highly complex interactions among inflammatory cells such as eosinophils, activated T cells, mast cells, and macrophages with structural tissue cells including the bronchial epithelium, endothelial cells and fibroblasts [1]. The chemotaxis and transmigration of eosinophils, eosinophil–epithelial cell interactions, and the subsequent eosinophil-mediated damage to bronchial epithelium by their highly toxic granule proteins such as eosinophilic cationic protein (ECP) together contribute to the pathogenesis of asthma [2,3]. Activated airway epithelial cells are potent sources of haematopoietic cytokines, interleukin (IL)-5 and granulocyte-macrophage colony-stimulating factor (GM-CSF), pro-inflammatory cytokines IL-1β, IL-6, IL-11, IL-16 and IL-18, and chemokines regulated upon activation normal T cell expressed and secreted (RANTES), eotaxin, macrophage inflammatory protein (MIP)-1α, monokine induced by interferon-γ (MIG), interferon-inducible protein-10 (IP-10), and IL-8 [1]. Eosinophils are also capable of producing and releasing a variety of proinflammatory cytokines such as IL-3, IL-4, IL-5, IL-6, IL-16, tumour necrosis factor (TNF)-α, and GM-CSF together with chemokines including eotaxin, IL-8, MIP-1α, monocyte chemoattractant protein (MCP) and RANTES [4,5]. The interaction of eosinophils and epithelial cells can also result in the induction of inflammatory mediators including cysteinyl leukotrienes [6]. Co-culture of IL-5-primed eosinophils and TNF-α-treated or IFN-γ-treated epithelial cells can enhance ECP release from eosinophil degranulation through CD18-dependent adhesion [7]. Therefore, the interaction between bronchial epithelial cells and eosinophils can up-regulate the responsiveness and survival of human eosinophils and epithelial cells through the release of cytokines and chemokines, thereby indicating a significant implication in the immunopathogenesis of allergic asthma.
Many previous reports have shown that the production of many chemokines can be regulated by both p38 mitogen-activated protein kinase (MAPK) and transcription factor nuclear factor (NF)-κB in eosinophils and epithelial cells [8–10]. NF-κB was also found to involve in the expression of many inflammatory cytokines and adhesion molecules during allergic inflammation [11,12]. Our previous studies have indicated that the activation of NF-κB and p38 MAPK plays a role in TNF-α-activated signalling pathways regulating the release of eosinophil chemokine eotaxin and the cell surface expression of intercellular adhesion molecule (ICAM)-1 of eosinophils [4,13,14].
Chemokines have been shown to control and direct the migration and activation of various leucocyte populations in asthma and lung inflammation [15,16]. In our clinical studies of patients with allergic asthma, we observed that the plasma level of proinflammatory cytokines IL-17, IL-18 and IL-12, and activated T cell chemokine thymus and activated-regulated chemokine (TARC) were elevated in these patients [17–19]. Recent studies using inhibitory antibodies and chemokine antagonists support the concept that interfering with this subset of chemokines and their receptors represent a new approach for immunotherapy of allergic diseases [20]. Although the MAPK signalling pathways have been implicated to play the critical role in the pathogenesis of asthma [21], the regulatory mechanisms for the production of chemokines upon interaction of eosinophils and epithelial cells have not been well studied. In the present study, we investigated the gene expression and release of neutrophil chemokine IL-8, monocyte chemoattractant MCP-1, Th1 cell chemokine IP-10, T cell chemoattractant RANTES and MIG, and the modulation of NF-κB and MAPK activity on the regulation of chemokines release upon the interaction of human blood eosinophils and bronchial epithelial cells.
Materials and methods
Inhibitors
IκB-α phosphorylation inhibitor, BAY 11–7082, and p38 MAPK inhibitor, SB 203580, were purchased from Calbiochem Corp, CA, USA. SB 203580 was dissolved in water. BAY 11–7082 (Calbiochem) [22] was dissolved in dimethyl sulphoxide (DMSO) and added to culture at a concentration of 1 µm. In all studies, the concentration of DMSO was 0·1% (v/v).
BEAS-2B cells
BEAS-2B is an adenovirus 12-SV40 virus hybrid (Ad12SV40) transformed human epithelial cell line, which was isolated from normal human bronchial epithelium obtained from autopsy of noncancerous individuals. It was purchased from the American Type Culture Collection (ATCC), VA, USA. BEAS-2B cells were maintained in D-MEM/F12 medium (Gibco Laboratories, NY, USA) supplemented with 10% defined fetal bovine serum (FBS) (Gibco) in 5% CO2−95% humidified air at 37°C. Before each experiment, BEAS-2B cells were cultured in 6- or 24-well plate until confluence.
Isolation of human blood eosinophils from buffy coat and eosinophil culture
Fresh human buffy coat obtained from the Hong Kong Red Cross Blood Transfusion Service was diluted 1 : 2 with phosphate-buffered saline (PBS) at 4°C and centrifuged using an isotonic Percoll solution (density 1·082 g/ml; Amersham and Pharmacia Biotech, Uppsala, Sweden) for 30 min at 1000× g. The eosinophil-rich granulocyte fraction was collected and washed twice with cold PBS containing 2% fetal calf serum. The cells were then incubated with anti-CD16 magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) at 4°C for 45 min and CD16-positive neutrophils were depleted by passing through a LS + column (Miltenyi Biotec) within a magnetic field. With this preparation, the drop-through fraction contained eosinophils with a purity of at least 98% as assessed by Hemacolor rapid blood smear stain (E. Merck Diagnostica, Darmstadt, Germany). The isolated eosinophils were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS (Gibco) and 20 m m Hepes (Gibco).
Endotoxin-free solutions
Cell culture medium was purchased from Gibco, free of detectable lipopolysaccharide (LPS) (<0·1 EU/ml). All other solutions were prepared using pyrogen-free water and sterile polypropylene plasticware. No solution contained detectable LPS, as determined by the Limulus amoebocyte lyase assay (sensitivity limit 12 pg/ml; Associates of Cape Cod, MA, USA).
Reverse transcription-polymerase chain reaction (RT-PCR)
After coculture, eosinophils were separated from adherent BEAS-2B cells by washing. Total RNA was extracted using Tri-Reagent (Molecular Research Center Inc, OH, USA). Extracted RNA was reverse transcribed into first-strand complementary DNA using First-Strand cDNA Synthesis Kit (Amersham Biosciences Corp, NJ, USA). PCR was performed in a reaction mixture containing 3 m m MgCl2, 200 µm dNTPs, 1 unit of AmpliTaq Gold DNA polymerase (Perkin Elmer, CA, USA), 50 pmol of 5′-and 3′ primers (Invitrogen, CA, USA) in PCR reaction buffer (1 min each at 94°, 56° and 72°C) for 30 cycles for β-actin, MCP-1, MIP-1α, MIG and IP-10 after an initial 12 min of denaturation at 94°C. Thirty cycles (2 min each at 94°, 56°C and 72°C) after an initial 12 min of denaturation at 94°C was adopted for RANTES. All RT-PCR were performed in the linear range of the PCR reaction according to the preliminary experiments. PCR Primers were as the following: IL-8 sense, 5′-CTGTGT GAAGGTGCAGTTTTGCC-3′ and antisense, 5′-CTCAGC CCTCTTCAAAAACTTCTCC-3′, yielding a 237-bp product [23]; MCP-1 sense, 5′-AATGCCCCAGTCACCTGCTGT TAT-3′ and antisense, 5′-GCAATTTCCCCAAGTCTCTGT ATC-3′, yielding a 427-bp product [24]; MIG sense, 5′-AAGAAGCACGTGGTAAAACA-3′ and antisense, 5′-TCTCGGTGGCTATCTTGTTA-3′, yielding a 214-bp product [24]; IP-10 sense, 5′-CCTGCTTCAAATATTTCCCT-3′ and antisense, 5′-CCTTCCTGTATGTGTTTGGA-3′, yielding a 229-bp product [24]; RANTES sense, 5′-ATATTC CTCGGACACCACAC-3′ and antisense, 5′-CACGTCCAGC CTGGGGAAGG-3′, yielding a 370-bp product [25]; β-actin sense, 5′-AGCGGGAAATCGTGCGTG-3′ and antisense, 5′-CAGGGTACATGGTGGTGCC-3′, yielding a 300-bp product [14]. After the amplification reaction using PTC-200 DNA EngineTM (MJ Research, Inc., MA, USA), PCR products were electrophoresed on 2% agarose gel in TAE buffer (pH 8·0) and stained with ethidium bromide. The electrophorectic bands were documented with Gene Genius Gel Documentation System (Syngene Inc, Cambridge, UK).
Assay of chemokines IL-8, MCP-1, MIG, IP-10 and RANTES in culture supernatant
The five chemokines in culture supernatant were measured simultaneously by cytometric bead array (CBA) using a four-colour FACSCalibur flow cytometer (Becton Dickinson, Palo Alto, CA, USA) [26,27]. In CBA, five bead populations with distinct fluorescence intensities had been coated with capturing antibodies specific for five different chemokines. These bead populations could be resolved in the fluorescence channels of the flow cytometer. After the beads had been incubated with 50 µl of culture supernatant, different chemokines in the supernatant were captured by their corresponding beads. The chemokine captured beads were then mixed with phycoerythrin-conjugated detection antibodies to form sandwich complexes. Following incubation, washing and acquisition of fluorescence data, the results were generated in graphical format using the BD CBA software. The concentrations of chemokines IL-8, RANTES, MCP-1, IP-10, and MIG were measured simultaneously using the human Chemokine CBA kit (BD Biosciences Pharmingen, CA, USA). The assay sensitivities of these 5 chemokines were 0·2, 1·0, 2·7, 2·8, and 2·5 ng/l, respectively. The coefficients of variation for all chemokine assays were less than 10%.
Nonradioactive immunoprecipitate kinase activity assay for p38 MAPK
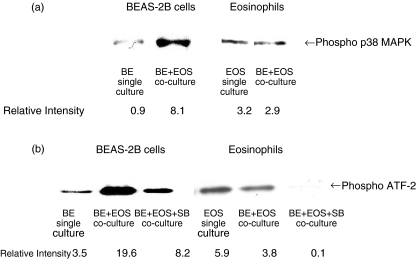
BEAS 2B cells were firstly grown in 6-well plate until confluence, then BEAS-2B cells were cultured with or without eosinophils (3 × 106/well) in 6-well plate for 30 min with the presence or absence of SB203580 (20 µm). Eosinophils were separated from adherent BEAS-2B cells by washing. Total cellular proteins were extracted for the detection of intracellular p38 MAPK activity using p38 MAP Kinase Assay Kit (Cell Signalling Technology Inc, MA, USA). Briefly, cells were lysed and the specific MAPK was immunoprecipitated. After completing the kinase enzymatic reaction of MAPK antibody immune complex, Western immunoblotting was used to detect the p38 MAPK product, phosphorylated ATF-2, using ECL chemiluminescent detection system according to the manufacturer's instructions.
Electrophoretic mobility shift assay (EMSA)
After coculture, eosinophils were separated from adherent BEAS-2B cells by washing. Cells were then harvested and washed, nuclear proteins were extracted with NE-PERTM nuclear and cytoplasmic extraction reagents (Pierce Chemical Co, IL, USA) according to the manufacturer's instructions. Equal amounts of nuclear protein extracts were subjected to a test for NF-κB protein/DNA binding using LightShiftTM chemiluminescent EMSA kit (Pierce) with a biotin end-labelled NF-κB oligonucleotide (5′-AGT TGA GGG GAC TTT CCC AGG C-3′) (Research Genetics Invitrogen Co, AL, USA). Briefly, nuclear extracts were incubated with biotin end-labelled NF-κB oligonucleotide for 20 min at room temperature to allow DNA/protein binding. The DNA/protein complexes were then resolved by a 6% native polyacrylamide gel electrophoresis and transferred to a Hybond-N + membrane (Amersham and Pharmacia Biotech). The biotin end-labelled DNA was detected using a streptavidin-horseradish peroxidase conjugate and a chemiluminescent substrate [14].
Western blot analysis
After coculture, eosinophils were separated from adherent BEAS-2B cells by washing. Cells were washed with ice cold PBS and lysed in 0·2 ml lysis buffer (20 m m Tris-HCl, pH 8·0, 120 m m NaCl, 1% Triton X-100, 10 m m EDTA, 1 m m EGTA, 0·05% 2-mercaptoethanol, 1X protease inhibitors). Cell debris was removed by centrifugation at 14 000 g for 15 min, and the supernatant was boiled in Laemmli sample buffer (Bio-Rad Laboratory, CA, USA) for 5 min. An equal amount of proteins was subjected to sodium dodecyl sulphate-10% polyacrylamide gel electrophoresis before blotting onto a PVDF membrane (Amersham and Pharmacia Biotech). The membrane was blocked with 5% skimmed milk in Tris-buffered saline with 0·05% Tween 20, pH 7·6 for 1 h at room temperature, and probed with primary rabbit antihuman phospho-p38 MAPK, antihuman phospho-IκB-α antibody (Cell Signalling Technology Inc, MA, USA) at 4°C overnight. After washing, membrane was incubated with secondary donkey antirabbit antibody coupled to horseradish peroxidase (Amersham and Pharmacia Biotech) for 1 h at room temperature. Antibody-antigen complexes were then detected using ECL chemiluminescent detection system according to the manufacturer's instructions (Amersham and Pharmacia Biotech) [4].
Statistical analysis
All data were expressed as mean ± SEM. Differences between groups were assessed by one way anova analysis. A probability P < 0·05 was considered significantly different. All analyses were performed using the Statistical Package for the Social Sciences (SPSS) statistical software for Windows, version 10·1.4 (SPSS Inc., Chicago, IL, USA).
Results
Effect of the interaction of BEAS-2B cells and eosinophils on the gene expression of chemokines of BEAS-2B cells and eosinophils
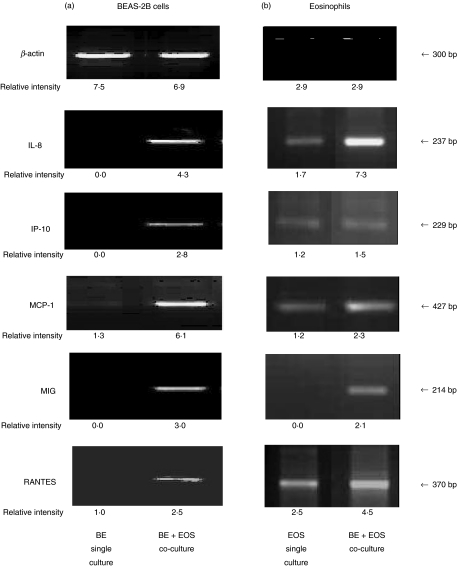
Figure 1 shows that BEAS-2B cells alone expressed little or undetectable mRNA gene expression of chemokine IL-8, IP-10, MCP-1, MIG and RANTES. However, coculture of BEAS-2B cells and eosinophils for 12 h could up-regulate the mRNA gene expression of chemokine IL-8, IP-10, MCP-1, MIG and RANTES of BEAS-2B cells. β-actin was used as positive control and remained constant in all treatments. Eosinophils only expressed little or undetectable mRNA chemokine gene expression except RANTES. The coculture of BEAS-2B cells and eosinophils for 12 h could up-regulate the mRNA gene expression of chemokine IL-8, MCP-1, MIG and RANTES of eosinophil cells.
Fig. 1.
Representative RT-PCR analysis of β-actin, IL-8, IP-10, MCP-1, MIG and RANTES mRNA expression in (a) BEAS-2B cells and (b) eosinophils. Total RNA was extracted from confluent BEAS-2B cells (6 × 105/well) or eosinophils (3 × 106/well) after coculture for 12 h in a 6-well plate, and then reverse transcribed and analysed by PCR. The β-actin housekeeping gene was used as the control. BE: BEAS-2B cells, EOS: eosinophils.
Release of IL-8, IP-10, MCP-1, MIG and RANTES upon the interaction of BEAS-2B cells and eosinophils
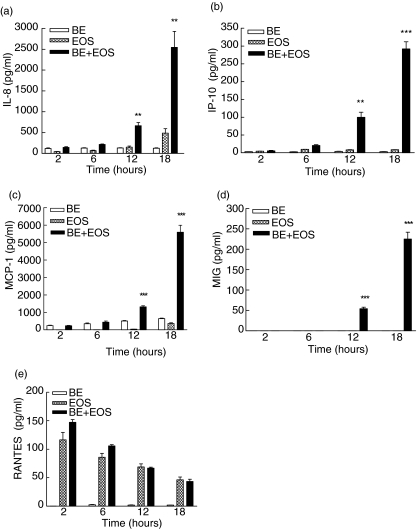
As shown in Fig. 2, the coculture of BEAS-2B cells and eosinophils exhibited synergistic effects on the release of IL-8, IP-10, MCP-1 and MIG but not RANTES after 12 and 18 h incubation (all P < 0·01), compared to that of BEAS-2B cells alone and eosinophils only. The mean value of RANTES in the coculture was found to be higher than that of BEAS-2B cells or eosinophils alone after 2 and 6 h incubation (Fig. 2e). A control experiment has been performed using transwell inserts (pore size 0·4 µm) to separate the BEAS-2B and eosinophils into two different compartments in the coculture using 24-well plate (Fig. 3a,b). Results showed that there were only very little amounts of IP-10, MIG and RANTES that could be detected in the BEAS-2B and eosinophils coculture supernatant using transwell inserts and the differences between the single and coculture using transwell inserts were not significant (all P > 0·05). However, there was significant elevation of IL-8 and MCP-1 in culture supernatant after 18 h coculture using transwell inserts (P < 0·001, Fig. 3a,b) although the amount was much less than that in coculture without transwell inserts as shown in Fig. 2.
Fig. 2.
Induction of the release of (a) IL-8 (b) IP-10 (c) MCP-1 (d) MIG and (e) RANTES upon the interaction of eosinophils and BEAS-2B cells. Confluent BEAS-2B cells (1·5 × 105/well) were cultured with or without eosinophils (5 × 105/well) for 2–18 h in a 24-well plate. Chemokine release in culture supernatant was determined using the human chemokine CBA kit and flow cytometry. Results are expressed as the arithmetic mean ± SEM from three independent experiments with different blood donors per single well on the tissue culture plate for each treatment. **P < 0·01, ***P < 0·001 when compared with the BEAS-2B and eosinophils control. BE: BEAS-2B cells, EOS: eosinophils.
Fig. 3.
Induction of the release of (a) IL-8 and (b) MCP-1 upon the coculture of eosinophils and BEAS-2B cells using transwell inserts. Confluent BEAS-2B cells (1·5 × 105/well) were cultured with or without eosinophils (5 × 105/well) for 18 h in 24-well plate using transwell inserts (pore size 0·4 µm) to separate the two cell types in two compartments. Chemokines released in culture supernatant was determined by human chemokine CBA kit using flow cytometry. Results are expressed as the arithmetic mean ± SEM from three independent experiments with different blood donors with single well on the tissue culture plate for each treatment. ***P < 0·001 when compared with the BEAS-2B and eosinophils control. BE: BEAS-2B cells, EOS: eosinophils.
Release of chemokines upon the interaction of paraformaldehyde-fixed BEAS-2B cells and eosinophils
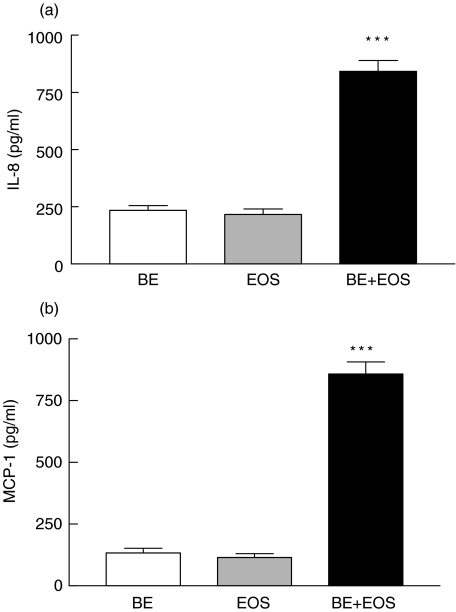
Figure 4a,b illustrates that the incubation of paraformaldehyde (1%) fixed BEAS-2B cells with eosinophils did not show any significant effect on the in vitro release of IL-8 and MCP-1 from eosinophils (all P > 0·05). However, fixed eosinophils could significantly induce BEAS-2B cells to release IL-8 and MCP-1 (P < 0·001). There were only very little amounts of IP-10, MIG and RANTES that could be detected in the coculture supernatant without any significant differences between different groups (all P > 0·05). Moreover, there was no chemokine release in fixed BEAS-2B cells only, fixed eosinophils only and coculture of fixed BEAS-2B cells and fixed eosinophils (Fig. 4a,b).
Fig. 4.
Induction of the release of (a) IL-8 and (b) MCP-1 upon the coculture of fixed or not fixed eosinophils and BEAS-2B cells. Confluent normal or paraformaldhyde (1%) fixed BEAS-2B cells (1·5 × 105/well) were cultured with or without normal or paraformaldhyde (1%) fixed eosinophils (5 × 105/well) for 18 h in 24-well plate. Chemokines released in culture supernatant was determined by human chemokine CBA kit using flow cytometry. Results are expressed as the arithmetic mean ± SEM from three independent experiments with different blood donors with single well on the tissue culture plate for each treatment. **P < 0·01, ***P < 0·001 when compared with the BEAS-2B and eosinophils control. BE: normal BEAS-2B cells, EOS: normal eosinophils, BE*: fixed BEAS-2B cells, EOS*: fixed eosinophils.
Activation of p38 MAPK activity in BEAS-2B cells and eosinophils in coculture
Figure 5a,b show that the interaction of BEAS-2B cells and eosinophils could rapidly induce the phosphorylation of p38 MAPK and ATF-2 at 30 min of BEAS-2B but not eosinophils, thereby indicating the activation of p38 MAPK in BEAS-2B cells. SB 203580 (20 µm) could suppress the phosphorylation of ATF-2 in both BEAS-2B cells and eosinophils, thereby confirming its inhibitory activity of p38 MAPK (Fig. 5b).
Fig. 5.
Activation of p38 MAPK activity in BEAS-2B cells and eosinophils. Confluent BEAS-2B cells (6 × 105/well) were cultured with or without eosinophils (3 × 106/well) in a 6-well plate for 30 min in the presence or absence of SB 203580 (20 µm). Total cellular proteins were extracted from BEAS-2B cells or eosinophils for the detection of (A) phosphorylated p38 MAPK protein level by Western blot analysis and (B) p38 MAPK activity by the detection of phosphorylated ATF-2 using a p38 MAP Kinase assay kit. Experiments were performed in three independent experiments with essentially identical results, and representative results are shown. BE: BEAS-2B cells, EOS: eosinophils, SB: SB 203580.
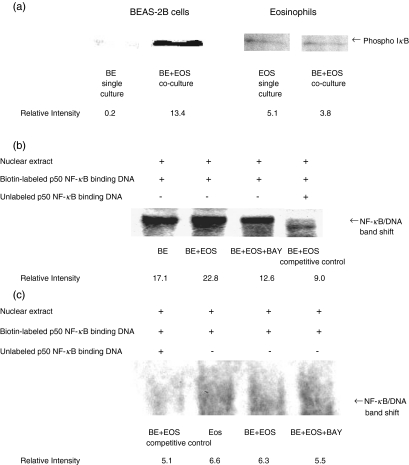
Activation of NF-κB activity in BEAS-2B cells and eosinophils in coculture
As shown in Fig. 6a, BEAS-2B cells without any treatment exhibited relatively low amount of phospho-IκB-α. Co-culture of BEAS-2B and eosniophils could activate the NF-κB activity in BEAS-2B cells by inducing the phosphorylation and hence the degradation of IκB-α at 30 min. Figure 6b illustrates that untreated BEAS-2B cells show a shifted band, thereby indicating a basal activity of NF-κB. Co-culture with eosinophils for 2 h was shown to activate NF-κB for subsequent gene transcription because there was a significant increase in intensity of band shift observed in BEAS-2B cells. Pretreatment of BAY 11–7082 (1 µm) for 1 h was found to inhibit the eosinophil induced band shift in BEAS-2B cells. Competitive control using excess unlabelled p50 NF-κB binding DNA confirmed the specificity of NF–κB–DNA interaction. Figure 6c illustrates that BEAS-2B cells could not have marked activation of NF-κB activity in eosinophils.
Fig. 6.
Effect of interaction between eosinophils and BEAS-2B cells on NF-κB activity in BEAS-2B cells and eosinophils. (a) Confluent BEAS-2B cells (6 × 105/well) were cultured with or without eosinophils (3 × 106/well) in a 6-well plate for 30 min. Total protein was then extracted from BEAS-2B or eosinophils. Phosporylated IκB was then analysed by Western blot. (b, c) Confluent BEAS-2B cells (6 × 105/well) were cultured with eosinophils (3 × 106/well) in a 6-well plate for 2 h with or without 1 h pretreatment and subsequent 2 h incubation of BAY 11–7082 (1 µm). Nuclear proteins were then extracted from (b) BEAS-2B cells or (c) eosinophils, and 5 µg protein was then subjected to EMSA. Experiments were performed in three independent experiments with essentially identical results, and representative results are shown. BE: BEAS-2B cells, EOS: eosinophils, BAY: BAY 11–7082.
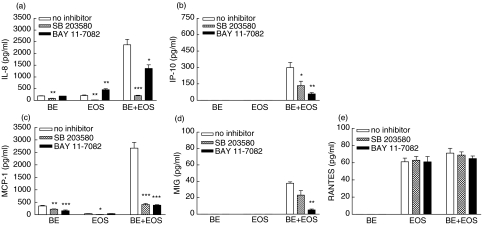
Effect of BAY 11–7082 and SB 203580 on the release of IL-8, IP-10, MCP-1, MIG and RANTES upon interaction between eosinophils and BEAS-2B cells
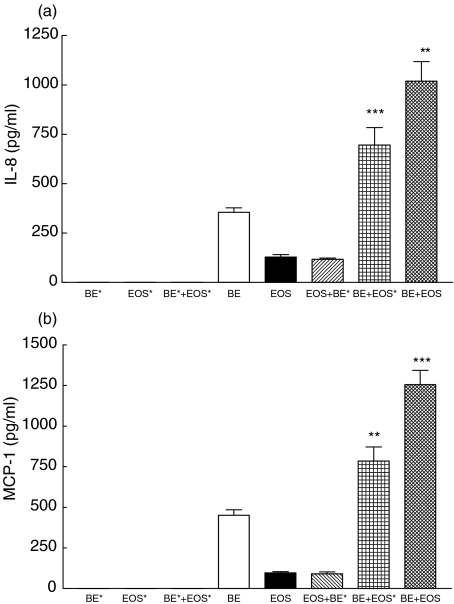
As shown in Fig. 7a–c, pretreatment of NF-κB inhibitor BAY 11–7082 (1 µm) or p38 MAPK inhibitor SB 203580 (20 µm) for 1 h could significantly suppress the induced IL-8, IP-10 and MCP-1 release upon the interaction of BEAS-2B cells and eosinophils (all P < 0·05). However, BAY 11–7082 but not SB 203580 could suppress MIG release from BEAS-2B-eosinophil coculture (Fig. 7d), while both BAY 11–7082 and SB 203580 could not suppress RANTES release from eosinophils only culture or BEAS-2B-eosinophil coculture (all P > 0·05, Fig. 7e). Moreover, both BAY 11–7082 and SB 203580 could suppress IL-8 and MCP-1 release from eosinophils and BEAS-2B cells, respectively (all P < 0·01, Fig. 7a,c), while only SB 203580 could suppress the IL-8 and MCP-1 release from BEAS-2B cells and eosinophils, respectively (all P < 0·05, Fig. 7a,c).
Fig. 7.
Effect of BAY 11–7082 and SB 203580 on the release of IL-8, IP-10, MCP-1, MIG and RANTES upon the interaction between eosinophils and BEAS-2B cells. Confluent BEAS-2B cells (1·5 × 105/well) were pretreated with BAY 11–7082 (1 µm) or SB 203580 (20 µm) for 1 h followed by culture with eosinophils (3 × 105/well) in a 24-well plate for a further 15 h in the presence of BAY 11–7082 or SB 203580. Chemokines released in the culture supernatant were determined using the human chemokine CBA kit and flow cytometry. Results are expressed as the arithmetic mean ± SEM from three independent experiments with different blood donors per single well on the tissue culture plate for each treatment. The eosinophil and BEAS-2B single culture, and coculture of eosinophils and BEAS-2B cells contained 0·1% DMSO as a DMSO control. *P < 0·05, **P < 0·01, ***P < 0·001 when compared with the control. BE: BEAS-2B cells, EOS: eosinophils, SB: SB203580, BAY: BAY 11–7082.
Discussion
Interaction of eosinophils with the bronchial epithelium represents a crucial mechanism of initiating local inflammation in allergic asthma. In this study, we examined inflammatory mediators released from the interaction of epithelial cells and eosinophils that mimic in vivo conditions in bronchial asthma. We adopted the multiplex CBA flow cytometric analysis for the quantification of five different chemokines in parallel for the better correlation of data. Figures 1 and 2 illustrate a remarkable increase in synthesis and release of a panel of leucocyte chemokines IL-8, IP-10, MCP-1 and MIG when bronchial epithelial cells interacted with eosinophils in vitro. As also found by us, incubation of paraformaldehyde (1%) fixed BEAS-2B cells with eosinophils did not show any significant effect on the in vitro release of chemokines from eosinophils but fixed eosinophils could activate chemkine IL-8 and MCP-1 release (Fig. 4). Moreover, single and coculture of fixed cells did not release any chemokines. Together with the induction of chemokine gene expression in BEAS−2B upon interaction with eosinophils (Fig. 1), it is believed that in vitro chemokine release can be induced mainly from BEAS-2B cells in the BEAS-2B-eosinophils coculture, at least partly, through the intercellular interaction. We did not find any significant elevation of RANTES in coculture of BEAS-2B-eosinophils (Fig. 2e) even the mean value of RANTES in the coculture was found to be higher than that of BEAS-2B cells alone or eosinophils alone after 2 and 6 h incubation (Fig. 2e). This may be due to the lower induction in RANTES gene expression of BEAS-2B cells (Fig. 1). Although BEAS-2B cell could also up-regulate the IL-8, MCP-1, MIG and RANTES gene expression in eosinophils, eosinophils may contribute in a lesser degree than BEAS-2B cells to the elevated production of chemokines in coculture since fixed BEAS-2 B cells could not induce eosinophils to secrete chemokines (Fig. 4). Moreover, BEAS-2B seems to have nonsignificant effect on the activation of p38 MAPK and NF-κB activities of eosinophils (Figs 5 and 6).
One possible mechanism is that the stimulatory effect of epithelial cell–eosinophil interactions on chemokine production can be due to the direct cell activation mediated through their surface adhesion molecules. A CD18/ICAM-1-dependent adhesion of eosinophils to bronchial epithelial cells has been demonstrated [28]. This adhesion was enhanced by the activation of epithelial cells with allergen-induced TNF-α [28]. Moreover, stimulation of human eosinophil respiratory burst, degranulation and release of leukotriene C4 by soluble factors, including platelet activating factor, have been demonstrated to be dependent on CD18-mediated cellular adhesion [29]. It has also been shown that CD14 molecule participates in LPS-induced cytokine release by bronchial epithelial cells [30]. In fact, we found that the coculture of bronchial epithelial cells with eosinophils could increase surface expression of ICAM-1 on bronchial epithelial cells, and the expression could be further elevated by LPS and TNF-α. Our control experiments also showed that there were only little amounts of IP-10, MIG and RANTES that could be produced in the BEAS-2B and eosinophils coculture supernatant using transwell insert compared with that of coculture without transwell insert and the differences between the single and coculture using transwell inserts were not significant. Therefore, the intercellular contact and up-regulated cell surface expression of adhesion molecules on both eosinophils and epithelial cells are crucial for their activation and chemokine IP-10, MIG and RANTES release. However, there was significant elevation of IL-8 and MCP-1 in coculture supernatant without any direct cell contact although the amount was much less than that of coculture without transwell inserts. This suggests that the induction of IL-8 and MCP-1 may mediate through mainly direct cell contact and other minor mechanisms such as the release of soluble mediators.
In our unpublished results, we have also found that the haematopoietic cytokine IL-3 and IL-5 could enhance eosinophil adhesion onto BEAS-2B cells in vitro and induce chemotaxis through a shared p38 MAPK and NF-κB signalling pathways. Previous studies also revealed that chemokine IL-8 release from epithelial cells was mediated through the p38 MAPK and NF-κB pathways [31,32]. TNF-α and LPS could initiate the ICAM-1 expression on epithelial cells through the activation of NF-κB [33]. We used BAY 11–7082 and SB 203580, the inhibitors for NF-κB and p38 MAPK activities, to elucidate the intracellular signalling mechanisms regulating the induction of chemokines. Following previous publications [4,14,22], we used the optimal concentration of BAY 11–7082 (1 µm) and SB 203580 (20 µm) with the highest inhibitory effect without any cell toxicity.
From our present results, the release of IL-8, IP-10, MCP−1 upon the interaction of eosinophils and epithelial cells were found to be mediated by the combined activation of p38 MAPK and NF-κB activities (Fig. 7). However, the release of MIG was mediated by NF-κB but not p38 MAPK while neither NF-κB nor p38 MAPK was responsible for the RANTES release (Fig. 7). The above discrepancies of the intracellular signalling mechanisms may be due to the differential activation of other different intracellular signalling mechanisms (e.g. Janus activated kinase (JAK)-signal transducer and activator of transcription (STAT)) and differential regulation of various transcription factors for the production of various chemokines. Protein kinase C (PKC) was recently found to regulate the NF-κB-dependent transcription in epithelial cells [35] therefore the role of PKC in chemokine such as MIG and RANTES release also required further investigation.
In summary, our present results have shown that the coculture of BEAS-2B cells and eosinophils could up-regulate the expression of a panel of leucocyte chemokines such as IL-8, IP-10, MCP-1 and MIG through intracellular NF-κB and p38 MAPK pathways in BEAS-2B cells, thereby providing new clues for the elucidation of immunopathological mechanisms of eosinophil-mediated allergic inflammation and the abnormally large number of leucocytes including neutrophils, lymphocytes, basophils and macrophages in lung of allergic pulmonary disorders [36]. As eosinophil interaction to bronchial epithelium and chemokine release are dynamically regulated by cytokine such as TNF-α [2,3], this process might be a target for therapeutic intervention in the treatment of allergic asthma. In view of the recent advances in the application of p38 MAPK and NF-κB inhibitors as potential anti-inflammatory agents in asthma [21,37], our findings therefore shed light for the future development of more effective agents for allergic and inflammatory diseases.
Acknowledgments
The study was supported by a Chinese University of Hong Kong Direct Grant for Research and a donation from Vigconic (International) Limited.
References
- 1.Chiappara G, Gagliardo R, Siena A, et al. Airway remodelling in the pathogenesis of asthma. Curr Opin Allergy Clin Immunol. 2001;1:85–93. doi: 10.1097/01.all.0000010990.97765.a1. [DOI] [PubMed] [Google Scholar]
- 2.Walsh GM. Eosinophil–epithelial cell interactions: a special relationship? Clin Exp Allergy. 2001;31:351–4. doi: 10.1046/j.1365-2222.2001.01032.x. [DOI] [PubMed] [Google Scholar]
- 3.Walsh GM. Eosinophil granule proteins and their role in disease. Curr Opin Hematol. 2001;8:28–33. doi: 10.1097/00062752-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Wong CK, Zhang JP, Ip WK, Lam CWK. Activation of p38 mitogen-activated protein kinase and NF-κB in tumor necrosis factor-α-induced eotaxin release of human eosinophils. Clin Exp Immunol. 2002;128:483–9. doi: 10.1046/j.1365-2249.2002.01880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giembycz MA, Lindsay MA. Pharmacology of the eosinophil. Pharmacol Rev. 1999;51:213–340. [PubMed] [Google Scholar]
- 6.Dent G, Ruhlmann E, Bodtke K, Magnussen H, Rabe KF. Up-regulation of human eosinophil leukotriene C4 generation through contact with bronchial epithelial cells. Inflamm Res. 2000;49:236–9. doi: 10.1007/s000110050585. [DOI] [PubMed] [Google Scholar]
- 7.Takafuji S, Ohtoshi T, Takizawa H, Tadokoro K, Ito K. Eosinophil degranulation in the presence of bronchial epithelial cells. Effect of cytokines and role of adhesion. J Immunol. 1996;156:3980–5. [PubMed] [Google Scholar]
- 8.Hashimoto S, Matsumoto K, Gon Y, et al. p38 MAP kinase regulates TNF alpha-, IL-1 alpha- and PAF-induced RANTES and GM-CSF production by human bronchial epithelial cells. Clin Exp Allergy. 2000;30:48–55. doi: 10.1046/j.1365-2222.2000.00641.x. [DOI] [PubMed] [Google Scholar]
- 9.Herlaar E, Brown Z. p38 MAPK signalling cascades in inflammatory disease. Mol Med Today. 1999;5:439–47. doi: 10.1016/s1357-4310(99)01544-0. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita N, Koizumi H, Murata M, Mano K, Ohta K. Nuclear factor kappa B mediates interleukin-8 production in eosinophils. Int Arch Allergy Immunol. 1999;120:230–6. doi: 10.1159/000024272. [DOI] [PubMed] [Google Scholar]
- 11.Holden NS, Catley MC, Cambridge LM, Barnes PJ, Newton R. ICAM-1 expression is highly NF-kappaB-dependent in A549 cells. Eur J Biochem. 2004;271:785–91. doi: 10.1111/j.1432-1033.2004.03982.x. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell TS, Christman JW. The role of nuclear factor-κB in cytokine gene regulation. Am J Respir Cell Mol Biol. 1997;17:3–9. doi: 10.1165/ajrcmb.17.1.f132. [DOI] [PubMed] [Google Scholar]
- 13.Ip WK, Wong CK, Lam CWK. Tumor necrosis factor-α-induced expression of intercellular adhesion molecule-1 on human eosinophilic leukemia EoL-1 Cells is mediated by the activation of nuclear factor-κB pathway. Clin Exp Allergy. 2003;33:241–8. doi: 10.1046/j.1365-2222.2003.01585.x. [DOI] [PubMed] [Google Scholar]
- 14.Wong CK, Ip WK, Lam CWK. IL-3, 5 and GM-CSF-induced adhesion molecule expression on eosinophils by p38 MAPK and NF-κ B. Am J Respir Cell Mol Biol. 2003;29:133–47. doi: 10.1165/rcmb.2002-0289OC. [DOI] [PubMed] [Google Scholar]
- 15.Lukacs NW. Role of chemokines in the pathogenesis of asthma. Nat Rev Immunol. 2001;1:108–16. doi: 10.1038/35100503. [DOI] [PubMed] [Google Scholar]
- 16.D'Ambrosio D, Mariani M, Panina-Bordignon P, Sinigaglia F. Chemokines and their receptors guiding T lymphocyte recruitment in lung inflammation. Am J Respir Crit Care Med. 2001;164:1266–75. doi: 10.1164/ajrccm.164.7.2103011. [DOI] [PubMed] [Google Scholar]
- 17.Wong CK, Ho CY, Ko FWS, et al. Proinflammatory cytokines (IL-17, IL-6, IL-18 and IL-12) and Th cytokines (IFN-γ, IL-4, IL-10 and IL-13) in patients with allergic asthma. Clin Exp Immunol. 2001;125:177–83. doi: 10.1046/j.1365-2249.2001.01602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung TF, Wong CK, Chan IHS, Ip WK, Lam CWK, Wong GWK. Plasma Concentration of Thymus and Activation-Regulated Chemokine (TARC) is Elevated in Childhood Asthma. J Allergy Clin Immunol. 2002;110:404–9. doi: 10.1067/mai.2002.126378. [DOI] [PubMed] [Google Scholar]
- 19.Leung TF, Wong CK, Lam CWK, Ip WK, Wong GWK, Chan IHS. Plasma TARC but not MDC Level is elevated in children with chronic stable asthma and decreased following corticosteroid treatment for acute exacerbation. Eur Respir J. 2003;21:616–20. [Google Scholar]
- 20.Luster AD. Antichemokine immunotherapy for allergic diseases. Curr Opin Allergy Clin Immunol. 2001;1:561–7. doi: 10.1097/00130832-200112000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Newton R, Holden N. Inhibitors of p38 mitogen-activated protein kinase: potential as anti-inflammatory agents in asthma? Biodrugs. 2003;17:113–29. doi: 10.2165/00063030-200317020-00004. [DOI] [PubMed] [Google Scholar]
- 22.Mori N, Yamada Y, Ikeda S, et al. Bay 11–7082 inhibits transcription factor NF-kappaB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood. 2002;100:1828–34. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]
- 23.Wan S, Yim AP, Wong CK. Expression of FHL2 and cytokine messenger RNAs in human myocardium after cardiopulmonary bypass. Int J Cardiol. 2002;86:265–72. doi: 10.1016/s0167-5273(02)00331-5. [DOI] [PubMed] [Google Scholar]
- 24.Jordan NJ, Kolios G, Abbot SE, et al. Expression of functional CXCR4 chemokine receptors on human colonic epithelial cells. J Clin Invest. 1999;104:1061–9. doi: 10.1172/JCI6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terada N, Maesako K, Hamano N, et al. RANTES production in nasal epithelial cells and endothelial cells. J Allergy Clin Immunol. 1996;98:S230–S237. doi: 10.1016/s0091-6749(96)70071-4. [DOI] [PubMed] [Google Scholar]
- 26.Wong CK, Lam CWK. Clinical applications of cytokine assays. Adv Clin Chem. 2003;37:1–46. doi: 10.1016/s0065-2423(03)37005-2. [DOI] [PubMed] [Google Scholar]
- 27.Tarnok A, Hambsch J, Chen R, Varro R. Cytometric bead array to measure six cytokines in twenty-five microliters of serum. Clin Chem. 2003;49:1000–2. doi: 10.1373/49.6.1000. [DOI] [PubMed] [Google Scholar]
- 28.Burke-Gaffney A, Hellewell PG. A CD18/ICAM-1-dependent pathway mediates eosinophil adhesion to human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1998;19:408–18. doi: 10.1165/ajrcmb.19.3.3179. [DOI] [PubMed] [Google Scholar]
- 29.Horie S, Kita H. CD11b/CD18 (Mac-1) is required for degranulation of human eosinophils induced by human recombinant granulocyte-macrophage colony-stimulating factor and platelet-activating factor. J Immunol. 1994;152:5457–67. [PubMed] [Google Scholar]
- 30.Striz I, Mio T, Adachi Y, Bazil V, Rennard S. The CD14 molecule participates in regulation of IL-8 and IL-6 release by bronchial epithelial cells. Immunol Lett. 1998;62:177–81. doi: 10.1016/s0165-2478(98)00046-7. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Cleary PP, Xu H, Li JD. Up-regulation of interleukin-8 by novel small cytoplasmic molecules of nontypeable Haemophilus influenzae via p38 and extracellular signal-regulated kinase pathways. Infect Immun. 2003;71:5523–30. doi: 10.1128/IAI.71.10.5523-5530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemeth ZH, Deitch EA, Szabo C, Fekete Z, Hauser CJ, Hasko G. Lithium induces NF-kappa B activation and interleukin-8 production in human intestinal epithelial cells. J Biol Chem. 2002;277:7713–9. doi: 10.1074/jbc.M109711200. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Chou C, Sun Y, Huang W. Tumor necrosis factor alpha-induced activation of downstream NF-kappaB site of the promoter mediates epithelial ICAM-1 expression and monocyte adhesion. Involvement of PKCalpha, tyrosine kinase, and IKK2, but not MAPKs, pathway. Cell Signal. 2001;13:543–53. doi: 10.1016/s0898-6568(01)00171-1. [DOI] [PubMed] [Google Scholar]
- 34.Matsuyama W, Wang L, Farrar WL, Faure M, Yoshimura T. Activation of Discoidin Domain Receptor 1 Isoform b with Collagen Up-Regulates Chemokine Production in Human Macrophages: Role of p38 Mitogen-Activated Protein Kinase and NF-kappaB. J Immunol. 2004;172:2332–40. doi: 10.4049/jimmunol.172.4.2332. [DOI] [PubMed] [Google Scholar]
- 35.Catley MC, Cambridge LM, Nasuhara Y, et al. Inhibitors of protein kinase C (PKC) prevent activated transcription: Role of events downstream of NF-kappa B DNA binding. J Biol Chem. 2004;279:18457–66. doi: 10.1074/jbc.M400765200. [DOI] [PubMed] [Google Scholar]
- 36.Rothenberg ME, Zimmermann N, Mishra A, et al. Chemokines and chemokine receptors: their role in allergic airway disease. J Clin Immunol. 1999;19:250–65. doi: 10.1023/a:1020531322556. [DOI] [PubMed] [Google Scholar]
- 37.Makarov SS. NF-kappaB as a therapeutic target in chronic inflammation: recent advances. Mol Med Today. 2000;6:441–8. doi: 10.1016/s1357-4310(00)01814-1. [DOI] [PubMed] [Google Scholar]