Abstract
This longitudinal study investigates the change of erythrocyte complement receptor (E-CR1) expression in patients with severe acute respiratory syndrome (SARS). Circulating E-CR1 expression was semiquantified by flow cytometric analyses in 54 SARS patients and in 212 healthy individuals as a control. Since E-CR1 expression is influenced by the genetic polymorphisms in the CR1 gene, a major genetic polymorphism located within intron 27 of the CR1 gene was simultaneously analysed by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP). The results showed that the expression level of E-CR1 (referred to as net fluorescence intensity values, NFI) was statistically correlated with the relevant genetic genotypes among the Chinese population including the healthy individuals (NFI: 5·14 ± 0·82, 3·57 ± 0·66 and 2·67 ± 0·32 for HH, HL and LL genotypes, respectively) and SARS patients (NFI: 3·52 ± 0·91 and 2·63 ± 0·70 for HH and HL genotypes, respectively). Interestingly, the expression density of E-CR1 was found to fall significantly during the initiation and progressive phases (weeks 1 and 2 after the disease onset) and gradually returned close to normal through their whole convalescent phase (beginning from weeks 2 or 3 to weeks 7 or 8) in SARS patients irrespective CR1 genotype. In conclusion, our findings, at least, suggest that E-CR1 is likely involved in immune pathogenesis of SARS disease.
Keywords: SARS, CR1, expression polymorphism, flow cytometry
Introduction
Severe acute respiratory syndrome (SARS) is a new emerging disease caused by a novel coronavirus (SARS-Cov) belonging to family Coronaviridae [1,2]. During the worldwide outbreaks of catastrophic SARS epidemic in 2003, SARS disease affected a cumulative number of 8098 subjects and claimed 774 deaths as recorded by the World Health Organization [3,4]. Clinically, SARS is characterized by persistent high fever (temperature >38°C), nonproductive cough or dyspnoea, rapidly progressing abnormal results on radiography and laboratory evidence of SARS-Cov RNA and existence of specific antibodies. Most of the SARS subjects exhibited a typical course of acute self-limited illness and around 10% died from SARS disease or associated complications [5]. To date, the considerable findings in early clinical manifestation, haematological, radiological, and microbilogical aspects have showed that SARS-Cov infection resulted in severe cytopathic changes [6–9]. The histological pathology in the lung of patients at autopsy, clinical manifestation and the abnormality showed by chest radiograph suggest that these changes are likely to be induced by the acute overwhelming immune response in vivo, but the underlying mechanism in the process of SARS-Cov infection is poorly understood yet [1, 5, 6].
A recent study indicated that complement receptor one (CR1, C3b receptor or referred to CD35 molecule) expressed on erythrocytes (E), can vary as much as 10-fold among the healthy individuals [10]. The CR1 gene is highly polymorphic with some single nucleotide polymorphism (SNP) sites which may contribute to variation of E-CR1 expression. Interestingly, Wilson et al. identified the genomic Hind III restriction fragment length polymorphism (RFLP) in intron 27 of the CR1 gene that, probably acting as a principle genetic polymorphism, had been found to correlate with E-CR1 expression in Caucasians. This gives rise to three genotypes including homozygosity of the HH (high) allele, heterozygosity of HL allele and homozygosity of the LL (low) allele that genetically regulate the high, intermediate and low expression of CR1, respectively [11]. E can bind complement opsonized microbe pathogens or immune complex (IC) through surface CR1 molecules and deliver them to tissue macrophages fixed at the reticuloendothelial system where they are eliminated [12]. As a result, CR1 on E (E-CR1) functions as one of the multiple components in host innate immunity and plays a crucial role against invading pathogens [13].
E lose CR1 as they age in the circulation and, in particular, this process is markedly accelerated in patients with a variety of inflammatory diseases characterized by chronic viral infections (HIV or HCV), lepromatous leprosy and specific autoimmune diseases like systemic lupus erythematosus (SLE) [11,14–17]. This suggests that E-CR1 molecules are likely to participate in the pathogenesis of immune-complex mediated diseases [18–21].
Beijing, as the most affected area by the SARS epidemic in early 2003, claimed 2521 cases and 191 deaths [22]. The viral infection may produce various haematological changes [6]. Our previous work and other findings motivated us to seek how E-CR1 molecules in peripheral blood change in patients with SARS-Cov infection. To address the question, we longitudinally recorded the alteration of circulating E-CR1 expression throughout the course of SARS illness in 54 SARS patients. In addition, we simultaneously analysed the distribution of the Hind III expression polymorphism in the CR1 gene among the SARS patients and healthy individuals. Our findings suggest E-CR1 molecules, as one of the important components in innate immunity, are involved in the process of the deadly SARS disease.
Materials and methods
Study patients
We employed 54 confirmed SARS patients (34 males, 20 females; age range 13–74 years; 79·6% (43/54) in age range 20–50 years) that were admitted to the Beijing 302 hospital during the period of March 5th and June 20th, 2003. All the patients fulfilled the accepted definition of SARS [2]. Briefly, the incubation period for all the patients was within the range of 2–14 days. In early prodromal period, all patients had fever ≥ 38°C over 48 h, a nonproductive cough or shortness of breath, new pulmonary infiltrates on chest radiography or high-resolution CT scans in the absence of an alterative diagnosis to explain the clinical presentation. Each patient was nursed in the isolation ward with other SARS patients.
The study protocol was approved by the Ethics Committee of this institution, and written informed consent was obtained from each participant. Fresh EDTA-anticoagulated peripheral blood (5·0 ml) was obtained from each of the SARS-diagnosed patients at enrolment and at weekly intervals. The genomic DNA samples were extracted from 0·2 ml of whole peripheral blood by using the QIAgen DNA purification kit (Qiagen, CA, USA) as the previous report [21]. The remaining blood was used for the analyses of E-CR1 by flow cytometry. We simultaneously recruited 212 healthy subjects who received a health examination in our hospital during the same period. Both the enrolled SARS patients and healthy individuals were excluded from HIV, HBV, HCV infections as well as free from the diseases such as influenza and pneumonia different from SARS. We randomly selected 39 sex- and aged-matched healthy individuals as a control for establishment of the reference values (NFI) of E-CR1. Both SARS patients and healthy subjects are from the Chinese Han ethnic origin.
Genotyping of CR1 alleles by PCR-RFLP
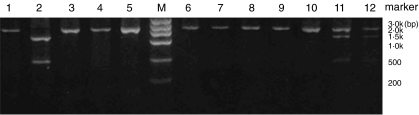
All the genomic DNA samples were used as the PCR templates in our study. We synthesized the PCR primers as previously reported by Cornillet et al. [14]. Primer 1: 5′-CCTTCAATGGAATGGTGCAT-3′ (4415nt − 4435nt): Primer 2: CCCTTGTAAGGCAAGTCTGG location 25nt −55nt upstream 3′ end of CR1 intron containing the Hind-3′ (cleavage site III), the full length of amplified product is a 1·8 kb cDNA fragment in size. CR1 genotypes were determined on the basis of fragments sizes of Hind III digested PCR products. Homozygous HH individuals exhibit only one fragment of 1·8 kb, heterozygotes (HL) show three fragments of 1·8 kb, 1·3 kb and 0·5 kb, respectively, and the mutant homozygous LL has two bands of 1·3 kb and 0·5 kb in sizes as shown in Fig. 1. The three different genotypes represent the high, intermediate and low expression levels of E-CR1, respectively.
Fig. 1.
Representative genotyping of Hind III fragments of CR1 gene using PCR-RFLP analysis in Chinese populations. Lanes 1,3–10 for HH genotype; lane 11–12 for HL genotype; lane 2 for LL genotype; M: DNA ladder marker.
Analysis of E-CR1 expression using flow cytometric assay
According to the previous reports [18,23], E-CR1 expression was semiquantitatively determined by using a flow cytometric analysis for all the SARS patients and healthy individuals, weekly if possible. Briefly, EDTA-anticoagulated venous blood was suspended in phosphate-buffered saline (PBS) and then layered onto a Ficoll-Hypaque gradient solution. After being centrifuged at 1000× g for 10 min, erythrocytes were gently collected in the bottom layer and washed three times with saline. 106 cells were suspended in 50 µl of PBS plus 1% bovine serum albumin (BSA) and incubated for 1 h at 4°C with 10 µl of fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antihuman CR1 antibody (E11 Clone, mouse IgG1) from BD-Pharmingen (Becton-Dickson, Mountain View, CA) [12,17]. As a control, 10 µl of mouse purified isotypic antibody conjugated with FITC or PBS were substituted for anti-CR1 antibody. After the samples had been washed, they were fixed with PBS containing BSA and 1% formaldehyde. Finally, the samples were subjected to immunofluorescent analysis in duplicate in FACScalibur (Becton-Dickson, Mountain View, CA, USA) within 12 h after fixation. The mean fluorescence intensity (MFI) of stained erythrocytes (MFIs) and that of controls (MFIc) were measured using the Cellquest software program (Becton Dickinson). The degree of E-CR1 expression was estimated as the ratio of MFIs to MFIc (MFIs/MFIc) and was expressed as the net fluorescence intensity (NFI) according to the previous reports [15,23]. To confirm the reproducibility of E-CR1 measurements, the fluorescence intensity of FITC-conjugated Calibrite Beads was measured before FACS analysis every time on a separate day, the coefficient of variance of MFI of Calibrite Beads was within 3%.
SARS laboratory testing for analyses of plasma SARS-Cov RNA and specific antibodies
The method was the same as described in our previous reports [22,24]. In brief, RNA was directly extracted from plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. The viral RNA was amplified through nested reverse-transcription polymerase chain reaction (RT-PCR). The products were analysed by electrophoresis in 2·5% agrose gel containing 0·5 µg/ml of ethidium bromide. The viral sequence was confirmed by DNA sequence analysis At the same time, plasma anti-SARS-CoV IgG antibodies was detected according to the instruction of commercial available enzyme-linked immunosorbent assay (ELISA) kit (Huada-Jibiai Biotech, Beijing). Indirect assay was employed by the kit using culture-obtained SARS-CoV antigen for the capture of antibodies. The OD value ≥ 0·105 was judged as positive. The routine analysis for urine and faeces samples, and complete differential blood count included total leucocytes, granulocytes, lymphocytes, monocytes, erythrocytes, haemoglobin, and platelets for SARS patients was simultaneously recorded, respectively.
Chest radiograph examination
In order to identify any abnormal changes occurring in the lungs, all patients underwent examination by chest radiography once daily for patients at the progressive disease stages or critical SARS status, and every 2–4 days for patients with stable or general disease status. According to a recent report [25], we retrospectively calculated the mean chest radiograph score (referred as MCR score representing the severity of pneumonia in SARS patients) on the basis of average percentage of area manifesting the ground glass opacification, consolidation or nodular shadow, in each lung with a maximal score of 10 (equivalent to 100% area involved).
Treatment protocols
All patients received ribavirin and steroid combination therapy. Rabavirin, a broad-spectrum antiviral agent, was empirically used to treat SARS, although its efficacy is not sure because the emergent outbreak of SARS did not permit physician to conduct any randomized controlled trials on the treatment of SARS. Also on the basis of early anecdotal experience and its radiologic resemblance to acute respiratory distress syndrome and other immunologically mediated lung diseases, different corticosteroid regimens were also employed for the treatment of SARS. The timing to commence corticosteroid and ribavirin therapy varied for each individual patient. The different steroid regimens were employed in clinic for SARS patients, the therapeutic dose of methylprednisolone includes ≤ 40 mg/day, 80–160 mg/day or ≥240 mg/day (intravenously once a day) based on the clinical manifestation, radiograph deterioration, PO2 decrease, high fever and ventilation of patients.
Statistical analysis
The distribution difference of CR1 allelic genotypes among the healthy subjects and the SARS patients was analysed using Pearson's χ2 test. The statistical difference of CR1 densities on E between the two groups was compared by using the SAS6·21 software program (t-test). The value of P < 0·05 represents a statistical difference.
Results
Distribution of CR1 genotyping in SARS patients and healthy subjects
The distribution of the CR1 Hind III polymorphism was analysed using PCR-RFLP for 54 SARS patients and 212 healthy subjects (Table 1). The percentages of HH, HL and LL genotypes were 66·67%, 31·48% and 1·85% for SARS patients, and 66·92%, 27·83% and 4·25% for healthy subjects, respectively. The H and L allele frequencies in both groups are consistent with the Hardy–Weinberg equilibrium. Statistical analysis showed that there was no significant difference between the healthy controls and SARS patients (P > 0·05). Therefore, these findings suggest that CR1 molecules are unlikely to have a direct influence on host genetic susceptibility of SARS-Cov infection entering target cells and viral replication.
Table 1.
Distribution of CR1 gene polymorphism among the SARS patients and the healthy subjects of Chinese Han ethnic origin.
| Group | No. cases | HH (%) | HL (%) | LL (%) | H/l allele frequencies |
|---|---|---|---|---|---|
| SARS patients | 54 | 36 (66·67) | 17 (31·48) | 1 (1·85) | 0·83/0·17* |
| Healthy subjects | 212 | 144 (67·92) | 59 (27·83) | 9 (4·25) | 0·82/0·18 |
Comparing the data between the two groups of SARS patients with healthy subjects, P > 0·05.
Comparison of E-CR1 expression levels among the healthy subjects and SARS patients
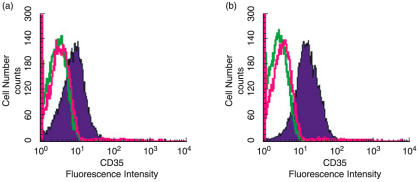
On admission into our unit, we observed that all the patients had a high fever (≥38°C) and mild or severe progressive consolidation change on serial chest radiographs in one or both sides of lungs. Using flow cytometric analysis, we semiquantitatively measured the E-CR1 expression levels in SARS patients, for which the NFI value of fluorescent density was used to indirectly indicate relative E-CR1 expression levels [15,23]. Figure 2 shows the representative differences of fluorescent densities of E-CR1 in a representative healthy individual and a SARS patient both of whom are with the HH genotype. In the samples from the SARS individual, MFIs and MFIc were 11·5 and 2·9, respectively. Thus NFI value of fluorescent density for the SARS patient was determined to be 3·96 (11·5/2·9). In the same way, NFI value for the healthy individual was determined to be 6·36 (14/2·2). Our results were summarized in Tables 2 and 3, which show that the measurements using flow cytometric analysis are stable and reproducible. The data in Table 2 seems to support the following notion:
Fig. 2.
Histograms of cell numbers and fluorescence intensities of erythrocyte CR1 stained with anti-CR1 antibody (shaded black line) or mouse purified IgG1 antibody (unshaded red line) or PBS (unshaded green line) from representative cases with HH genotypes at the sex- and age-match. (a) Erythrocytes from SARS patients in progressive stage; (b) Erythrocytes from a healthy subject.
Table 2.
Net fluorescence intensity (NFI) of E-CR1 molecules is associated with the corresponding CR1 genotype among both healthy subjects and the SARS patients during the disease progression stage.
| Group | Genotype | Cases (n) | NFI range of CR1 on E | Average NFI values (mean ± sd) |
|---|---|---|---|---|
| Healthy subjects (n = 52) | HH | 36 | 4·27–6·85 | 5·14 ± 0·82* |
| HL | 14 | 2·82–5·31 | 3·57 ± 0·66* | |
| LL | 2 | 2·45, 2·90 | 2·67 ± 0·32 | |
| SARS patients (n = 54) | HH | 36 | 2·44–5·25 | 3·52 ± 0·91*† |
| HL | 17 | 1·99–3·58 | 2·63 ± 0·70*† | |
| LL | 1 | 2·99 | 2·99 |
Comparison of HH genotype with HL genotype among the healthy subjects (P < 0·01) or among the SARS patients (P < 0·05);
Comparison of healthy subjects with SARS patients for the HH genotype (P < 0·01) or for HL genotype (P < 0·05).
Table 3.
Association of SARS status with net fluorescence intensity (NFI) of E-CR1 expression at 7th day after the onset of illness.
| Group | Genotype | Cases (n) | NFI range of CR1 | Average NFI values (mean ± sd) |
|---|---|---|---|---|
| Critical SARS (n = 20) | HH | 12 | 2·44–4·20 | 3·06 ± 0·69 |
| HL | 8 | 1·99–3·55 | 2·47 ± 0·51 | |
| LL | 0 | No patient | No patient | |
| General SARS (n = 34) | HH | 24 | 2·46–5·26 | 3·98 ± 0·87 |
| HL | 9 | 2·27–3·58 | 2·82 ± 0·57 | |
| LL | 1 | 2·99 | 2·99 |
the HH, HL, and LL genotypes of the CR1 expression polymorphism correlated with the high, intermediate and low average expression quantities of the CR1 allele in both healthy and SARS patients though the variations of E-CR1 expression partly overlapped among the individuals with the HH and HL types as well as those with HL and LL types.
there was a clinically significant decrease in the average NFI values of fluorescent densities in SARS patients compared with those found in healthy subjects (P < 0·01), even though the individuals had the same HH and HL genotypes. However, there is an exception, no difference was found between the healthy controls and SARS case with the LL genotype which may be due to the small sample sizes (one or two subjects) present in these groups.
By comparing the magnitude of decrease in E-CR1 expression levels among the SARS patients between the critical and general status, we found that the critical SARS patients exhibited a lower average NFI value of E-CR1 compared to those found in the general SARS patients at day 7 after onset of illness (3·06 ± 0·69 versus 3·98 ± 0·87, P < 0·05 for HH genotypes; 2·47 ± 0·51 versus 2·82 ± 0·57, P = 0·08 for HL genotypes) (Table 3).
Even though the number of enrolled SARS patients in different groups as shown in Tables 2 and 3 is limited, our observations, at least in part, suggest that the magnitude of decrease in E-CR1 was consistent with the severity of SARS disease as related to lung damage with progressive changes as shown by chest radiograph examinations.
Among the 54 patients, 52 cases survived the disease through a hospitalization for 27–60 days (average hospitalization duration 40·5 ± 12·6 days). However, 2 of the 54 patients (males, and 73 years old, respectively) were critical and succumbed to the disease due to development of acute respiratory distress syndrome (ARDS) and aspergillosis as previously reported [26], resulting in a mortality rate of 3·7% (2/54). Even though the patients who died were identified as HH genotype, they had a dramatic decline level of E-CR1 expression compared to healthy subjects with the HH genotype (NFI: 2·44–2·52 versus 5·14 ± 0·82). In addition, the 2 patients also were older, more 50 years old, and probably had some severe underlying diseases like hypertension and diabetes [1,5] which may also contributed to the severity of SARS-Cov infection.
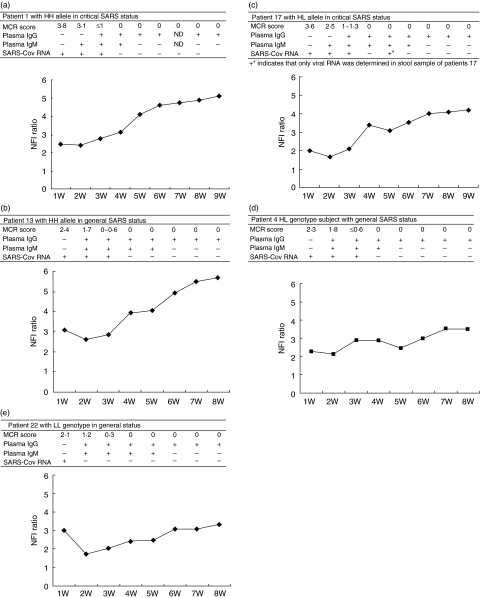
Longitudinal investigation of 5 representative individual SARS patients
The quantitative analyses of E-CR1 were conducted weekly for each of the 54 patients during the entire period of their hospitalization from progressive to the convalescence stage. To obtain profiles of circulating E-CR1 molecules, clinical status (pneumonia severity), the viral RNA and specific antibodies, 5 representative individual SARS patients with HH, HL and LL genotypes were serially analysed in combination through the entire duration of the disease (Fig. 3). SARS-Cov RNA and specific antibodies were detected in the plasma in all the patients analysed (coexistence of the viral RNA and the antibodies was uniquely observed in patient 1, 13 and 17). The serum IgG became positive in 100% of the samples within 25 days following onset of the disease. We observed a decrease in the fluorescent densities of E-CR1 at the onset of clinical illness, which persisted from the 7th to the 15th day with variations occurring during the entire progressive stage accompanied with abnormal chest radiographs and the appearance of SARS-Cov in vivo. The lower E-CR1 levels slowly started increasing during the seventh or eighth week of the illness while the chest radiographs or high-resolution CT scans shown complete resolution in approximately three weeks after the onset of symptoms for patients in convalescence. The E-CR1 levels returned to normal for all of the patients within 9–14 weeks from the onset of illness.
Fig. 3.
Schematic diagram of dynamic alteration of surface E-CR1 density, laboratory data and radiographic parameters in five representative SARS patients are summarized. (a) HH genotype individual with critical SARS status. (b) HH genotype individual with general SARS status; (c) HL genotype subject with critical SARS status; (d) HL genotype individuals with general SARS status; (e) LL genotype individual with general status.For each patient with SARS, upper table shows the pneumonia severity (MCR score is referred as mean chest radiograph score as described in Materials and Methods), detection of SARS coronavirus RNA and specific IgG antibodies in plasma each week. The lower figure shows the dynamic alteration of circulating E-CR1 by longitudinal observation. Symbols ‘–’ and ‘+’ indicate negative and positive result, respectively; ND represents no data. NFI ratio stands for net fluorescent intensity (NFI) ratio as described in Materials and Methods. The plasma IgG and IgM were determined using ELISA as described in Methods and Materials.
Discussion
The analysis of genetic mutations responsible for CR1 stability and expression level on E, its distribution, and dynamic alteration would provide a alternative approach to understanding the mechanism which regulates the severity of the inflammatory tissue damage and pathogenesis of SARS-Cov infection [14,17]. Our current observations confirmed a significant correlation between the Hind III RFLP and the relative expression of E-CR1 in Chinese Han ethnic population sample. This is consistent with an earlier report of 100 Southern Chinese and Taiwanese blood donors [27] and is also similar to results found in Caucasian populations. A recent study in Thailand also found a correlation between the Hind III RFLP and E-CR1 [28]. These findings further support the notion that variations of E-CR1 among healthy individuals are inherited and regulated by different HH, HL and LL genotypes [17,27]. We found that there was no significant difference in the frequency of HH, HL and LL genotypes between the SARS patients and healthy subjects in the Chinese Han population sample. Our limited data suggests that all the subjects have similar susceptibility to SARS viral infection. Unlike other complement regulatory proteins such as membrane control protein (MCP) and decay-accelerating factor (DAF), CR1 levels may have no direct influence on the ability of the virus to enter human target cells, and the viral replication process.
Different declining levels of E-CR1 have been found in patients not only with SLE, chronic HIV, and HCV infection, but also in patients with malaria, malignant tumour, and chronic diseases [15, 20, 21, 28, 29, 30, 31]. Interestingly, we also found a significant decrease of E-CR1 in SARS patients, which may suggest an acquired deficiency of host innate immunity [13]. SARS-Cov viral loads may be an important factor in the decrease of E-CR1 since the early rapid decline and sequential persistent lower levels of E-CR1 occurred during the periods when SARS-Cov is positively detected in patient's plasma, sputum, throat swab and/or stool samples. In this way, SARS viral infection may induce the reduction or damage of innate immunity.
As shown in Tables 2 and 3, the fluorescence intensity values of E-CR1 molecules varied over a broad range and some of them even partly overlapped between the individuals with HH, HL and LL alleles. One possibility is that the CR1 gene is highly polymorphic with a large number of single nucleotide polymorphism (SNP) sites [27], which may also influence the expression of E-CR1 independent of the H and L allele. The fluorescent density values of E-CR1 varies over a wide range for a definite genotype such as the HH, HL or LL alleles in various individuals, which may be influenced by the existing haplotypes as reported by Xiang et al. [17].
Finding of a dramatically decreased E-CR1 expression during the SARS disease progression in all the SARS patients compared with the healthy controls implies that the E-CR1 molecules are engaged in the pathogenesis of SARS. This is probably occurring during the process of forming antibody-SARS antigens and complement immune complexes at the same time when the host produces the specific antibodies to the SARS antigens. The evidence that coexistence of the specific antibodies and the direct viral RNA was found in some patients supports this hypothesis. Therefore, the aetiology of the E-CR1 reduction may be associated with the removal of the immune complex (IC) by tissue macrophages during which CR1 is proteolytically cleaved from the surface of E though its underlying mechanism is still unknown.
We would like to mention that our data should be carefully interpreted because of: (1) the limitation of a small sample size for some groups; (2) the factors including the case ageing and the steroid therapy may have also influenced the variation of E-CR1 expression in the SARS patients [25]. The different steroid regimens (doses ranging 80 mg –480 mg each day) were empirically administrated for the treatment of SARS based on pneumonia severity as determined by chest radiograph examinations. In addition, we were unable to design a randomized controlled investigation due to the rapid emergence of the SARS epidemic.
It is noteworthy that despite the clinical recovery and pulmonary damage resolution, the lower E-CR1 levels still were longitudinally sustained for a 7–8-week period and increased continuously and slowly to near normalization. Therefore, our findings suggest that the E-CR1 density undergoes an acute but reversal decrease, which correlates with the initiation, progression and convalescence stages of SARS disease [13, 15, 32, 33]. The density of the E-CR1 molecules in patients during the convalescence stage increased to alomost as high a level as found in healthy subjects. This may be because the old erythrocytes with decreased density of CR1 molecules were gradually replaced by newly produced erythrocytes having normal density of E-CR1 molecules. This in turn increased the percentage of erythrocytes with normal density of E-CR1 molecules in the total peripheral population contributing to the slow rise of peripheral E-CR1 levels in SARS patients at convalescence stage. In fact, reticulocytes have shown to increase E-CR1 levels as compared to cells that have been in the circulatory system for a period of time (J.-M. Moulds, unpublished observation).
Collectively, our findings show that an acute but reversible acquired reduction of E-CR1 may represent a significant alteration in SARS disease, suggesting that E-CR1 molecules may be one of the important components in host innate immunity, which probably participates in the process of the deadly SARS disease. This knowledge continues to increase our understanding of the immune pathogenesis of SARS.
Acknowledgments
We thank the patients who participated in this study. The work was supported by Sino-UK Collaboration Foundation for SARS Immunopathogenesis Study (Number: H030230100130) and the grants from Beijing Natural Science Foundation (Number: 7034051) and Emergent Foundation for SARS Treatment and Prevention (Number: 03F017).
References
- 1.Ksiazek TG, Erdman D, Goldsmith CS, et al. SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris JS, Lai ST, Poon LL, et al. SARS Study Group Coronavirus as possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes JM. The SARS response-building and assessing an evidence-based approach to future global microbial threats. J Am Med Assoc. 2003;290:3251–3. doi: 10.1001/jama.290.24.3251. [DOI] [PubMed] [Google Scholar]
- 4.Torpy JM, Lynm C, Glass RM. Severe acute respiratory Syndrome (SARS) J Am Med Assoc. 2003;290:3318. doi: 10.1001/jama.290.24.3318. [DOI] [PubMed] [Google Scholar]
- 5.Peiris JS, Chu CM, Cheng VC, et al. HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–62. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Openshaw PJ. What does the peripheral blood tell you in SARS? Clin Exp Immunol. 2004;136:11–2. doi: 10.1111/j.1365-2249.2004.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–8. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera AH, Xiang L, Martin SG, Lewis J, Wilson JG. Analysis of complement receptor type 1 (CR1) expression on erythrocytes and of CR1 allelic markers in Caucasian and African American populations. Clin Immunol Immunopathol. 1998;87:176–83. doi: 10.1006/clin.1998.4529. [DOI] [PubMed] [Google Scholar]
- 11.Wilson JG, Murphy EE, Wong WW, Klickstein LB, Weis JH, Fearon DT. Identification of a restriction fragment length polymorphism by a CR1 cDNA that correlates with the number of CR1 on erythrocytes. J Exp Med. 1986;164:50–9. doi: 10.1084/jem.164.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig ML, Bankovich AJ, Taylor RP. Visualization of the transfer reaction: tracking immune complexes from erythrocyte complement receptor 1 to macrophages. Clin Immunol. 2002;105:36–7. doi: 10.1006/clim.2002.5266. [DOI] [PubMed] [Google Scholar]
- 13.Pascual M, Danielsson C, Steiger G, Schifferli JA. Proteolytic cleavage of CR1 on human erthrocytes in vivo: evidence for enhanced cleavage in AIDS. Eur J Immunol. 1994;24:702–8. doi: 10.1002/eji.1830240332. [DOI] [PubMed] [Google Scholar]
- 14.Cornillet P, Philbert F, Kazatchkine MD, Cohen JH. Genomic determination of the CR1 (CD35) density polymorphism on erythrocytes using polymerase chain reaction amplification and Hind restriction enzyme digestion. J Immunol Meth. 1991;136:193–7. doi: 10.1016/0022-1759(91)90006-2. [DOI] [PubMed] [Google Scholar]
- 15.Kazatchkine MD, Jouvin MH, Wilson JG, Fischer E, Fischer A. Human diseases associated with C3 receptor deficiencies. Immunol Lett. 1987;14:191–5. doi: 10.1016/0165-2478(87)90100-3. [DOI] [PubMed] [Google Scholar]
- 16.Moulds JM, Zimmerman PA, Doumbo OK, et al. Molecular identification of Knops blood group polymorphisms found in long homologous region D of complement receptor 1. Blood. 2001;97:2879–85. doi: 10.1182/blood.v97.9.2879. [DOI] [PubMed] [Google Scholar]
- 17.Xiang L, Rundles JR, Hamilton DR, Wilson JG. Quantitative Alleles of CR1: coding sequence analysis and comparison of haplotypes in two ethnic groups. J Immunol. 1999;163:4939–45. [PubMed] [Google Scholar]
- 18.Kanto T, Hayashi N, Takehara T, et al. Low expression of erythrocyte complement receptor type 1 in chronic hepatitis C patients. J Med Virol. 1996;50:126–34. doi: 10.1002/(SICI)1096-9071(199610)50:2<126::AID-JMV5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 19.Stoute JA, Odindo AO, Owuor BO, Mibei EK, Opollo MO, Waitumbi JN. Loss of red blood cell-complement regulatory proteins and increased levels of circulating immune complexes are associated with severe malarial anemia. J Infect Dis. 2003;187:522–5. doi: 10.1086/367712. [DOI] [PubMed] [Google Scholar]
- 20.Tausk FA, McCutchan A, Spechko P, Schreiber RD, Gigli I. Altered erythrocyte C3b receptor expression, immune complexes, and complement activation in homosexual men in varying risk groups for acquired immune deficiency syndrome. J Clin Invest. 1986;78:977–82. doi: 10.1172/JCI112688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang FS, Hong WG, Cao Y, et al. Population survey of CCR5 Δ32, CCR5 m303, CCR2b 64I and SDF1 3′A allele frequencies in indigenous Chinese ethnic groups, and in. HIV-1 seropositive and HIV-1 sero-negative (high-risk) groups. J Acquir Immune Defic Syndr. 2003;32:124–30. doi: 10.1097/00126334-200302010-00002. [DOI] [PubMed] [Google Scholar]
- 22.Xu D, Zhang Z, Chu F, Li YG, Jin L, Zhang LX, Gao F, Wang FS. Variation analysis of spike glycoprotein gene of SARS coronavirus transmitted in one hospital of Beijing area. Emerg Infect Dis. 2004;10:789–94. doi: 10.3201/eid1005.030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen JH, Aubry JP, Jouvin MH, Wijdenes J, Bancherau J, Kazatchkine M, Revillard JP. Enumeration of CR1 complement receptors on erythocytes using a new method for detecting low density cell surface antigens by flow cytometry. J Immunol Meth. 1987;99:53–8. doi: 10.1016/0022-1759(87)90031-7. [DOI] [PubMed] [Google Scholar]
- 24.Xu DP, Zhang Z, Wang FS. Existence of SARS coronavirus quasispecies in single patients. N Engl J Med. 2004;350:1366–7. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- 25.Ho JC, Ooi GC, Mok TY, et al. High-dose pulse versus nonpulse corticosteroid regimens in severe acute respiratory syndrome. Am J Respir Crit Care Med. 2003;168:1449–56. doi: 10.1164/rccm.200306-766OC. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Ding Y, Li X, Yang L, Zhang W, Kang W. Fatal aspergillosis in a patient with SARS who was treated with corticosteroids. N Engl J Med. 2003;349:507–8. doi: 10.1056/NEJM200307313490519. [DOI] [PubMed] [Google Scholar]
- 27.Moulds JM, Brai M, Cohen J, et al. Reference typing report for complement receptor 1 (CR1) Exp Clin Immunogenet. 1998;15:291–4. doi: 10.1159/000019084. [DOI] [PubMed] [Google Scholar]
- 28.Inada Y, Lange M, McKinley GF, Sonnabend JA, Fonville TW, Kanemitsu T, Tanaka M, Clark WS. Hematologic correlates and the role of erythrocyte CR1 (C3b receptor) in the development of AIDS. AIDS Res. 1986;2:235–47. doi: 10.1089/aid.1.1986.2.235. [DOI] [PubMed] [Google Scholar]
- 29.Nagayasu E, Ito M, Akaki M, Nakano Y, Kimura M, Looareesuwan S, Aikawa M. CR1 density polymorphism on erythrocytes of falciparum malaria patients in Thailand. Am J Trop Med Hyg. 2001;64:1–5. doi: 10.4269/ajtmh.2001.64.1.11425154. [DOI] [PubMed] [Google Scholar]
- 30.Lach-Trifilieff E, Marfurt J, Schwarz S, Sadallah S, Schifferli JA. Complement receptor 1 (CD35) on human reticulocytes: normal expression in systemic lupus erythematosus and HIV-infected patients. J Immunol. 1999;162:7549–54. [PubMed] [Google Scholar]
- 31.Cockburn IA, Mackinnon MJ, O'Donnell A, et al. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci U S A. 2004;101:272–7. doi: 10.1073/pnas.0305306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dervillez X, Oudin S, Libyh MT, et al. Catabolism of the human erythrocyte C3b/C4b receptor (CR1, CD35): vesiculation and/or proteolysis? Immunopharmacology. 1997;38:129–40. doi: 10.1016/s0162-3109(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 33.Imrie HJ, Jones DR. Complement coating of erythrocytes is reduced following their interaction with neutrophils in vitro without loss of complement receptor 1 (CR1) Clin Exp Immunol. 1997;109:217–22. doi: 10.1046/j.1365-2249.1997.4151312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]