Abstract
Nuclear hormone receptors are ligand-dependent transcriptional regulators that modulate chromatin structure. However, the precise molecular mechanisms by which receptors recruit chromatin-remodeling activity are not fully elucidated. We show that in the absence of its ligand-binding domain, the glucocorticoid receptor (GR) is able to interact with both nuclear receptor coactivators and the BRG1 chromatin-remodeling complex in vivo. Individually, the GR makes direct interactions with BRG1-associated factor 60a (BAF60a) and BAF57, but not with BRG1, BAF155, or BAF170. Further, BAF60a possesses at least two interaction surfaces, one for GR and BRG1 and a second for BAF155 and BAF170. A GR mutant, GR(R488Q), that fails to interact with BAF60a in vitro has reduced chromatin-remodeling activity and reduced transcriptional activity from the promoter assembled as chromatin in vivo. Stable expression of a BAF60a truncation mutant, BAF60a4-140, caused chromatin-specific loss of GR functions in vivo. In the presence of the BAF60a mutant, the GR fails to interact with the BRG1 complex and consequently is also deficient in its ability to activate transcription from chromatin. Thus, in addition to previously identified BAF250, BAF60a may provide another critical and direct link between nuclear receptors and the BRG1 complex that is required for promoter recruitment and subsequent chromatin remodeling.
Eukaryotic genes are highly organized into chromatin, which may restrict the access of regulatory factors to promoter control sequences (4, 20, 37, 46-48). The repetitive basic unit of chromatin is the nucleosome, which is an octamer of core histones 2A, 2B, 3, and 4. This histone octamer is wrapped by approximately 147 bp of DNA. In addition, the linker histone H1, which includes multiple subtypes in mammals, binds to the DNA between two adjacent nucleosomes, called linker DNA (16). The C-terminal histone fold domain of each core histone forms the structural body around which the DNA is wrapped (3, 25). In contrast, the histone N-terminal tails, which are rich in basic residues, are thought to extend outward and interact with adjacent nucleosomes. This interaction allows nucleosomal arrays to self-associate into a higher-order chromatin fiber (26).
Nuclear hormone receptors (NHRs) represent a superfamily of transcription factors with conserved structural and functional domains (28). As a member of the nuclear receptor superfamily, the domain structure of the glucocorticoid receptor (GR), which includes a DNA binding domain (DBD), a ligand binding domain (LBD), and a hypervariable N-terminal domain, has been extensively investigated (28). NHRs are potent transactivators with two transcriptional activation functions (AFs), namely, the ligand-independent AF-1 within the N-terminal domain and the ligand-dependent AF-2 centered at helix 12 of the LBD (18). The LBD has proven to be a particularly promiscuous interacting surface for a variety of coactivator and corepressor complexes that are critical for NHR transcriptional function (18). Ligands that bind members of the NHR family include steroids, thyroid hormones, vitamin D3, retinoids, and a growing number of lipophilic molecules (7). These hormones, acting via their respective NHRs, regulate various metazoan physiological processes, such as reproduction, development, homeostasis, and metabolism, by controlling gene expression. Binding of hormone to the NHR transforms the receptor conformation and creates an interface that allows the receptor to interact with chromatin-remodeling complexes and coactivator molecules (8). In particular, numerous protein-protein interactions between the LBD and coregulatory molecules and chromatin-remodeling proteins have been documented (8, 30). More recently, in vitro experiments have revealed an interaction between the N-terminal AF-1 of GR and the entire yeast SWI/SNF complex (42).
The yeast SWI/SNF complex is the prototype chromatin-remodeling complex and was initially described as a homogeneous multiprotein complex (24, 36, 49). In contrast, the mammalian SWI/SNF complex has been shown to be heterogeneous, with complexes that contain a BRG1 or brm ATPase in addition to another 7 to 14 BRG1-associated factors (BAFs) (45). The functional core of the BRG1 complex includes BRG1 or brm, BAF170, BAF155, and INI1, which has been defined in vitro by reconstitution of chromatin-remodeling activity with recombinant proteins (38). What role(s) the additional BAFs may play in mediating the diverse functions of the BRG1 complex in human cells remains unknown.
With the steroid hormone-activated mouse mammary tumor virus (MMTV) promoter used as a model, it has been reported that GR-mediated transactivation of MMTV is a bimodal, two-step process (1, 18). Hormone-dependent interaction of GR with the BRG1 complex is required to allow chromatin remodeling, an obligatory step prior to activation of MMTV transcription from a chromatin template (11). We have now explored the mechanisms that underlie GR recruitment of the BRG1 complex to remodel chromatin by using a C-terminal truncation mutant protein devoid of the LBD that has been shown previously to be transcriptionally active (18). We find that this protein, GR1-556, spanning the N-terminal, DBD, and hinge regions of the GR, interacts with BAF60a and BAF57, but not BRG1, BAF170, or BAF155. Moreover, BAF60a interacts with BRG1 via its N terminus and with BAF170 and BAF155 through its C terminus.
Within the context of the complete receptor, we show that a mutation in the receptor’s DBD GR(R488Q), which is unable to interact with the N-terminal BAF60a, activates transcription from a transiently transfected reporter but not a chromatin-integrated reporter gene, indicating an essential role for the GR-BAF60a interaction in remodeling chromatin. Furthermore, stable introduction of a mutant BAF60a lacking its C terminus into human osteosarcoma cells functionally disrupts GR-activated chromatin remodeling and prevents subsequent transcription from chromatin. This loss of GR activity results from the loss of the hormone-dependent interaction between the GR and BRG1 complex. Mechanistic analysis of GR(R488Q) and BAF60a4-140 mutants together suggests that the interaction between the GR and the N-terminal BAF60a is required to transmit chromatin-remodeling signal for efficient transcriptional activation. Thus, BAF60a may function as a critical interaction surface for recruitment and signal transmission by NHR to remodel the promoter chromatin in vivo.
MATERIALS AND METHODS
Cell lines and plasmids.
2963.1 (PR+) and A1-2 (PR+, GR+) are T-47D-derived cell lines with chromatin integration of chloramphenicol acetyltransferase (CAT) and luciferase reporters driven by MMTV promoters, as described previously (2, 33). The chromatin structure of nucleosome B (Nuc-B), which contains the proximal glucocorticoid response elements of MMTV promoter, remodels hormone independently in 2963.1 cells, whereas remodeling of Nuc-B in A1-2 cells depends on glucocorticoid induction (2, 33). The 2963.1/GR556 cell line is derived from stable transfection of pSV-GR1-556 plasmid into 2963.1 cells (31). Cotransfection with MMTV-long terminal repeat (LTR) luciferase reporter and a G418 selection cassette, pRSV.neo, created the SW-13/M3-2 clone. Chromatin remodeling of the Nuc-B in SW-13/M3-2 depends on transfection of BRG1 and GR plus hormone treatment. The U-2 OS-GR-derived UL3 cell line also contains stable chromatin integration of the MMTV luciferase reporter (12). Glucocorticoid treatment is sufficient to induce chromatin remodeling of the Nuc-B in UL3 cells. Plasmids pBS-hBAF60a, pBS-hBAF155, pBS-hBAF170, and pBS-hBAF57 were described previously (43, 44). The plasmids expressing GST-GR1-556 were constructed by inserting the cognate coding sequence of rat GR from pSG5-GR (9) into pGEX-2T (Amersham-Pharmacia). The plasmids expressing GST-BAF60a4-128 and GST-BAF60a129-435 were constructed by inserting the coding sequence of hBAF60a residues 4 to 128 and 129 to 435 into pGEX-4T2 (Amersham-Pharmacia). The plasmids expressing H6GR1-556 and H6BAF60a4-140 were constructed by inserting coding sequences of rat GR residues 1 to 556 and hBAF60a residues 4 to 140 into pcDNA4-HisMax (Invitrogen), respectively.
Protein expression and purification of Escherichia coli.
Strain BL21 harboring plasmids expressing GST-GR1-556, GST-BAF60a4-128, or GST-BAF60a128-435 was grown to an optical density at 600 nm of 1.0, followed by 0.1 mM isopropyl-β-d-thiopyranogalactoside induction for 1 h at 30°C. Cells were collected by centrifugation at 10,000 × g for 5 min. Cells were resuspended in B-PER solution (5 ml/g [wet weight] of cells) (Pierce) containing protease inhibitor cocktail (Sigma) with 5 mM dithiothreitol (DTT) and lysed by mixing to homogenate at 4°C for 20 min. Cell debris were removed by centrifugation at 27,000 × g at 4°C for 15 min. Glutathione S-transferase (GST) fusion proteins were purified from the supernatant with a B-PER GST fusion protein purification kit (Pierce). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and GelCode Blue staining (Pierce).
GST pull-down.
GST or fusion protein (10 μg) was loaded onto 5 μl of immobilized glutathione beads (Pierce) for each reaction. The beads were incubated with 10 μl of TNT-wheat germ extract-expressed target protein lysate in 0.5 ml of pull-down buffer (50 mM HEPES-KOH [pH 7.9], 150 mM NaCl, 0.1% [vol/vol] Tween-20, 10% [vol/vol] glycerol, 1 mM EDTA, 0.5 mM DTT) at 4°C for 16 h. Beads were washed five times with pull-down buffer. Associated proteins were eluted with 20 μl of Laemmli SDS sample buffer and analyzed by SDS-10% PAGE. Gel was stained with GelCode Blue (Pierce), dried, and visualized by autoradiography.
CAT and luciferase assays.
CAT activity and luciferase activities were determined as described previously following hormone treatments (11). For transient template assays, SW-13/M3-2 cells were transfected with 3 μg of MMTV-CAT plasmid plus 1 μg of pSG5-GR and 1 μg of pJB5-BRG1 expression plasmids, and UL3 and 60N.17 cells were transfected with 5 μg of MMTV-CAT plasmid with 15 μl of FuGENE6 (Roche) for 24 h. Aliquots of each sample were assayed for CAT and luciferase activities, and activities were normalized to the amount of protein in the lysate. The results were summarized from at least three trials of triplicate transfection; error bars were used to represent standard deviations.
Chromatin remodeling.
The in vivo chromatin hypersensitivity was assessed as described previously (11). In brief, nuclei were isolated after hormone treatment and subjected to limited digestion by SstI (which cleaves within the Nuc-B of MMTV promoter) to measure the hypersensitivity of the specific chromatin locus. Genomic DNA was then purified and completely digested by HaeIII (which cleaves further upstream from the SstI site). The digestion efficiencies of SstI in vivo and HaeIII in vitro are analyzed by reiterative primer extension with a 32P-labeled primer specific for the MMTV-LTR (5′-TCT GGA AAG TGA AGG ATA AGT GAC GA-3′) and Taq polymerase. If the SstI site in Nuc-B is cut in vivo, the extension stops there; otherwise, the stop occurs at the HaeIII site and results in a longer product. The extended products were analyzed on 8% sequencing gels and quantified with a PhosphorImager (Amersham Biosciences). Chromatin remodeling is determined by calculating the in vivo digestion efficiency of SstI as follows: 100% × band intensity of SstI/(SstI + HaeIII).
Immunoprecipitation.
Cells were washed with phosphate-buffered saline, pelleted, and lysed in IP buffer (20 mM HEPES-KOH [pH 7.9], 250 mM NaCl, 10% [vol/vol] glycerol, 0.1% [vol/vol] Tween 20, 0.2 mM EDTA, 2 mM DTT) containing 20 μg of aprotinin/ml and 20 μg of leupeptin/ml by using a micro tissue grinder. After centrifugation at 20,800 × g for 15 min, the supernatants were separated and measured for protein concentration by the Bradford assay. Whole-cell extract (1 mg) was diluted into 0.5 ml of IP buffer containing 1% donkey serum (Sigma) and precleared once with 40 μl of protein G-agarose beads (Sigma) at 4°C for 1 h. Anti-BRG1 antibody (10 μg; N-15, Santa Cruz), was incubated with the lysate at 4°C overnight and then incubated with protein G-agarose for 1 h. After five washes with IP buffer, the immunoprecipitated complexes were eluted with 2× Laemmli SDS-PAGE sample buffer and boiled for 3 min. Coimmunoprecipitated proteins and input whole-cell extract (10%) were detected by Western blotting with antibodies against GR (against rat GR1-325), BRG1 (H88, Santa Cruz), BAF155 (44), and BAF60a (against BAF60a4-64).
RESULTS
The GR interacts with BAF60a and BAF57.
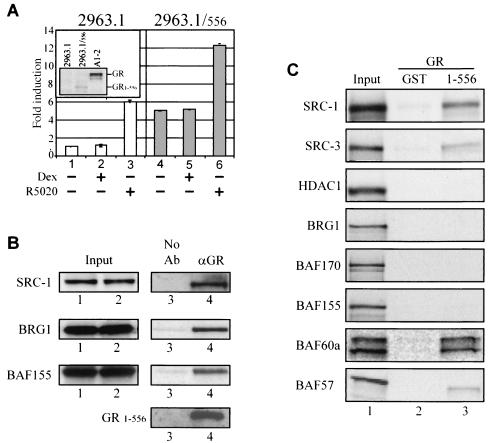
Previous studies on GR-regulated chromatin remodeling in mammalian cells have focused on hormone-dependent pathways (11, 12, 34). However, recent experiments in yeast demonstrated that the GR amino-terminal region could interact with the SWI/SNF complex (42). To explore activity of the AF-1 domain of the GR from a chromatin-assembled promoter in mammalian cells, experiments with the GR truncation mutant GR1-556 that lacks the entire LBD were initiated (18). In initial experiments, the truncated GR1-556 was stably expressed in T-47D/2963.1 cells, which express endogenous PR but not the GR (Fig. 1A, inset). Although transcription from the MMTV-CAT is induced by the synthetic progestin (R5020), the MMTV promoter has been previously shown to adapt a persistently open chromatin structure in T-47D/2963.1 cells (33). In the absence of R5020, expression of GR1-556 raised the MMTV activity to a level similar to that observed with 2963.1 cells in the presence of R5020 (Fig. 1A, lane 4 versus lane 3). Furthermore, the GR1-556 appears to be additive with PR in activating MMTV transcription compared to the parental line (Fig. 1A, lane 6 versus lane 3). As expected for a GR mutant lacking the LBD, addition of glucocorticoid did not elevate the activity of either the parental cell line, which lacks a GR, or the 2963.1/556 subline (Fig. 1A, compare lanes 1 and 2 with lanes 4 and 5).
FIG. 1.
GR1-556 interacts with SRC-1 and BRG1 complex in vivo. (A) 2963.1 cells, which express endogenous PR, and derived 2963.1/556 cells were treated with ethanol (−), dexamethasone (Dex) (10−7 M), or R5020 (10−8 M) for 24 h. The CAT activity of cell lysate was analyzed by kinetic assays and normalized with total protein. The insert shows expression of GR1-556 in 2963.1/556 cells but not in the parental 2963.1 cells or A1-2 cells which express the GRwt. (B) SRC-1 associates with GR1-556 in 2963.1/556 cells. GR1-556-associated complexes were immunoprecipitated from whole-cell extracts of 2963.1/556 cells with no antibody (lane 3) or BUGR2 anti-GR antibody (lane 4) and immunoblotted with antibodies against SRC-1, BRG1, BAF155, and GR (BUGR2). The cognate inputs of SRC-1, BRG1, and BAF155 are shown in lanes 1 and 2 for the no-antibody (No Ab) and anti-GR antibody (αGR) immunoblots, respectively. (C) Interaction of GR1-556 with SRC-1, SRC-3, BRG1, BAF170, BAF155, BAF60a, and BAF57. GST and GR fusion proteins were used to pull down SRC-1, SRC-3, HDAC1, BRG1, and BAF170, 155, 60a, or 57. Input lysate (10%) was loaded as control.
The in vivo interacting partners for GR1-556 were investigated by coimmunoprecipitation assays to understand the mechanisms by which GR1-556 activated the MMTV promoter in an open chromatin environment (33). Consistent with the constitutive activation seen in these cells, the GR1-556 protein interacted with the steroid receptor coactivator SRC-1 in the absence of hormone (Fig. 1B). In addition, we also observed GR1-556 interactions with chromatin-remodeling proteins BRG1 and BAF155 in the absence of hormone (Fig. 1B) (11). This suggests that the GR lacking the entire LBD was capable of a similar spectrum of protein interactions seen previously for the hormone-activated full-length receptor (11).
The direct interactions of GR1-556 with steroid receptor coactivators and the BRG1 complex were then evaluated in vitro by using a GST-fused GR1-556 protein in pull-down assays with in vitro-translated coactivators SRC-1, SRC-3, and individual BAF proteins. In agreement with the in vivo coimmunoprecipitation results (Fig. 1B), the GR1-556 directly interacts with coactivators SRC-1 and SRC-3, but not histone deacetylase protein 1 (HDAC1), which is often found as part of a corepressor complex (Fig. 1C). Similar pull-down conditions were then applied to test whether the GR1-556 interacts with core catalytic BRG1 complex, BRG1, BAF170, and BAF155 (Fig. 1C) (21, 38). Surprisingly, GR1-556 failed to interact with these molecules, suggesting that although these molecules can be detected via coimmunoprecipitation experiments, those interactions are indirect (Fig. 1C) (11, 12). In contrast, BAF60a and, to a lesser extent, BAF57 interact directly with GR1-556 (Fig. 1C, lane 3) (44). When the receptor is further truncated to produce GR1-152 and GR1-325 proteins, these mutants show reduced interaction with BAF60a and BAF57, suggesting that the region between GR residues 326 and 556, which encompasses the DBD and hinge domains, is critical for GR1-556 interaction with BAF60a and BAF57 (data not shown).
BAF60a displays discrete interaction surfaces for the GR and core BRG1 complex.
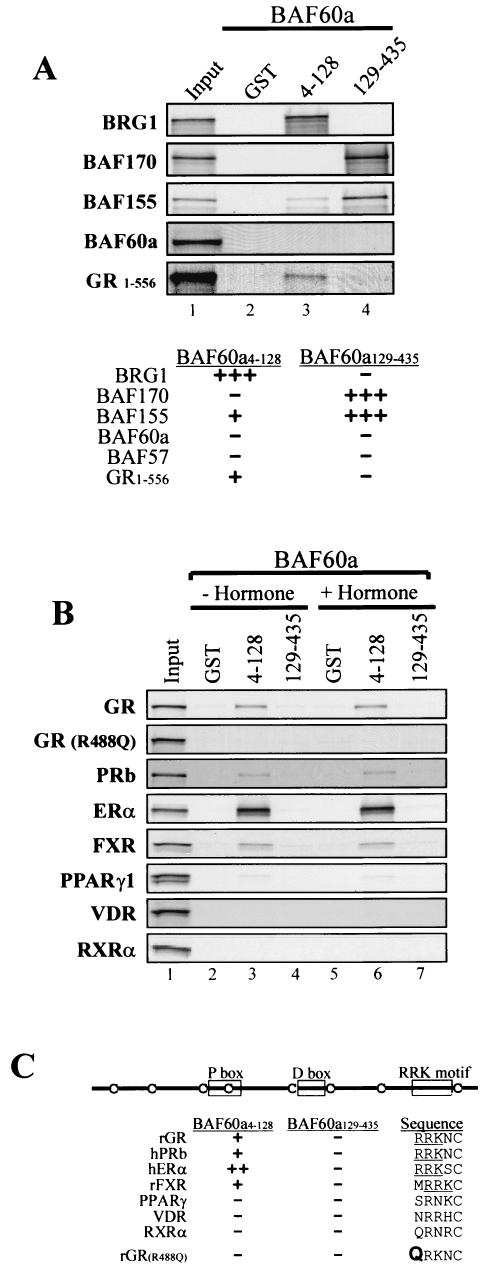
Given that GR1-556 did not interact with the enzymatic core of the BRG1 complex, we next asked if and how the BAF60a communicated between the GR and other members of the complex. The GST-fused BAF60a N-terminal (BAF60a4-128) and C-terminal (BAF60a129-435) domains were tested in pull-down assays with BRG1, BAF170, BAF155, BAF57, and intact BAF60a. The N-terminal BAF60a interacts strongly only with BRG1, interacts to a lesser degree with BAF155, and interacts not at all with BAF170, whereas the C-terminal BAF60a interacts strongly with BAF170 and BAF155 but not with BRG1 (Fig. 2A). Finally, neither the N- nor C-terminal BAF60a interacted with BAF57 (P.-W. Hsiao and T. K. Archer, unpublished data) or BAF60a itself (Fig. 2A). Given the polarity of the interactions of BRG1, BAF170, and BAF155 with BAF60a, we examined the interactions of the BAF60a fragments with GR1-556 and the full-length GR. The results demonstrate that GR1-556 is similar to BRG1 in that it interacts with the N terminus of BAF60a but not the C terminus (Fig. 2A). Significantly, the same specificity of interactions is seen with the intact GR as that seen for GR1-556, and this interaction is not influenced by the addition of hormone (Fig. 2B).
FIG. 2.
BAF60a links steroid hormone receptors and the BRG1 complex. (A) GST and BAF60a fusion proteins were used to pull down BRG1, BAF170, BAF155, BAF60a, and GR1-556, as described in the legend for Fig. 1C. The relative strengths of the interactions are indicated below the autoradiogram. (B) In vitro pull-down assays de-scribed in the legend for panel A were performed with GR, mutant GR(R488Q), PRb, ERα, FXR, PPARγ1, VDR, and RXRα. Hormones were applied at 1 μM dexamethasone for GR and 10 nM R5020 (PRb), 0.1 μM 17β-estradiol (ERα), 10 μM chenodeoxycholic acid (FXR), 10 μM 15d-prostaglandin J2 (PPARγ1), 10 nM EB1089 (VDR), or 10 μM 9-cis retinoic acid (RXRα) and compared to results for the vehicle without hormone (− Hormone). (C) Sequence alignment of the nuclear receptors used in the pull-down assays to illustrate the RRK motif in the DBD.
The GR is a member of the nuclear receptor superfamily, which shares both structural homology and conservation in mechanism of action (28). Thus, like the GR, the BRG1 complex has been implicated in transcriptional activation of the estrogen receptor (ER) (14, 19). In the next set of experiments, we examined if BAF60a might also mediate the interactions of other nuclear receptors with the BRG1 complex. As shown, N-terminal BAF60a also interacts with progesterone receptor b (PRb), ER alpha (ERα), and farnesoid receptor (FXR), and minimally with peroxisome proliferator-activated receptors gamma (PPARγ), suggesting conservation of mechanism with the GR (Fig. 2B). In contrast, neither N- nor C-terminal BAF60a interacts with vitamin D receptor (VDR) or retinoid-X receptor alpha (RXRα), suggesting that interaction with the complex via BAF60a might represent a level of selectivity or specificity among the nuclear receptors (Fig. 2B).
Examination of the receptor DBD amino acid sequences revealed that the receptors that interacted with BAF60a contained an RRK motif located at helix 2 of the DBD (Fig. 2C). The GR, as with most nuclear receptors, has been extensively mutated to define the functions of individual domains (13, 15, 31, 39). Of particular interest was a mutation within the RRK motif of the GR, GR(R488Q), that resulted in loss of activation from a yeast episome without significant reduction of its affinity for DNA or activation on transiently transfected plasmids (39). A prediction of this potential chromatin-specific effect would be a failure of the mutant GR to interact with the remodeling complex via BAF60a. In agreement with our prediction, the GR(R488Q) mutant failed to interact with BAF60a in our assay (Fig. 2B). Together, this result suggests that GR residue 488 resides within an essential surface for GR interaction with the N-terminal BAF60a. Therefore, in the next set of experiments, the GR(R488Q) mutant was inspected for hormone-dependent transcriptional activation and chromatin remodeling in vivo.
Chromatin-specific loss-of-function mutation in the GR.
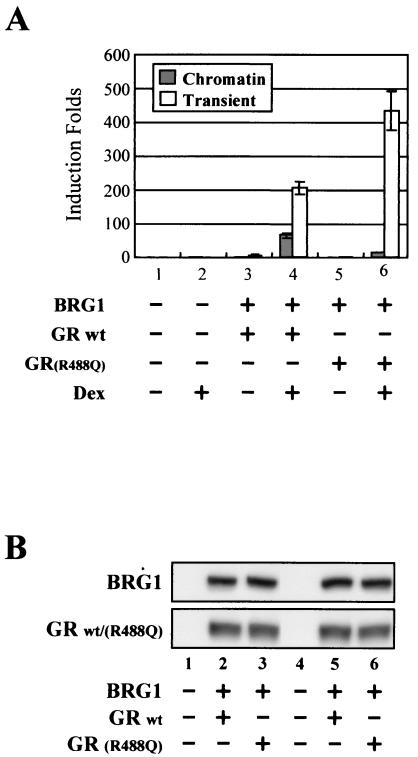
To analyze the chromatin-specific activity of the GR(R488Q) mutant, we chose an SW-13-based cell line that does not express GR but harbors a chromatin-integrated MMTV-luciferase reporter gene (SW-13/M3-2). However, because these cells lack BRG1, chromatin remodeling of Nuc-B as well as hormone-induced luciferase expression in SW-13/M3-2 cells is dependent on the introduction of both GR and BRG1 (Fig. 3A, lanes 1 to 4; Fig. 4A, lane 2 versus lane 4). To compare the transcriptional activation of the wild-type GR (GRwt) and the GR(R488Q) mutant from chromatin versus transiently transfected or nonchromatin templates in SW-13/M3-2 cells, GR plasmids were transfected along with BRG1 and MMTV-CAT reporter plasmid. Hormone treatment of cells expressing GRwt activates MMTV transcription from both chromatin and transient templates in the same cells (Fig. 3A, lane 4 versus lane 3), whereas the GR(R488Q) mutant results in preferential activation from transient template of the same promoter (Fig. 3A, lane 6 versus lane 5). Interestingly, in the presence of both transient and chromatin templates, the GR(R488Q) mutant, while exhibiting reduced activity on the chromatin template, activates transient template to a greater extent than GRwt (Fig. 3A, lane 6 versus lane 4). As shown in Fig. 3B, expression levels of GRwt and the GR(R488Q) mutant in the same transfection were equivalent; therefore, the chromatin-specific loss of transcriptional activity may result from the inability of GR(R488Q) to interact with BAF60a, as seen earlier. Whereas the reduction of activity on the chromatin template is consistent with our hypothesis, the enhanced activity on the transient template was unanticipated and will require further study.
FIG. 3.
GR(R488Q) mutant fails to interact with BAF60a in vitro and is unable to activate transcription from a chromatin template in vivo. (A) GRwt and GR(R488Q) were transfected with BRG1 and MMTV-CAT reporter into SW-13/M3-2 cells. Cells were treated posttransfection with ethanol (−) or dexamethasone (10−7 M) (+) for 24 h. CAT and luciferase activities in the same cell lysate were plotted according to dexamethasone induction. (B) Expression of transfected BRG1, GRwt, and GR(R488Q) was examined by Western blotting.
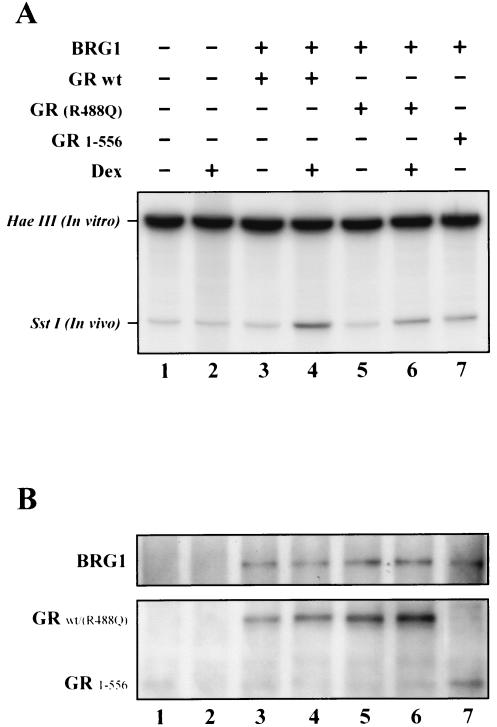
FIG. 4.
Both GR1-556 and the GR(R488Q) mutant exhibit decreased chromatin remodeling. (A) Chromatin remodeling stimulated by GRwt, GR(R488Q), or GR1-556 was examined by SstI hypersensitivity. SW-13/M3-2 cells were transfected with BRG1 and GRwt, GR(R488Q), or GR1-556 for 24 h. Nuclei were isolated from the transfected cells after treatment with dexamethasone (10−7 M) (+) or control (ethanol) (−) and were immediately subjected to limited digestion by SstI to assess hypersensitivity of Nuc-B within the chromatin promoter. (B) The BRG1 complex interaction with GRwt, GR(R488Q), or GR1-556 in transfected SW-13/M3-2 cells was examined by coimmunoprecipitation with BRG1 antibody from 500 μg of whole-cell extract followed by Western blot analysis with antibodies against BRG1 and GR.
GR-mediated chromatin remodeling was analyzed by restriction enzyme hypersensitivity assays in SW-13/M3-2 cells after transfection of GRwt, GR(R488Q), or GR1-556 with BRG1 (Fig. 4). The GRwt induces the hypersensitivity of the Nuc-B upon hormone treatment (Fig. 4A, lane 4 versus lane 3). Consistent with the reduced promoter activation seen earlier, the GR(R488Q) mutant has a reduced activity of chromatin remodeling in response to hormone (Fig. 4A, lane 6 versus lane 5). Similarly, the GR1-556 mutant exhibited reduced chromatin-remodeling activity compared to the GRwt (Fig. 4A, lane 7 versus lane 4). These results imply that GR-dependent chromatin remodeling is compromised by interruption of GR interaction with the N terminus of BAF60a or by losing GR-LBD interaction(s) with the BRG1 complex. We next directly investigated BRG1 complex interactions with the GR mutants by coimmunoprecipitation assays. As the results show, BRG1 interacts with GRwt in addition to GR(R488Q) and GR1-556 mutants (Fig. 4B). These data suggest that interruption of GR interaction with the N terminus of BAF60a reduces chromatin remodeling (Fig. 4A) while maintaining interactions with the entire BRG1 complex (Fig. 4B), presumably via other domains and subunits.
BAF60a mutant inhibits GR-mediated remodeling and transcription from chromatin.
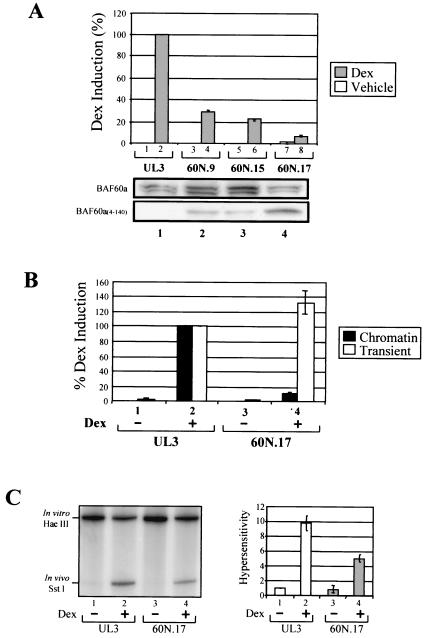
The physiological significance of BAF60a in mediating this network of interactions between the GR and the BRG1 complex was further evaluated by expressing a BAF60a C-terminal truncation mutant (BAF60a4-140) in human osteosarcoma (UL3) cells, which express GRwt and harbor chromatin-integrated MMTV luciferase reporter (12). Given that the BAF60a mutant can interact with the GR and BRG1 but not with BAF170 or BAF155 (Fig. 2), one might predict that the function of the core complex, namely, BRG1, BAF155, and BAF170, might be disrupted. In agreement with this prediction, cell lines expressing various levels of BAF60a4-140 show a clear inhibition of GR activity (Fig. 5A). Indeed, the degree to which GR activity is impaired corresponds to the expression level of BAF60a4-140 in the various clones. This suggests that the BAF60a mutant, lacking the C-terminal domain, acts as a dominant-negative inhibitor of MMTV transcription in vivo.
FIG. 5.
Expression of BAF60a4-140 blocks GR activation. (A) Upper panel, UL3 cells and derived BAF60a4-140-expressing clones 60N.9, 60N.15, and 60N.17 were treated with ethanol (−) or 10−8 M dexamethasone (+) for 24 h. Chromatin-assembled MMTV-luciferase expression was plotted as the percentage of dexamethasone (Dex)-induced activity in UL3 cells. Lower panel, BAF60a and BAF60a4-140 were detected in nuclear extracts from UL3 cells and BAF60a4-140-derived clones by immunoblotting with antibody against BAF60a4-64. (B) Expression of BAF60a4-140 inhibits GR activation of the chromatin template, but not the transient template. An MMTV-CAT plasmid was transfected into UL3 and 60N.17 cells. Cells were treated posttransfection with dexamethasone (10−8 M) (+) or ethanol (−) for 24 h. CAT and luciferase activities in the same cell lysate were plotted using the dexamethasone-induced activity of UL3 cells as 100%. (C) Chromatin structure of the MMTV promoter in UL3 and 60N.17 cells was examined by SstI hypersensitivity after treatment with ethanol (−) or 10−8 M dexamethasone (+) for 1 h. The SstI hypersensitivity of each lane is plotted relative to that of lane 1 from the right panel. Error bars represent standard deviations of three trials.
We next analyzed the mechanistic role of BAF60a4-140 in the glucocorticoid response in vivo by evaluating GR activity from transiently and stably introduced MMTV reporters in the UL3 parental and BAF60a mutant daughter cells, as exemplified by 60N.17 (Fig. 5). Comparison of the parental UL3 cells and 60N.17 cells revealed no effect on GR-activated transcription from a transient template, while transcription from chromatin is inhibited in the same cells (Fig. 5B). Consistent with the reduced gene expression from the chromatin template, GR-activated chromatin remodeling, as determined by restriction enzyme hypersensitivity, is also reduced in the cells expressing BAF60a4-140 (Fig. 5C). Unlike the repressed chromatin, the DNA structure of transiently transfected plasmid is constitutively hypersensitive to restriction enzymes, and chromatin remodeling is not required for activation of a transient template (1). This suggests that while GR-mediated chromatin remodeling functions are impaired, the interaction of the BAF60a4-140 mutant with GR (see below) does not interfere with the transcriptional activity of the receptor.
Mutant BAF60a prevents GR interaction with the BRG1 complex in vivo.
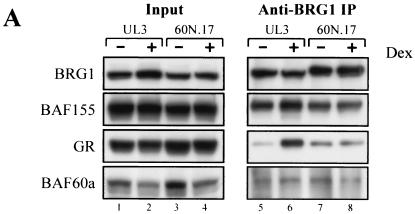
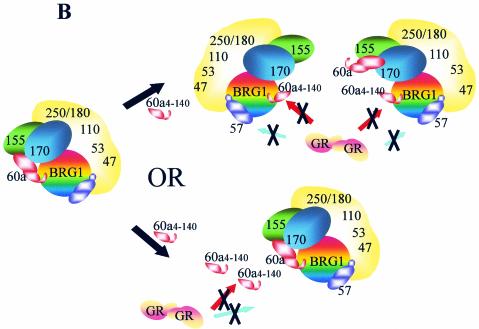
In order to determine how BAF60a4-140 inhibits GR-activated chromatin remodeling, we investigated the protein-protein interactions with the BRG1 complex in both UL3 and 60N.17 cells. The BRG1 complex in UL3 and 60N.17 cells treated with vehicle or hormone was immunoprecipitated with an antibody against BRG1. In agreement with earlier studies, the GR shows a hormone-dependent interaction with BRG1 in UL3 cells such that the GR is enriched in BRG1 complex upon hormone treatment (Fig. 6A, compare lanes 5 and 6). In contrast, hormone treatment does not enrich GR in the BRG1 complex in 60N.17 cells (Fig. 6A, compare lanes 7 and 8). This suggests that expression of BAF60a4-140 may either directly compete BAF60a for the GR or indirectly affect the interaction surface of BRG1 complex in 60N.17 cells. Due to the lack of the BAF60a C terminus, which is required to interact with BAF170 and BAF155, the BAF60a4-140 mutant protein may be integrated into a mutant complex with or without full-length BAF60a (Fig. 6B). While not being able to distinguish these possibilities, in either case, the hormone-dependent GR interaction with the BRG1 remodeling complex is significantly compromised.
FIG. 6.
BAF60a4-140 inhibits GR interaction with the BRG1 complex. (A) GR interaction with the BRG1 complex was examined by coimmunoprecipitation with an anti-BRG1 antibody. Input (100 μg of whole-cell extract) and the coimmunoprecipitated complex (from 1 mg of whole-cell extract) were analyzed by Western blotting with antibodies against BRG1, BAF155, GR, and BAF60a. (B) Models for BAF60a4-140 inhibition of GR interaction with the BRG1 complex. The BAF60a4-140 mutant may integrate into BRG1 complex alone or together with BAF60a to generate a nonfunctional complex. Alternatively, the BAF60a4-140 mutant may directly block the ability of the GR to interact with the complex.
DISCUSSION
Numerous studies have demonstrated the profound role that chromatin structure plays in regulating gene expression (4, 10). The fact that chromatin could prevent transcription factors from accessing cognate binding sites was addressed with the characterization of molecular machines that could alter chromatin structure and change the accessibility of these sites (22, 37). This in turn led to the concept of activator-mediated recruitment of these complexes to promoters, a process particularly well studied with respect to nuclear receptors (11, 19, 32, 41, 42).
The recruitment of the BRG1 complex by the GR is required for hormone-stimulated transcription in mammalian cells (11). Although GR recruitment of the BRG1 complex is hormone dependent, GR1-556, lacking the entire LBD, can interact with the complex in a hormone-independent manner (Fig. 1). Previous in vitro and in vivo experiments suggest the GR-BRG1 complex interaction is accomplished, in part, via a direct interaction between the C terminus of BAF250 and the GR (34). However, there is not an obligatory requirement for BAF250, as GR activates chromatin remodeling and transcription in cells that lack BAF250 (12). To evaluate whether there are other BAFs involved in the underlying mechanisms of the GR-BRG1 complex interaction in mammalian cells, we initiated in vitro pull-down assays with GR1-556 (15).
Although nuclear receptor coactivators were initially identified via hormone-dependent interactions with the receptor LBD (30), a number of coactivators have recently been shown to interact with AF-1 domains. Specifically, it has been shown that SRC-1 interacts with phosphorylated ERβ AF-1 and AR AF1 (27, 40), p300 interacts with AF-1 of ERα and ERβ (23), GRIP1 interacts with AR AF-1 (27), and DRIP150 interacts with GR AF-1 (17). Further, experiments in yeast suggest that sequences in the GR N-terminal domain are sufficient to interact with the yeast SWI/SNF complex (42, 49). Together, these findings are consistent with our observations that GR1-556 recruits coactivators and the BRG1 chromatin-remodeling complex in vivo.
The in vitro assays demonstrated that the GR1-556 interaction with the BRG1 complex is through BAF60a and BAF57 rather than through the core components, namely, BRG1, BAF170, or BAF155, of the complex. They also demonstrate that BAF60a may function as a scaffold or connector for the remodeling complex by linking BRG1 with BAF170 and BAF155. This could have the effect of placing GR in close proximity with the core ATPase activity of the complex. Interestingly, while expression of the BAF60a4-140 is sufficient to block interactions between the GR and the BRG1 complex, it does not appear to significantly affect the interactions between BRG1 and BAF155 (Fig. 6A). This may suggest that the direct interactions demonstrated in vitro between BAF60a C terminus and BAF155 are not essential for the integrity of the BRG1 complex in vivo. Alternatively, this may imply that in the presence of BAF60a4-140, the full-length BAF60a is still able to interact with BAF155 and BAF170, perhaps resulting in an altered BRG1 complex with both BAF60a4-140 and BAF60 (see below) (Fig. 6B).
The observation that BAF60a acts as a bridge between the GR and the BRG1 complex in human cells is consistent with experiments in yeast, where SWP73, the yeast homologue of BAF60a, is required for GR- and GR1-556-activated transcription (6). We observe two lines of evidence that point to the physiological significance of GR and the N-terminal BAF60a interactions. First, the GR(R488Q) mutant, which is unable to interact with the N terminus of BAF60a, has impaired chromatin-remodeling activity and shows deficient transactivation from the chromatin-assembled template (Fig. 3 and 4). Second, expression of N-terminal BAF60a mutant protein disrupts GR interaction with the BRG1 complex, and therefore BAF60a4-140 functions as a dominant-negative mutant (Fig. 5). These results are significant in light of a recent report that shows that BAF57 interacts with ERα, requiring the ER DBD for the association (5). However, while we know that BAF57 also interacts with the GR, the significance of any BAF57-mediated GR/BRG1 complex interactions remains to be investigated. In addition, BAF250 is known to interact with the LBD of the GR (34), suggesting that in vivo, the GR has at least three BAF proteins with which to associate. Therefore, multiple interaction surfaces may be present within the BRG1 complex and may independently be able to interact with the individual domains of GR. Nevertheless, a critical role for the BAF60a/GR interaction is suggested by the ability of the BAF60a4-140 mutant to disrupt the GR-BRG1 interactions and chromatin remodeling (Fig. 5). This result lends support for one of two potential explanations for the ability of BAF60a4-140 to act as a dominant-negative mutant. Coexpression of BAF60a4-140 with endogenous BAF60a might result in a hybrid or heterogeneous BRG1 complex that could interact with the GR either through the endogenous BAF60a or BAF57 and BAF250. The results in Fig. 6A seem to support an alternative view where BAF60a4-140 prevents the interaction with the complex (Fig. 6). Unfortunately, it does not rule out the possibility that the GR fails to interact due to an alteration in the putative hybrid BRG1 complex.
Viewed from the perspective of the GR, it is clear that the interaction of GR with the BRG1 complex in itself is not sufficient to efficiently activate transcription from chromatin templates (Fig. 3). While GR interaction with BRG1 complex was seen in both GR(R488Q) and GR1-556 mutants, a decrease in chromatin remodeling was observed with both mutants (Fig. 4). However, transcriptional activation from a chromatin template is severely impaired, reflecting the diminished level of chromatin remodeling (Fig. 3 and 6). Thus, we suggest that multiple interactions between the receptors and BAF subunits contribute to the activation on chromatin remodeling. The binding data presented demonstrate that both hormone-dependent and -independent interactions are observed between the GR and the BRG1 complex. Previous observations and our current data show that for full-length GR, the interactions with the remodeling complex require hormone in vivo (Fig. 5). In contrast, the truncated receptor that lacks the LBD (GR1-556) exhibits hormone-independent interactions with the complex. Further, both the full-length and truncated receptors show hormone-independent interactions in vitro. Considering both the nature of the interactions with the BRG1 complex and the subcellular localization of the full-length and truncated receptors, we can reconcile the current and previous observations. In vivo, while full-length GR depends on hormone to translocate into the nucleus, GR1-556 migrates to the nucleus independently of hormone (35). In vitro, no hormone-dependent transitions are required for either the GR1-556 or the GRwt to bind, suggesting that the interactions with the BRG1 complex are distinct from the hormone-dependent changes in the LBD structure required for coactivator binding. The extent to which hormone-dependent and -independent interactions contribute to in vivo transcriptional activation from chromatin by other NHRs that are predominantly nuclear, such as the ER and PR, remains to be determined.
The functional significance of the BAF60a-GR interactions is also consistent with previous work that demonstrated that the PR could sequester the BRG1 complex in the presence of anti-progestins (11). Here, we demonstrate that N-terminal BAF60a also interacts with PRb, suggesting that both the PR and GR may compete for binding to this region (Fig. 2B). Moreover, we observed only a subset of nuclear receptors that interact with the N-terminal BAF60a, suggesting it to be a common, but not universal, mechanism of interaction among nuclear receptors. An examination of the sequences from the limited number of receptors assayed suggested that a three-amino-acid RRK motif located in the second zinc finger of the receptor DBD correlated with receptor-BAF60a interactions (Fig. 2C). In support of this idea, a GR mutated at the first R residue, QRK, failed to interact with BAF60a and appeared to be a chromatin-specific mutant with respect to transcription. This suggests that it might be possible to subdivide the nuclear receptor superfamily into BAF60a-dependent and -independent families. In addition, we propose that the flexible helix 2 structure of GR DBD, where the RRK motif is located, is a BAF60a binding surface.
The in vitro binding experiments show that both the GR and BRG1 bind to the same region of BAF60a, namely, N-terminal residues 4 to 128 (Fig. 2). In addition, BAF60a appears to interact with the GR at least in part via the RRK motif in the second zinc finger of the receptor DBD by using its amino terminus, while its C terminus can interact with BAF155 and 170 (Fig. 2). This raises the possibility that BRG1 protein, with its ATPase domain, will be in close proximity to the DBD of the GR and hence adjacent to the hormone response element within the chromatin template. The binding data also suggest a geometric model where BAF60a plays a critical role in allowing the GR to interact with the BRG1 complex by making possible interactions among at least four molecules, namely, BRG1, BAF170, BAF155, and the GR (Fig. 6B).
We have made extensive use of cell lines transfected both stably and transiently with the MMTV promoter to investigate contributions of a defined chromatin structure to GR regulation of transcription. In this light, it is interesting that the impairment of GR interactions with the BRG1 complex by mutation of either the GR, using GR(R488Q), or BAF60a, using BAF60a4-140, leads to opposing effects on the transient and chromatin templates. While we lack an explanation for this divergent response, the rapid recycling of the GR may support a model wherein the GR, prevented from interacting with the BRG1 complex, may be a more potent activator on a template that does not require remodeling. Support for such a view comes from recent experiments that suggest that protein-protein interactions may constrain receptor mobility and localization (29).
Based on the preceding studies, we propose that via different surfaces, the GR interacts with noncore subunits of the BRG1 complex, including BAF250, BAF57, and BAF60a. A direct GR DBD region interaction with N-terminal BAF60a appears to be required to recruit the BRG1 complex and to promote efficient chromatin remodeling. In turn, BAF60a, through N- and C-terminal domains, interlaces BRG1 with BAF170 and BAF155, which forms the core remodeling complex. In addition, GR, via an N-terminal portion, can also recruit coactivators such as SRC-1 and SRC-3 for activating transcription. These interactions do not preclude the well-established role for the LBD in recruiting both chromatin remodeling machines and coactivators but rather suggest an additional and complementary set of interactions. The interactions of BAF60a with BRG1, BAF170, BAF155, and the GR indicate the first evidence for specific geometrical arrangement of BAF subunits within the BRG1 complex and provide insight into the mechanisms by which a subset of nuclear receptors recruits chromatin-remodeling complexes. In this way, the pattern of expression of noncore components of chromatin-remodeling machines may provide a mechanism to regulate the delivery of these multiprotein complexes to specific promoters in vivo.
Acknowledgments
We thank H. Karimi Kinyamu (NIEHS) and Bonnie J. Deroo (NIEHS) for critical reading of the manuscript and Keith R. Yamamoto (UCSF, San Francisco) for providing the GR (R488Q) plasmid and Lise Ballerup (Leo Pharmaceuticals, Denmark) for EB1089.
REFERENCES
- 1.Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. [DOI] [PubMed] [Google Scholar]
- 2.Archer, T. K., E. Zaniewski, M. L. Moyer, and S. K. Nordeen. 1994. The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol. Endocrinol. 8:1154-1162. [DOI] [PubMed] [Google Scholar]
- 3.Arents, G., R. W. Burlingame, B. C. Wang, W. E. Love, and E. N. Moudrianakis. 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88:10148-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, B. R., R. S. Levinson, K. R. Yamamoto, and R. D. Kornberg. 1996. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10:2131-2144. [DOI] [PubMed] [Google Scholar]
- 7.Chawla, A., J. J. Repa, R. M. Evans, and D. J. Mangelsdorf. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866-1870. [DOI] [PubMed] [Google Scholar]
- 8.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 9.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher, T. M., and J. C. Hansen. 1996. The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr. 6:149-188. [DOI] [PubMed] [Google Scholar]
- 11.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 12.Fryer, C. J., H. K. Kinyamu, I. Rogatsky, M. J. Garabedian, and T. K. Archer. 2000. Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J. Biol. Chem. 275:17771-17777. [DOI] [PubMed] [Google Scholar]
- 13.Giguere, V., S. M. Hollenberg, M. G. Rosenfeld, and R. M. Evans. 1986. Functional domains of the human glucocorticoid receptor. Cell 46:645-652. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, D. M., R. Losson, and P. Chambon. 1992. Ligand dependence of estrogen receptor induced changes in chromatin structure. Nucleic Acids Res. 20:4525-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godowski, P. J., S. Rusconi, R. Miesfeld, and K. R. Yamamoto. 1987. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature 325:365-368. [DOI] [PubMed] [Google Scholar]
- 16.Hill, D. A. 2001. Influence of linker histone H1 on chromatin remodeling. Biochem. Cell Biol. 79:317-324. [PubMed] [Google Scholar]
- 17.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsiao, P. W., B. J. Deroo, and T. K. Archer. 2002. Chromatin remodeling and tissue-selective responses of nuclear hormone receptors. Biochem. Cell Biol. 80:343-351. [DOI] [PubMed] [Google Scholar]
- 19.Ichinose, H., J. M. Garnier, P. Chambon, and R. Losson. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95-100. [DOI] [PubMed] [Google Scholar]
- 20.Jones, K. A., and J. T. Kadonaga. 2000. Exploring the transcription-chromatin interface. Genes Dev. 14:1992-1996. [PubMed] [Google Scholar]
- 21.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, Y., T. Kitamoto, Y. Masuhiro, M. Watanabe, T. Kase, D. Metzger, J. Yanagisawa, and S. Kato. 2000. p300 mediates functional synergism between AF-1 and AF-2 of estrogen receptor alpha and beta by interacting directly with the N-terminal A/B domains. J. Biol. Chem. 275:15645-15651. [DOI] [PubMed] [Google Scholar]
- 24.Laurent, B. C., and M. Carlson. 1992. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 6:1707-1715. [DOI] [PubMed] [Google Scholar]
- 25.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 26.Luger, K., and T. J. Richmond. 1998. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8:140-146. [DOI] [PubMed] [Google Scholar]
- 27.Ma, H., H. Hong, S. M. Huang, R. A. Irvine, P. Webb, P. J. Kushner, G. A. Coetzee, and M. R. Stallcup. 1999. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 19:6164-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruvada, P., C. T. Baumann, G. L. Hager, and P. M. Yen. 2003. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J. Biol. Chem. 278:12425-12432. [DOI] [PubMed] [Google Scholar]
- 30.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 31.Miesfeld, R., P. J. Godowski, B. A. Maler, and K. R. Yamamoto. 1987. Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science 236:423-427. [DOI] [PubMed] [Google Scholar]
- 32.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mymryk, J. S., D. Berard, G. L. Hager, and T. K. Archer. 1995. Mouse mammary tumor virus chromatin in human breast cancer cells is constitutively hypersensitive and exhibits steroid hormone-independent loading of transcription factors in vivo. Mol. Cell. Biol. 15:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishi, M., H. Ogawa, T. Ito, K. I. Matsuda, and M. Kawata. 2001. Dynamic changes in subcellular localization of mineralocorticoid receptor in living cells: in comparison with glucocorticoid receptor using dual-color labeling with green fluorescent protein spectral variants. Mol. Endocrinol. 15:1077-1092. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and I. Herskowitz. 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68:573-583. [DOI] [PubMed] [Google Scholar]
- 37.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 38.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 39.Schena, M., L. P. Freedman, and K. R. Yamamoto. 1989. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 3:1590-1601. [DOI] [PubMed] [Google Scholar]
- 40.Tremblay, A., G. B. Tremblay, F. Labrie, and V. Giguere. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell 3:513-519. [DOI] [PubMed] [Google Scholar]
- 41.Urnov, F. D., and A. P. Wolffe. 2001. A necessary good: nuclear hormone receptors and their chromatin templates. Mol. Endocrinol. 15:1-16. [DOI] [PubMed] [Google Scholar]
- 42.Wallberg, A. E., K. E. Neely, A. H. Hassan, J. A. Gustafsson, J. L. Workman, and A. P. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G. R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 46.Wolffe, A. P., and J. C. Hansen. 2001. Nuclear visions: functional flexibility from structural instability. Cell 104:631-634. [DOI] [PubMed] [Google Scholar]
- 47.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 48.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 49.Yoshinaga, S. K., C. L. Peterson, I. Herskowitz, and K. R. Yamamoto. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598-1604. [DOI] [PubMed] [Google Scholar]