Abstract
Type 1 allergic reactions, such as hay fever and allergic asthma, triggered by grass pollen allergens are a global health problem that affects ≈20% of the population in cool, temperate climates. Ryegrass is the dominant source of allergens because of its prodigious production of airborne pollen. Lol p 5 is the major allergenic protein of ryegrass pollen, judging from the fact that almost all of the individuals allergic to grass pollen show presence of serum IgE antibodies against this protein. Moreover, nearly two-thirds of the IgE reactivity of ryegrass pollen has been attributed to this protein. Therefore, it can be expected that down-regulation of Lol p 5 production can significantly reduce the allergic potential of ryegrass pollen. Here, we report down-regulation of Lol p 5 with an antisense construct targeted to the Lol p 5 gene in ryegrass. The expression of antisense RNA was regulated by a pollen-specific promoter. Immunoblot analysis of proteins with allergen-specific antibodies did not detect Lol p 5 in the transgenic pollen. The transgenic pollen showed remarkably reduced allergenicity as reflected by low IgE-binding capacity of pollen extract as compared with that of control pollen. The transgenic ryegrass plants in which Lol p 5 gene expression is perturbed showed normal fertile pollen development, indicating that genetic engineering of hypoallergenic grass plants is possible.
During the flowering period in spring and summer, grasses produce prolific amounts of pollen containing allergenic proteins known to cause allergic reactions such as hay fever and allergic asthma in humans. Grass pollen allergy afflicts up to 20% of the population in cool, temperate climates (1). Of the grasses, ryegrass is the major cause of this disease, because ryegrass produces prodigious amounts of pollen. Molecules in ryegrass pollen that provoke allergenic reactions have been identified and characterized (2–4), and two different proteins, Lol p1 and Lol p 5, have been described as the major allergens (2). Lol p 5, a 31-kDa protein, is less abundant but more allergenic than Lol p 1. More than 90% of patients allergic to grass pollen have IgE recognizing this allergen (2, 4).
Molecular analysis of Lol p 5 has shown that it is rich in alanine (23%) and proline (13%). It contains a putative signal peptide of 25 amino acids, indicating that Lol p 5 is first synthesized as precursor in the cytosol and transported to the amyloplast for posttranslational modifications. Although there has been significant effort in the identification and cloning of group 5 allergens from several grasses (2, 3, 5–7), their biological function in the plant is still not known. Sequence analysis of Lol p 5 indicates that a repeated sequence of Pro-Ala-Thr generally occurs in proteins having a structural function, and Pro and Ala richness is a characteristic of several of the known cell-wall structural proteins (8). Various functions have been proposed for Lol p 5, including roles during pollen development, pollen-tube growth, and pollen-stigma recognition, as well as starch mobilization during pollen germination. However, no experimental proof is available for any of the suggested functions, and it is not known whether normal pollen development can proceed in the absence of this protein.
Studies with antibodies and nucleic acid probes indicated that Lol p 5 is expressed exclusively in pollen (2, 9), and, within the pollen grain, Lol p 5 has been localized in the starch granules (2). When ryegrass pollen comes in contact with water, the pollen grains burst, releasing thousands of these starch granules. These starch granules have been proposed as the micronic fractions or asthma-triggering particles that enter the human airway to induce an IgE-mediated response in asthmatic patients. Because of clinical significance, most of the studies on Lol p 5 have been focused on its diagnostic and therapeutic aspects. On the other hand, it would be desirable to breed plants without this allergy-causing protein. Advances in genetic engineering techniques allow us to introduce genes into plant cells enabling us to create and select plant cultivars that could not be obtained by traditional breeding methods.
In this study, we report the generation of ryegrass devoid of Lol p 5 in its pollen by specifically down-regulating its expression by the antisense RNA approach. The transgenic ryegrass plants in which the Lol p 5 gene activity is perturbed show normal fertile pollen development, thus indicating the feasibility of genetically engineered hypoallergenic ryegrass.
MATERIALS AND METHODS
Plant Materials.
A commercial genotype of Lolium rigidum L. supplied by Valley Seeds (Melbourne, Australia) was used. Seeds were stored at 4°C until used.
Gene Construct and Microprojectile Bombardment.
A pollen-specific promoter, Ory s1, was used to drive the antisense expression of Lol p 5. The 1,507-bp 5′ upstream region of Ory s1 was fused with 0.94 kilobases (nucleotides 1–943; ref. 2) of the Lol p 5 cDNA clone in reverse orientation and cloned into a derivative of the pUC vector. The antisense orientation of Lol p 5 DNA was confirmed by double-stranded sequencing. This vector also carried a plant selectable marker gene, the neomycin phosphotransferase-II (NPT-II) gene. Particle bombardment was performed with a helium-driven DuPont Biolistic Delivery System PDS-1000 (Bio-Rad) at 1,100 psi by using 1.0-μm gold particles following the manufacturer’s instructions.
Regeneration of Transgenic Plants.
Ryegrass seeds were surface sterilized in 13% (vol/vol) sodium hypochlorite with vigorous shaking for 20 min. After rinsing five times in sterile distilled water, seeds were placed onto callus-induction medium (MS medium containing 20 g/liter sucrose, 5 mg/liter 2,4-dichloro phenoxyacetic acid, and 6 g/liter agar; ref. 10; Sigma). The pH of the tissue culture medium used in this study was adjusted to 5.8 before autoclaving at 121°C for 20 min. The culture plates were sealed with 3M surgical tape and were incubated at 25°C with a 16-h day length [General Electric cool white fluorescent tubes produced 45 μE⋅m−2⋅s−1 (1 einstein = 1 E = 1 mol of photons) at the level of the Petri dish]. For bombardment, 4-week-old calli were used. For bombardment, calli were spread out in the middle of a Petri dish containing agar-solidified MS medium. After bombardment, the calli were allowed to recover for 2–3 days at room temperature before they were transferred to a selection medium (callus-induction medium with 25 mg/liter kanamycin and 25 mg/liter geneticin) for 4 weeks. Calli were transferred to regeneration medium (MS medium with 2 mg/liter 6-benzyl aminopurine). Shoots appeared within 6–8 weeks on this medium. Regenerated shoots were rooted on MS medium without hormone. Transformed plants were selected on solidified MS medium (10) without sucrose containing 25 mg/liter kanamycin and 35 mg/liter geneticin, transferred to soil, and grown under greenhouse conditions (18 h of light at 20°C).
Confirmation of Transformation.
Leaf-disc assay. A leaf-disc assay was used as a diagnostic marker for NPT-II expression. For this assay, leaves from green regenerated shoots were removed carefully from root induction medium. By using aseptic techniques, the leaves were cut into small sections approximately 4–5 mm in length. The leaf sections, underside down, were pressed carefully into the surface of agar (0.7%) containing 25 mg/liter kanamycin and 25 mg/liter geneticin. Leaf sections from wild-type tissue-culture regenerated plants were used as controls. The plates were sealed with 3M micropore tape and incubated under dim light at 25°C.
Plant DNA isolation.
Genomic DNA was extracted from leaf tissues by using the cetyltrimethylammonium bromide method of Doyle and Doyle (11). DNA preparations were quantified with a GeneQuant DNA/RNA calculator (Amersham Pharmacia).
PCR analysis.
PCR was performed in a volume of 20 μl containing 50 ng of genomic DNA; 10 mM Tris⋅HCl (pH 8.3); 50 mM KCl; 2 mM MgCl2; 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 25 mM of primer (primer 1, 5′-GAGGCTATTCGGCTATGACTG-3′; primer 2, 5′-GGAGCGGCGATACCGTAAAGC-3′; internal primers of the NPT-II gene designed to amplify the 690-bp DNA fragment), and 1 unit Taq DNA polymerase (GIBCO/BRL) by using a Perkin–Elmer GeneAmp PCR System 2400 thermocycler. The PCR program used had an initial strand separation step at 94°C for 2 min; followed by 35 cycles with denaturation at 94°C for 30 s, annealing at 60°C for 1 min, elongation at 72°C for 2 min; and a final extension step at 72°C for 10 min. The amplification products were separated by electrophoresis on 0.8% agarose gels. Molecular masses were estimated by using HindIII–EcoRI-digested λ-DNA as a standard.
PCR-amplified products from the gel were transferred onto Hybond N+ nylon membrane (Amersham Pharmacia) and hybridized with a 32P-labeled DNA probe specific for the NPT-II gene as described by Sambrook et al. (12).
RNA Isolation.
Total RNA was extracted from pollen of transgenic and wild-type control plants by using the SNAP kit (Invitrogen) following the manufacturer’s instructions. RNA preparations were quantified with a GeneQuant DNA/RNA calculator (Amersham Pharmacia).
RNA Blot Analysis.
Total RNA (5 μg) was electrophoresed and transferred onto Hybond N+ nylon membrane (Amersham Pharmacia). Lol p 5 sense and antisense RNA was detected with [α-32P]UTP-labeled single-stranded RNA probes generated by in vitro transcription from the Lol p 5 cDNA clone.
Pollen Viability Test.
The fluorochromatic reaction was used as a test for pollen viability. For this test, fresh pollen grains from wild-type and transformed plants were incubated in a drop of fluorescein diacetate solution [Sigma; 0.2 mg/ml final concentration in 10% (vol/vol) sucrose] and viewed with an epifluorescence microscope under UV excitation.
Scanning Electron Microscopy.
Pollen grains from dehiscing anthers were collected, fixed in 2% (vol/vol) glutaraldehyde, dehydrated through an ethanol series, and viewed with a scanning electron microscope (Philips Electronic Instruments, Mahwah, NJ).
SDS/PAGE and Protein Blotting.
Total proteins were extracted by grinding mature anthers in PBS (10 mM phosphate buffer, pH 7.2/150 mM NaCl) containing 1 mM PMSF followed by centrifugation (Beckman–Spinco GS-15R) at 14,000 rpm for 10 min. Soluble proteins in the supernatant were quantified by Bio-Rad assay (Bio-Rad). Proteins were separated on SDS/15% PAGE gels by using the Mini-Protean II system (Bio-Rad). The gels were stained with Coomassie brilliant blue R250.
For Western blotting, the SDS/PAGE-separated proteins were transferred onto a nitrocellulose membrane (Schleicher & Schuell). The blots were probed with antibodies essentially as described by Singh et al. (2). Polyclonal antibodies were diluted 1:50 by using 0.5% BSA in PBS; monoclonal antibodies and sera of patients allergic to grass pollen were used at dilutions of 1:5. Binding of polyclonal antibodies was detected with anti-rabbit Ig-conjugated alkaline phosphatase (Promega), whereas binding of monoclonal antibodies was detected with anti-mouse Ig-conjugated alkaline phosphatase (Promega). The color reaction was developed by using 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate as substrates. Binding of human IgE with sera of patients allergic to grass pollen was detected by using 125I-labeled anti-human IgE (Bioclone, Marrickville, Australia).
Immunolocalization.
Mature anthers from transgenic and wild-type plants were fixed in 4% (vol/vol) paraformaldehyde in 50 mM Pipes buffer (pH 8.0) overnight. Samples were dehydrated in a graded ethanol and xylene series and embedded in paraffin. Sections 8-μm thick were attached to poly-l-lysine coated slides, deparaffinized with xylene, and rehydrated through graded ethanol series. Sections were incubated in 5% (vol/vol) skim milk in PBS for 1 h, washed twice with PBS, and incubated overnight with monoclonal antibodies diluted 1:5 with 1% BSA in PBS. Binding of monoclonal antibodies was detected with anti-mouse Ig-conjugated alkaline phosphatase (Promega). The color reaction was developed by using 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate as substrates.
In Vitro Pollination.
In vitro pollination was performed as described by Elgersma et al. (13). Ovaries with pistils were excised and placed upright on agar (0.6%) in Petri dishes. Fresh pollen was dusted on the stigmas. After 2 h at room temperature, pistils were fixed in formalin/acetic acid/alcohol, rinsed in water, and stained with 1% aniline blue. Pistils were gently squashed and examined by fluorescent microscopy.
RESULTS
Transformation and Regeneration of Transgenic Ryegrass.
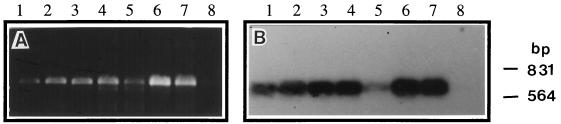
Embryogenic calli induced from seeds of ryegrass were used for plant regeneration. The effect of different concentrations of 2,4-dichloro phenoxyacetic acid for callus induction was evaluated to produce high-quality regenerable calli (data not shown). An efficient regeneration system for production of viable green shoots from a commercial cultivar of ryegrass with 6-benzyl aminopurine was also developed. The GUS reporter gene was used initially to assay transient expression in callus. Microprojectile bombardment conditions were optimized on the basis of the frequency of transient gene expression in callus tissue. For bombardment, 3- to 4-week old calli were found to be ideal targets. For selection of stably transformed tissue and regenerated plants, the concentration of the antibiotic required to inhibit growth and to kill control untransformed calli was determined. Plants were regenerated from callus lines that grew in the presence of selection medium. To avoid escapes, i.e., untransformed shoots, a transformant selection step was introduced in which green regenerated shoots were exposed to a selection medium containing kanamycin and geneticin but without a carbohydrate (sucrose) source. Within 2–4 weeks, most of the untransformed shoots turned white, whereas transformed shoots remained green on this medium and continued to grow. Transfer of regenerated green shoots to transformant selection medium was very effective at screening out false positives. NPT-II expression was assayed on small leaf pieces placed on agar medium containing 25 mg/liter kanamycin and 35 mg/liter geneticin without sucrose. Untransformed leaf pieces do not swell on this medium and turn white within 7 days; leaf pieces from transformed plants expand in size and remain green. PCR amplification of genomic DNA from transgenic plants identified a DNA fragment of the expected 690-bp size (Fig. 1A). This fragment was identical to the NPT-11 gene, as shown by probing with 32P-labeled probe specific to the NPT-11 gene (Fig. 1B). No amplification was observed in untransformed ryegrass. Confirmation of stable integration of DNA in regenerated plants was also done by Southern analysis (data not shown). The plants were transferred successfully to greenhouse conditions.
Figure 1.
(A) PCR analysis of transgenic plants of ryegrass showing amplification of a 690-bp fragment. (B) Southern blot of PCR gel probed with 32P-labeled NPT-II-specific probe. Lanes 1–7, transgenic plant; lane 8, nontransformed ryegrass control.
Analysis of Transgenic Plants Carrying Antisense Lol p 5.
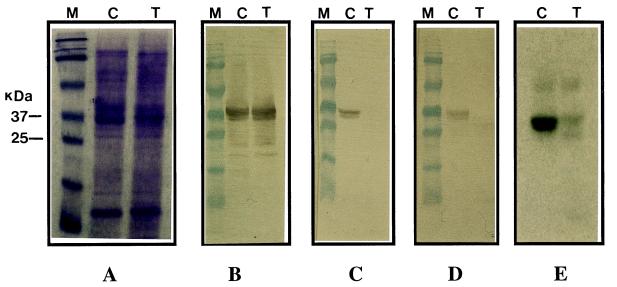
Transgenic plants were analyzed for allergenic protein content by probing the pollen protein blot with Lol p 5-specific monoclonal and polyclonal antibodies. Total protein extracted from mature anthers of transgenic and wild-type plants was analyzed by SDS/PAGE, and a representative result is shown in Fig. 2A. No difference was observed in the overall soluble pollen protein profile of transgenic and wild-type control plants. Use of Lol p 5-specific monoclonal and polyclonal antibodies showed presence of Lol p 5 protein in pollen extract from wild-type plants. In contrast, no such immunoreactive protein could be detected in pollen extract of the transgenic plants (Fig. 2 C and D). Incubation with anti-Lol p 1-specific monoclonal antibody revealed that expression of Lol p 1 was not affected in pollen of transgenic plants (Fig. 2B), indicating specific inhibition of Lol p 5 production.
Figure 2.
Western blot analysis of ryegrass pollen proteins. The position of molecular mass markers are shown at left. In all cases, 10 μg of pollen protein was loaded per lane. (A) SDS/PAGE of total proteins stained with Coomassie brilliant blue R250. (B–E) Western blots probed with anti-Lol p 1 monoclonal antibody (B), anti-Lol p 5 monoclonal antibody (C), anti-Lol p 5 polyclonal antibodies (D), and IgE antibodies from serum of a patient allergic to grass pollen (E). M, mass marker; C, nontransformed control; T, transgenic ryegrass plant.
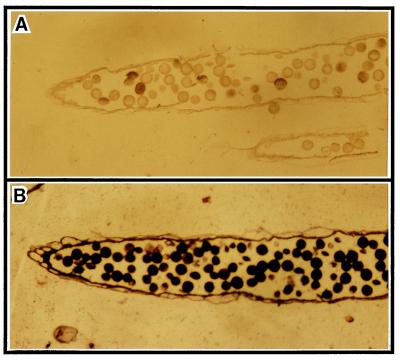
Further immunolocalization studies with anthers of transgenic plants confirmed that Lol p 5 expression was not detectable in pollen, whereas Lol p 1 protein is present (Fig. 3).
Figure 3.
Immunocytochemical analysis of transgenic ryegrass. Sections of mature anthers probed with anti-Lol p 5 monoclonal antibody (A) and anti-Lol p 1 monoclonal antibody (B). Lol p 5 could not be detected in pollen of the transgenic plant, whereas Lol p 1 (used as internal control) remained unchanged.
Northern blot analysis of pollen from transgenic plants showed the presence of the expected ≈950-bp size of the antisense transcript (data not shown), indicating that the repression in allergen synthesis is caused by the expression of the introduced antisense gene.
Evaluation of IgE-Binding Capacity.
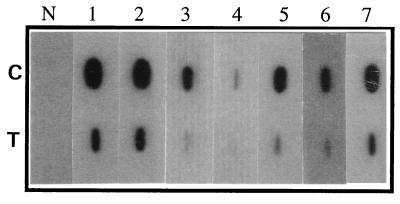
Allergens induce production of IgE antibodies in the serum of susceptible individuals, and allergenicity of a protein can be determined by measuring its IgE-binding capacity (14). To evaluate IgE-binding capacity of the transgenic plants, a western blot of the pollen extract was incubated with the serum of a patient with a grass allergy (Fig. 2E). A significant reduction in IgE-binding capacity of pollen extract from transgenic plants was observed. Further, we used a slot-blot assay to analyze sera from a number of patients with grass pollen allergies for IgE reactivity to the pollen of the transgenic plants. A dramatic reduction in overall IgE reactivity of pollen extract from transgenic plants was observed in all the sera tested (Fig. 4).
Figure 4.
Evaluation of allergic activity of the transgenic pollen as measured by IgE reactivity of pollen extracts in slot-blot analysis. Upper row, untransformed control pollen (C); Lower row, transgenic pollen (T). Lanes 1–7 probed with sera from seven patients allergic to ryegrass pollen; N, serum from a nonallergic patient.
Pollen Viability and Pollination Studies.
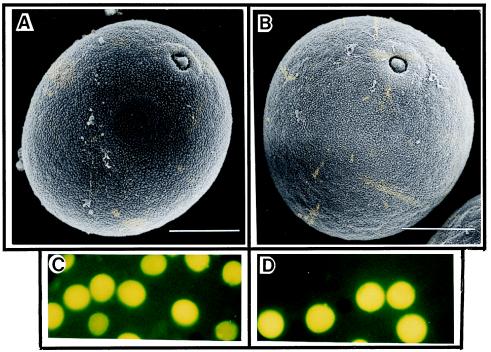
There were no differences observed in the morphology of the transgenic plants, which grew normally and produced inflorescences comparable to wild-type control plants (Fig. 5). A fluorochromatic test of the pollen from transgenic and wild-type control plants showed that pollen viability was more than 95% (Fig. 6 C and D). In addition, scanning electron microscopic observations revealed that pollen of transgenic plants was indistinguishable from wild-type pollen (Fig. 6 A and B).
Figure 5.
Transgenic ryegrass showing normal inflorescence development and anthesis.
Figure 6.
Scanning electron microscopy (A and B) and pollen viability test (C and D) of control and transgenic ryegrass pollen. (A and C) Pollen from control plant. (B and D) Pollen from transgenic plant. No difference between pollen of control and transgenic ryegrass was observed. (A and B, bars = 10 μm.)
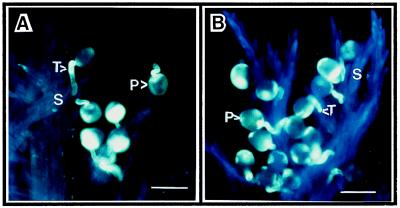
As ryegrass is known to be self-incompatible, pollen from transgenic plants was used to cross-pollinate wild-type stigmas. Fig. 7 shows pollen germination and penetration of the stigmatic tissues, indicating that pollen of the transgenic plants retained its normal function.
Figure 7.
Pollen germination, stigma penetration, and pollen-tube growth of control and transgenic ryegrass pollen. (A) Pollen from control plant. (B) Pollen of transgenic plant. (A and B, bars = 50 μm.) No difference between pollen of control and transgenic ryegrass was observed. S, stigma; P, pollen; T, pollen tube.
DISCUSSION
Grass pollen is the most potent source of allergens worldwide. Lol p 5 or group 5 is among the most important and widespread grass pollen allergen, because it reacts with IgE antibodies of more than 90% of patients allergic to grass pollen, contains most of the grass-pollen-specific IgE epitopes, and elicits strong biological responses. Isolation and characterization of cDNA clones encoding Lol p 5 indicated the presence of a leader peptide and a major processed protein of 31 kDa with pI 9.0 (2). Lol p 5 is synthesized in cytosol as a preallergen, transported during development to amyloplasts, and localized in starch granules (2). These starch granules are released after osmotic rupture from pollen and function as micronic carriers of allergen, which are small enough to penetrate the human airways to trigger IgE-mediated response in asthmatic patients. IgE antibodies play a crucial role in grass pollen allergy through their binding to the cell surface of mast cells and basophils via high-affinity Fc-receptors. Binding of the allergen to specific IgE bound to the membrane of mast cells triggers the release of various inflammatory mediators responsible for the clinical symptoms of such allergenic diseases as hay fever. Significant efforts have been made in developing diagnostic (14–16) and therapeutic reagents (17–22) for designing new and more effective immunotherapeutic strategies for treatment of allergic diseases. An alternative approach to this problem would be to reduce the amount of allergen content in the source plant. Traditional plant breeding and selection methods have not been successful in decreasing or eliminating these proteins. Recent advances in plant genetic engineering have made the introduction of new genes possible in monocot plants, including grasses. The most successful and therefore favored gene-delivery system for monocots involves direct introduction of DNA into intact cells by using high-velocity microprojectile bombardment. We have used this gene-delivery system for introducing antisense construct targeted to the Lol p 5 gene in ryegrass.
Antisense RNA with a complementary sequence of messenger RNA has been used to inhibit gene expression in prokaryotic and eukaryotic organisms (23), and the antisense strategy has also been reported to be practically applicable to transgenic crop plants. Moreover, the antisense approach has been successful in down-regulating an anther-specific gene, Bcp 1, by using a tissue-specific promoter for male sterility studies (24, 25). Prerequisite for successful repression of a gene is the availability of the suitable promoter to drive antisense expression (23, 26). The Ory s1 promoter, originally isolated from rice, has been shown to be pollen specific, and its expression pattern correlates with that of Lol p 5 (27). In this study, we used the Ory s1 promoter to down-regulate Lol p 5 expression in the pollen of ryegrass and report the production of hypoallergenic grass that shows no Lol p 5 production (as indicated by a lack of Lol p 5-specific antibody binding to pollen extract). In addition, our results also suggest that Lol p 5 is not essential for normal pollen development.
The biological role of Lol p 5 in grass pollen is not clear. Though comparison of Lol p 5 with sequence databases revealed no similarity to other proteins of known function, various roles have been suggested for this protein, such as involvement in pollen development and pollen-stigma recognition processes (28, 29). As most grasses are known to be self-incompatible; involvement of Lol p 5 in reactions leading to self-incompatibility has also been proposed. During the present study, self-incompatibility was not overcome in transgenic plant after self-pollination, indicating that Lol p 5 is not involved in this process. In addition, we also did not see any effect on pollen morphology and pollen viability (Fig. 6), suggesting that Lol p 5 may not be essential for pollen development. However, the allergenicity of the ryegrass pollen was remarkably reduced (Fig. 2), as indicated by the extremely low IgE-binding capacity of the pollen from the transgenic plants (Fig. 4).
Our results show that Lol p 5 allergen synthesis in pollen could be suppressed by introducing antisense constructs with microprojectile bombardment without affecting normal pollen development and, therefore, may have potential in breeding hypoallergenic plant cultivars.
Acknowledgments
Our sincere thanks to Mr. Donald Coles of Valley Seeds for support during this study and to Prof. Scott Russell for scanning electron microscopy and critical review of the manuscript. Financial assistance from the Australian Research Council is also gratefully acknowledged.
References
- 1.Wuthrich B. Int Arch Allergy Appl Immunol. 1989;90:3–10. doi: 10.1159/000235067. [DOI] [PubMed] [Google Scholar]
- 2.Singh M B, Hough T, Theerakulpisut P, Avjioglu A, Davies S, Smith P M, Taylor P, Simpson R J, Ward L D, McCluskey J, et al. Proc Natl Acad Sci USA. 1991;88:1384–1388. doi: 10.1073/pnas.88.4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong E K, Griffith I J, Knox R B, Singh M B. Gene. 1993;134:235–240. doi: 10.1016/0378-1119(93)90099-o. [DOI] [PubMed] [Google Scholar]
- 4.Smith, P. M., Ong, E. K., Knox, R. B. & Singh, M. B. (1994) 31, 491–498. [DOI] [PubMed]
- 5.Silvanovich A, Astwood J, Zhang L, Olsen E, Kisil F, Sehon A, Mohapatra S, Hill R. J Biol Chem. 1991;266:1204–1210. [PubMed] [Google Scholar]
- 6.Vrtala S, Sperr W, Reimitzer I, van Ree R, Laffer S, Muller W-D, Valent P, Lechner K, Rumpold H, Kraft D, et al. J Immunol. 1993;151:4773–4781. [PubMed] [Google Scholar]
- 7.Bufe A, Becker W M, Schramm G, Peterson A, Mamat U, Schlaak M. J Allergy Clin Immunol. 1994;94:173–181. doi: 10.1016/0091-6749(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 8.Cassab G I, Varner J E. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:321–353. [Google Scholar]
- 9.Griffith I J, Smith P M, Pollock J, Theerakulpisut P, Avjioglu A, Davies S, Hough T, Singh M B, Simpson R J, Ward L D, et al. FEBS Lett. 1991;279:210–215. doi: 10.1016/0014-5793(91)80151-r. [DOI] [PubMed] [Google Scholar]
- 10.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 11.Doyle J J, Doyle J I. Focus. 1990;12:13–15. [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Elgersma A, Stephenson A G, den Nijs A P M. Sex Plant Reprod. 1989;2:225–230. [Google Scholar]
- 14.Valenta R, Kraft D. Curr Opin Immunol. 1995;7:751–756. doi: 10.1016/0952-7915(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 15.Valenta R, Duchene M, Vrtala S, Birkner T, Ebner C, Hirschwehr R, Breitenbach M, Rumpold H, Scheiner O, Kraft D. J Allergy Clin Immunol. 1991;88:889–894. doi: 10.1016/0091-6749(91)90245-j. [DOI] [PubMed] [Google Scholar]
- 16.Valenta R, Steinberger P, Duchene M, Kraft D. Immunol Cell Biol. 1996;74:187–194. doi: 10.1038/icb.1996.26. [DOI] [PubMed] [Google Scholar]
- 17.Briner T J, Kuo M C, Keating K M, Roger B L, Greenstem J L. Proc Natl Acad Sci USA. 1993;90:7608–7612. doi: 10.1073/pnas.90.16.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norman P S. Curr Opin Immunol. 1993;5:968–973. doi: 10.1016/0952-7915(93)90114-8. [DOI] [PubMed] [Google Scholar]
- 19.Valenta R, Ball T, Vrtala S, Duchene M, Kraft D, Scheiner O. Biochem Biophys Res Commun. 1994;199:106–108. doi: 10.1006/bbrc.1994.1201. [DOI] [PubMed] [Google Scholar]
- 20.Takai T, Yokota T, Yasue M, Nishiyama C, Yuuki T, Mori A, Okudaira H, Okumura Y. Nat Biotechnol. 1997;15:754–758. doi: 10.1038/nbt0897-754. [DOI] [PubMed] [Google Scholar]
- 21.Vrtala S, Hirtenlehen K, Vangelista L, Pastore A, Eichler H-G, Sperr W R, Valent P, Ebner C, Kraft D, Valenta R. Clin Invest. 1997;99:1673–1681. doi: 10.1172/JCI119330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M B, de Weerd N, Bhalla P L. Int Arch Allergy Immunol. 1999;119:75–85. doi: 10.1159/000024181. [DOI] [PubMed] [Google Scholar]
- 23.Stam M, Mol J N M, Kooter J M. Ann Bot. 1997;79:3–12. [Google Scholar]
- 24.Xu H, Knox R B, Taylor P E, Singh M B. Proc Natl Acad Sci USA. 1995;92:2106–2110. doi: 10.1073/pnas.92.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhalla P L, Smith N. Mol Breed. 1998;4:531–541. [Google Scholar]
- 26.Tada Y, Nakase M, Adachi T, Nakamura R, Shimada H, Takayoshi M, Fujimura T, Matsuda T. FEBS Lett. 1996;391:341–345. doi: 10.1016/0014-5793(96)00773-9. [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Goulding N, Zhang Y, Swoboda I, Singh M B, Bhalla P L. Sex Plant Reprod. 1999;12:125–126. [Google Scholar]
- 28.Bufe A, Schramm G, Keown M B, Schlaak M, Becker W M. FEBS Lett. 1995;363:6–12. doi: 10.1016/0014-5793(95)00259-c. [DOI] [PubMed] [Google Scholar]
- 29.Preuss D, Lemineux B, Yen G, Davis R W. Genes Dev. 1993;7:974–985. doi: 10.1101/gad.7.6.974. [DOI] [PubMed] [Google Scholar]