Abstract
In budding yeast, the Elm1 kinase is required for coordination of cell growth and cell division at G2/M. Elm1 is also required for efficient cytokinesis and for regulation of Swe1, the budding yeast homolog of the Wee1 kinase. To further characterize Elm1 function, we engineered an ELM1 allele that can be rapidly and selectively inhibited in vivo. We found that inhibition of Elm1 kinase activity during G2 results in a phenotype similar to the phenotype caused by deletion of the ELM1 gene, as expected. However, inhibition of Elm1 kinase activity earlier in the cell cycle results in a prolonged G1 delay. The G1 requirement for Elm1 kinase activity occurs before bud emergence, polarization of the septins, and synthesis of G1 cyclins. Inhibition of Elm1 kinase activity during early G1 also causes defects in the organization of septins, and inhibition of Elm1 kinase activity in a strain lacking the redundant G1 cyclins CLN1 and CLN2 is lethal. These results demonstrate that the Elm1 kinase plays an important role in G1 events required for bud emergence and septin organization.
Wee1-related kinases delay entry into mitosis by phosphorylating and inhibiting mitotic cyclin-dependent kinases (20, 38). In fission yeast, Wee1 is thought to play a critical role in coordination of cell growth and cell division by delaying entry into mitosis until a critical size has been reached (34, 35). Swe1, the budding yeast homolog of Wee1, is also required for coordination of cell growth and cell division at G2/M, indicating that the basic functions of fission yeast Wee1 have been conserved in budding yeast (24). As in fission yeast wee1− mutants, swe1Δ cells enter mitosis prematurely, leading to the birth of abnormally small daughter cells.
Recent work has identified an intricate signaling network that is required for regulation of Swe1 and for coordination of cell growth and cell division at G2/M (1, 2, 10, 18, 26, 29, 30, 39, 41, 43). This network includes the kinases Elm1, Gin4, Cla4, and Hsl1 as well as a number of proteins required for regulation of these kinases, including Nap1, Cdc42, Hsl7, and the septins. Inactivation of this signaling network can cause cells to undergo a prolonged G2/M delay while polarized cell growth continues, leading to the formation of highly elongated cells that are much larger than normal. Deletion of the SWE1 gene eliminates the G2/M delay and the elongated cell phenotype caused by inactivation of the signaling network, suggesting that the network negatively regulates Swe1 activity to allow entry into mitosis (2, 29, 30, 41). The mechanisms by which the signaling network negatively regulates Swe1 at G2/M are poorly understood. Some studies have concluded that Swe1 is targeted for degradation at G2/M and that components of the signaling network regulate Swe1 stability (31, 32, 40). Other studies, however, have concluded that Swe1 is stable throughout G2/M and that the signaling network regulates the phosphorylation state of Swe1 (24, 41).
Several different kinds of experiments have shown that the Elm1 kinase plays an important role in mitotic events. For example, elm1Δ shows strong genetic interactions with the mitotic cyclins and Elm1 is required for proper localization of the septins, for efficient cytokinesis, and for activation of the Gin4 kinase during mitosis (8, 9, 41). elm1Δ cells undergo a prolonged G2/M delay at the short-spindle stage and become highly elongated. Deletion of the SWE1 gene largely rescues the elongated cell phenotype and mitotic delay observed in elm1Δ cells (9, 18, 41). Furthermore, Elm1 is required for full hyperphosphorylation of Swe1 in vivo (41). Taken together, these observations suggest that Elm1 is a negative regulator of Swe1. However, elm1Δ swe1Δ cells still mislocalize the septins, are inviable at 37°C, and show severe defects in cytokinesis (9, 41). It therefore seems likely that Elm1 carries out functions in addition to the functions required for regulation of Swe1.
Much of our understanding of Elm1 and other proteins required for regulation of Swe1 has come from analysis of the phenotypes caused by gene deletions (1, 2, 10, 18, 26, 29, 30, 39, 41, 43). Although these studies have been informative, analysis of mutant phenotypes caused by gene deletions can be misleading. Cells carrying gene deletions must be grown for many generations before they can be studied, which can make it difficult to distinguish primary versus secondary effects. In addition, accumulation of suppressors and other forms of adaptation can mask phenotypes.
To circumvent the problems of gene deletions, we generated an analog-sensitive version of the Elm1 kinase that can be rapidly and selectively inhibited in vivo by the adenine analog 1 naphthol-methyl PP1 (1NM-PP1) (4, 7). We found that inhibition of Elm1 kinase activity during G2 causes a phenotype similar to the elm1Δ phenotype. Surprisingly, however, inhibition of Elm1 kinase activity early in the cell cycle reveals that Elm1 has G1 functions that were missed in studies with elm1Δ cells. Inhibition of Elm1 kinase activity in early G1 causes prolonged delays in G1 cyclin transcription, bud emergence, and polarization of the septins. In addition, inhibition of Elm1 kinase activity is lethal in cells lacking the G1 cyclins Cln1 and Cln2. These observations show that Elm1 is a member of a growing group of proteins that are required for the normal timing of entry into mitosis and for viability in cln1Δ cln2Δ cells (3, 14, 48).
MATERIALS AND METHODS
Strains and culture conditions.
Except where noted, all cells were grown in yeast extract-peptone-dextrose (YEPD) medium. All strains were in the W303 strain background (leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1). The pGAL-CLN2 plasmid in strain SA49 was a gift of Ray Deshaies. The additional features of these strains are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| AS1 | MATaelm1Δ::TRP1 bar1Δ | 41 |
| AS20 | MATaelm1Δ::URA3 bar1Δ | This study |
| AS79 | MATaelm1-as bar1Δ | This study |
| AS88 | MATaelm1-as CLN2-3XHA::LEU2 bar1Δ | This study |
| DK186 | MATabar1Δ | 1 |
| AS105 | MATacln1Δ::HIS3 cln2Δ::LEU2 elm1-as bar1Δ | This study |
| AS96 | MATacln1Δ::HIS3 cln2Δ::LEU2 bar1Δ | This study |
| SA49 | MATaelm1-as pGAL-CLN2(CEN/URA3) | This study |
Generation of analog-sensitive and catalytically inactive alleles of ELM1.
The ELM1 gene was amplified from yeast genomic DNA (primers: GCGGGATCCTTCTTGAAGTAGCTATTAAG and GCGTCTAGACAATATTTATCGATGAAGTT) and cloned into the BamH1 and XbaI sites of Ycplac111 to yield pDK96B. The analog-sensitive ELM1 mutation was made by mutating pDK96B to change threonine 200 to a glycine (pAS18) (primers: CTGGATAGTCGGTAATTGGTGCA and TGCACCAATTACCGACTATCCAG). To integrate the elm1-as allele into the ELM1 locus, PCR was used to amplify the elm1-as gene, using the same primers used in the initial amplification. Four 100-μl PCR mixtures were pooled and transformed into AS20, an elm1 deletion strain marked with URA3. To select for integration events that replaced the URA3 gene with the elm1-as allele, the transformation was plated onto YEPD, allowed to grow at 30°C for 2 days, and then replica plated onto plates containing 5-fluoro-orotic acid (5-FOA). Transformants that grew on 5-FOA and exhibited wild-type cell morphology were then tested for sensitivity to 1NM-PP1.
Cell cycle arrests, Western blotting, Northern blotting, halo assays, antibodies, and treatment with 1NM-PP1.
Cell cycle arrests with α-factor and Western blotting were carried out as previously described (26). For cell cycle time course experiments, cells were arrested for 3 h with 1 μg of α-factor/ml. Cells were released from the arrest by washing three times with YEPD medium, and 1NM-PP1 was added at the indicated times. The 1NM-PP1 was added from a 12 mM stock in dimethyl sulfoxide (DMSO), and control cells were mock treated with DMSO. The zero time point corresponds to the first wash with YEPD medium. Over 200 cells were scored for each time point when determining the percentages of cells that had undergone bud emergence or septin polarization. For Western blotting, samples were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer containing 2 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 65 mM Tris-HCl (pH 6.8), 3% sodium dodecyl sulfate, 5% β-mercaptoethanol, and 10% glycerol. Halo assays were carried out by pipetting 10 μl of 1 mM 1NM-PP1 onto sterile Whatman 3MM filter disks on a lawn of cells plated onto YEPD plates. Northern blot experiments were carried out as previously described (26). The anti-Elm1 antibody was made by immunizing rabbits with a glutathione S-transferase-Elm1 fusion protein.
For one experiment (see Fig. 5), strain SA49 was grown overnight in synthetic medium lacking uracil and containing 2% glycerol-2% ethanol as a carbon source. Cells at an optical density of 0.5 were then washed in YEPD medium containing 2% glycerol-2% ethanol, and α-factor was added to reach a concentration of 0.5 μg/ml. After 3.5 h at room temperature, cells were washed three times with YEPD medium containing 2% glycerol-2% ethanol and 25 μM 1NM-PP1 was added to half of the cells at 15 min after the first wash.
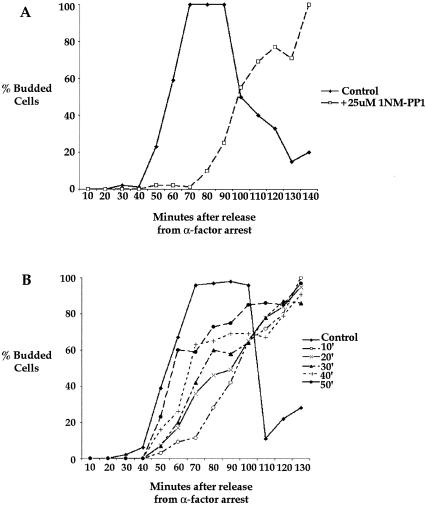
FIG. 5.
Expression of CLN2 from the GAL1 promoter does not rescue the defect in bud emergence caused by inhibition of elm1-as kinase activity. (A) elm1-as cells (strain SA49) that express CLN2 from the GAL1 promoter were released from an α-factor arrest and treated with 25 μM 1NM-PP1 or mock treated with DMSO 15 min after release from the arrest. The percentages of cells undergoing bud emergence were then determined at 15-min intervals and plotted as a function of time. (B) Examples of cells at the 105-min time point in panel A.
RESULTS
Generation of an analog-sensitive elm1 kinase.
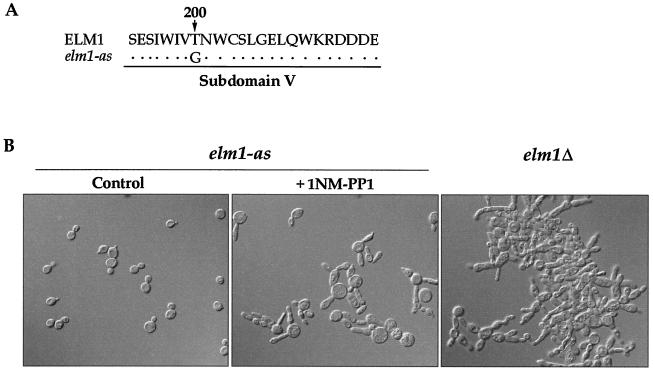
To generate a version of the Elm1 kinase that can be specifically inhibited in vivo, we changed a threonine in subdomain V of the Elm1 kinase domain to glycine (Fig. 1A) (4, 7, 45). Cells carrying the engineered ELM1 kinase displayed none of the morphological defects characteristic of a loss of ELM1 function; however, the cells formed colonies at a slightly slower rate than wild-type cells, suggesting that the mutation has a slight effect on ELM1 function. We refer to this analog-sensitive allele as elm1-as.
FIG. 1.
The phenotype caused by inhibition of elm1-as activity. (A) Threonine 200 in subdomain V was changed to a glycine to generate the elm1-as mutant. (B) Log-phase elm1-as cells (strain AS79) growing at 30°C in YEPD medium were mock treated with DMSO (Control) or treated with 25 μM 1NM-PP1 for 8 h. elm1Δ cells (strain AS1) are shown for comparison.
We treated log phase elm1-as cells with various concentrations of 1NM-PP1, a synthetic adenine-like molecule that specifically inhibits analog-sensitive kinases (5). As a control, we mock treated the elm1-as strain. We found that treatment of cells with 25 μM 1NM-PP1 resulted in an elongated bud phenotype similar to the phenotype caused by elm1Δ (Fig. 1B). Treatment of wild-type cells with the same concentration of inhibitor had no effect (45). Treatment of elm1-as cells with lower concentrations of 1NM-PP1 also produced elongated buds; however, a longer period of time was required for observation of the elongated bud phenotype. Western blotting experiments showed that the Elm1 protein is stable after treatment of elm1-as cells with 1NM-PP1, demonstrating that the mutant phenotype is not due to loss of the Elm1 protein (data not shown). We also treated the elm1-as cells with two other PP1 analogs (2NM-PP1 and 1Na-PP1) and found that 1NM-PP1 is the most potent inhibitor of elm1-as kinase activity (6, 7).
We found that inhibition of elm1-as kinase activity caused a slightly different phenotype than deletion of the ELM1 gene (Fig. 1B). Cells carrying a deletion of the ELM1 gene were elongated and grew as large clumps of interconnected cells. elm1-as cells grown overnight in the presence of 1NM-PP1 were also elongated; however, the cells did not form large clumps. In addition, inhibition of elm1-as kinase activity resulted in the appearance of large rounded cells that were observed at a higher frequency than in elm1Δ cells. The different phenotypes caused by elm1Δ and inhibition of elm1-as kinase activity might reflect the existence of kinase-independent functions of Elm1 that are not affected by inhibition of elm1-as activity. Alternatively, elm1-as may retain a small amount of activity in the presence of 1NM-PP1. To help distinguish these possibilities, we generated a mutation in the catalytic domain of ELM1 by changing the conserved lysine in subdomain II (lysine 117) to an alanine. In many other kinases, this mutation has been shown to produce a catalytically inactive kinase domain (23). We found that cells carrying elm1K117A have the same phenotype as elm1Δ cells, suggesting that the different phenotype caused by inhibition of elm1-as kinase activity may be due to residual kinase activity in the presence of 1NM-PP1 (data not shown).
Deletion of the SWE1 gene largely eliminated the elongated cell phenotype caused by inhibition of elm1-as kinase activity, as previously reported for the elongated cell phenotype caused by elm1Δ (data not shown).
Inhibition of elm1-as kinase activity causes a delay in bud emergence.
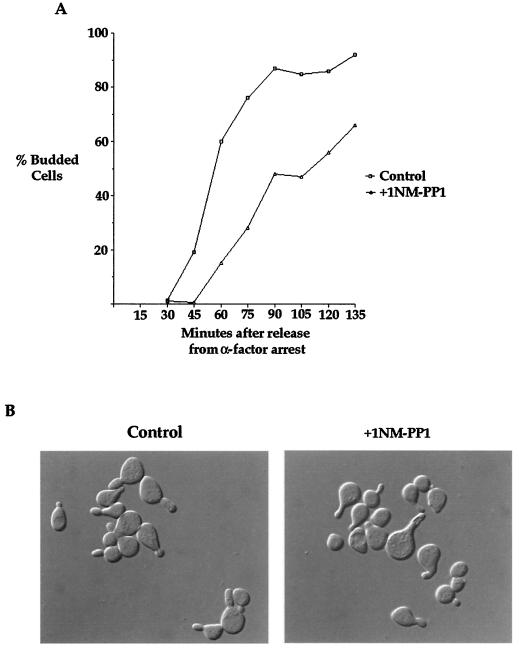
To more carefully characterize the phenotype caused by inhibition of elm1-as kinase activity, we arrested cells in G1 with the mating pheromone α-factor and released them into fresh medium containing 1NM-PP1. We then took samples every 10 min and observed the effects of the presence of 1NM-PP1 upon bud emergence and bud growth. The results of this experiment were striking (Fig. 2A). With the mock-treated control, we found that 50% of the cells were budded by 60 min and nearly 100% of the cells were budded by 70 min. In contrast, the analog-treated cells exhibited a 40-min delay in bud emergence, with 50% of the budding occurring at 100 min and 100% of the budding occurring at 140 min. The delay in bud emergence caused by inhibition of Elm1 kinase activity was unexpected, since previous studies carried out using elm1Δ cells gave no evidence of a budding delay (8, 9, 41).
FIG. 2.
Inhibition of elm1-as kinase activity causes a prolonged delay in bud emergence. (A) elm1-as cells (strain AS79) were released from an α-factor arrest and treated with 25 μM 1NM-PP1 or mock treated at 10 min after release from the arrest. Samples were taken every 10 min for 140 min, and the percentages of budded cells were calculated. (B) elm1-as cells (strain AS79) were released from an α-factor arrest and treated with 25 μM 1NM-PP1 at 10, 20, 30, 40, and 50 min after release from α-factor arrest. Samples were taken every 10 min for 130 min, and the percentages of budded cells were determined.
To determine when Elm1 kinase activity is required for bud emergence, we arrested the elm1-as strain in G1 with α-factor, released the cells from the arrest, and then added 25 μM 1NM-PP1 at 10, 20, 30, 40, and 50 min after release from the arrest. We then determined the percentage of cells with buds at each time point (Fig. 2B). We found that addition of inhibitor up to 40 min after release from α-factor resulted in a significant delay in bud emergence, demonstrating that Elm1 kinase activity is still required for normal timing of bud emergence at least 30 to 40 min after release from α-factor.
Elm1 kinase activity is required for proper organization of the septins during G1.
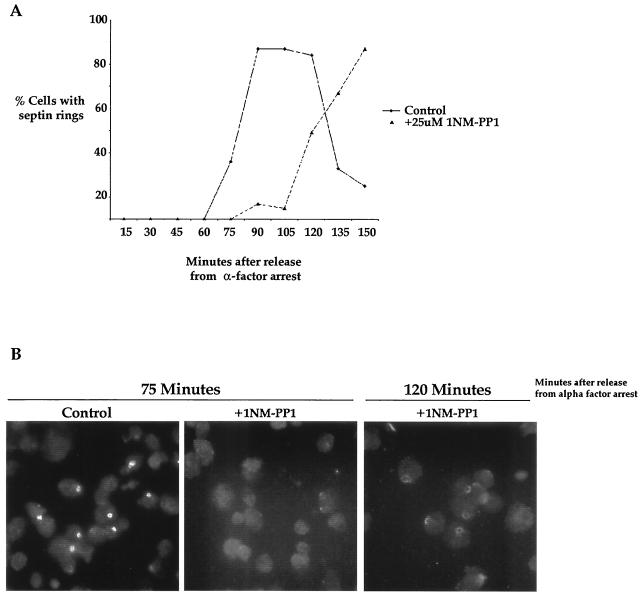
Previous work has shown that bud emergence is preceded by polarization of septin filaments to the site of bud emergence (36). To determine whether the delay in bud emergence is correlated with a delay in polarization of the septins, we treated elm1-as cells with 1NM-PP1 10 min after release from an α-factor arrest and then used antibody staining to determine the percentage of cells with polarized septins at 10-min intervals (Fig. 3A). We observed a delay in polarization of the septins that correlated with the delay in bud emergence, demonstrating that the requirement for Elm1 kinase activity occurs before polarization of the septins. When the septin ring eventually does appear in the inhibitor-treated cells, it is poorly focused and does not stain as intensely as the septin rings in the control cells, indicating that Elm1 kinase activity is required for the proper organization of the septins (Fig. 3B).
FIG. 3.
Inhibition of Elm1 kinase activity causes a delay in septin localization and aberrant septin organization. (A) elm1-as cells were released from an α-factor arrest and treated with 25 μM 1NM-PP1 or mock treated at 10 min after release from the arrest. Samples were taken every 15 min, and the percentages of cells with polarized septin staining were determined after staining with an anti-Cdc11 polyclonal antibody. (B) elm1-as cells were released from an α-factor arrest and treated with 1NM-PP1 as described for Fig. 3A. Examples of the septin staining observed at the indicated times are shown.
Elm1 kinase activity is required before transcription of the G1 cyclin Cln2.
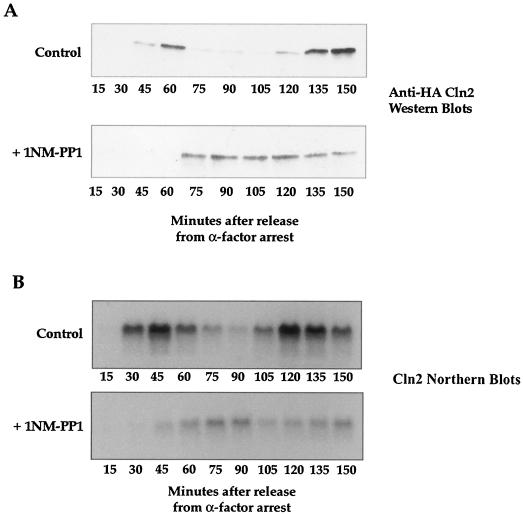
Synthesis of the G1 cyclins is a key event required for bud emergence (12, 22, 37). We therefore determined whether the G1 cyclin Cln2 protein is synthesized with normal kinetics after treatment of elm1-as cells with 1NM-PP1. We made an elm1-as strain that had a 3XHA-tagged Cln2 and then assayed synthesis of the Cln2 protein after release from α-factor arrest and treatment with 1NM-PP1 at 10 min (Fig. 4A). In the mock-treated cells, the synthesis of Cln2 directly correlated with bud emergence, first appearing at 45 min, peaking at 60 min, and then reappearing at 135 min as the cells entered a second cell cycle. In the analog-treated strain, however, we found that the Cln2 protein was not synthesized until 75 min, which correlated with the delay in bud emergence shown in Fig. 2. Cln2 protein levels remained fairly constant throughout the remainder of the time course, which may have been due to the fact that cells treated with 1NM-PP1 undergo bud emergence over a broad time range (Fig. 2B).
FIG. 4.
The requirement for Elm1 kinase activity occurs before transcription of the G1 cyclin CLN2. (A) Western blots showing the behavior of a Cln2-3XHA fusion protein in 1NM-PP1-treated and mock-treated elm1-as cells (strain AS88) as a function of time after release from an α-factor arrest. 1NM-PP1 was added at 10 min after release from the arrest. (B) Cln2 Northern blots from an experiment identical to the one whose results are shown in panel A.
To determine whether the delay in the appearance of the Cln2 protein was due to a transcriptional delay, we performed Northern blot experiments to monitor Cln2 message levels. Again, we released cells from an α-factor arrest and added 1NM-PP1 at 10 min. We found that there was a delay in the synthesis of the Cln2 message that correlated with the delay in the synthesis of the Cln2 cyclin (Fig. 4B). These results show that the requirement for Elm1 kinase activity occurs before transcription of Cln2 during early G1.
Expression of the Cln2 from a conditional promoter does not rescue the bud emergence defect in elm1-as cells.
Since synthesis of Cln2 is thought to play a role in driving bud emergence, the delay in bud emergence observed in elm1-as cells may have been due to the delay in Cln2 synthesis (12, 22, 37). To test this possibility, we generated an elm1-as strain that expresses Cln2 from the GAL1 promoter. The elm1-as GAL1-CLN2 cells were arrested with α-factor in medium containing glycerol and ethanol as a carbon source to allow rapid induction of Cln2 expression upon the addition of galactose. The cells were then released from the arrest into medium containing galactose, and 1NM-PP1 was added to half of the culture at 15 min. Control cells initiated bud emergence at 45 min and then became elongated, as previously reported for cells expressing CLN2 from the GAL1 promoter (Fig. 5A) (27). In the cells treated with 1NM-PP1, buds began to emerge approximately 15 min later. However, it was somewhat difficult to assay bud emergence, because the buds that formed had poorly defined bud necks and in some cases appeared to represent an elongation of the shmoo. Examples of buds in the control and the 1NM-PP1-treated cells at 105 min are shown in Fig. 5B. Thus, expression of Cln2 did not fully rescue the delay in bud emergence caused by inhibition of elm1-as kinase activity. Expression of Cln2 also failed to rescue the defect in septin organization caused by inhibition of elm1-as kinase activity (data not shown).
Inhibition of Elm1 kinase activity after bud emergence results in a prolonged mitotic delay.
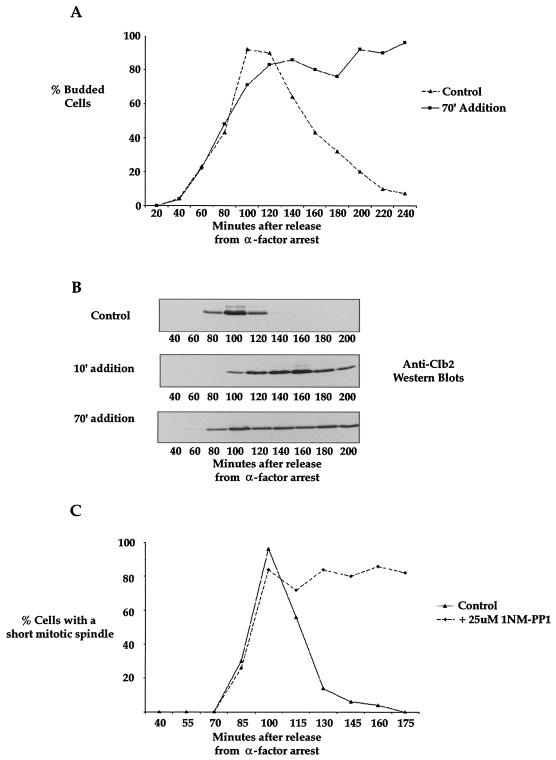
To determine whether inhibition of Elm1 kinase activity later in the cell cycle results in mitotic defects, we added 1NM-PP1 to the elm1-as strain 70 min after release from the α-factor block and then determined the percentage of cells with buds as a function of time. This experiment was performed for a longer period of time to allow growth of elongated buds, and we added α-factor to the culture at 70 min to prevent a second round of bud emergence if the cells exited mitosis. We found that when 1NM-PP1 was added at 70 min, more than 90% of the cells remained arrested with buds at 240 min, suggesting that addition of 1NM-PP1 after bud emergence results in a prolonged mitotic delay (Fig. 6A). At the end of the time course, 37% of the cells had elongated buds. The fact that only 37% of the cells had elongated buds suggests that cells need to go through multiple cell cycles before all of the cells have elongated buds, as observed for cells grown overnight in the presence of 1NM-PP1 (Fig. 1B).
FIG. 6.
Inhibition of Elm1 kinase activity late in the cell cycle causes a mitotic arrest with high Clb2 levels and a short spindle. (A) elm1-as cells (strain AS79) were released from α-factor arrest and treated with 1NM-PP1 70 min after release. Samples were taken every 15 min, and the percentage of budded cells was determined at each time point. (B) elm1-as cells (strain AS79) were released from α-factor arrest and treated with 1NM-PP1 at 10 or 70 min after release. Samples were taken every 20 min, and Western blotting was used to assay levels of the Clb2 mitotic cyclin. (C) elm1-as cells (strain AS79) were released from α-factor arrest and treated with 1NM-PP1 70 min after release. Samples were taken every 15 min, and the percentage of cells with a short mitotic spindle was determined at each time point.
As a further means of assaying mitotic progression, we determined the kinetics of the appearance of the Clb2 mitotic cyclin after treatment of elm1-as cells with 1NM-PP1. We released elm1-as cells from an α-factor arrest and added 1NM-PP1 at 10 min or 70 min after the release. Again, we added α-factor back to the culture at 70 min to ensure that the cells did not enter a second cell cycle. We took samples every 20 min and assayed Clb2 protein levels by Western blotting (Fig. 6B). In the mock-treated control, the Clb2 protein exhibited wild-type kinetics, with protein levels rising at 75 min, peaking at 90 min, and decreasing at 120 min. However, when treated with 1NM-PP1 10 min after release from α-factor arrest, the appearance of the Clb2 protein was delayed and Clb2 protein levels remained high throughout the remainder of the time course. When 1NM-PP1 was added 70 min after release from α-factor arrest, the Clb2 protein appeared with normal kinetics and remained at high levels throughout the remainder of the time course. These results suggest that inhibition of Elm1 kinase activity after bud emergence results in a prolonged mitotic delay with high Clb2 levels.
To determine at what stage of mitosis the elm1-as cells arrest, we added 1NM-PP1 to the cells at 70 min and then took samples every 15 min and stained cells with anti-tubulin antibodies. We found that the inhibition of elm1-as kinase activity after bud emergence causes cells to undergo a prolonged delay at the short-spindle stage of mitosis (Fig. 6C). Previous work has shown that elm1Δ cells also undergo a prolonged delay at the short-spindle stage (41).
Loss of Elm1 function in cln1Δ cln2Δ cells is lethal.
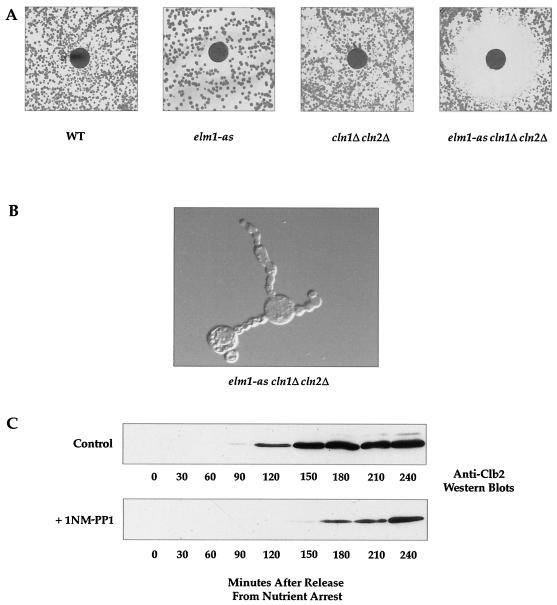
Budding yeast cells carry three G1 cyclins called CLN1, CLN2, and CLN3. Cells from which any two of these cyclin genes have been deleted are viable, whereas deletion of all three is lethal, indicating that the G1 cyclins are functionally redundant (11, 47). Previous work found that loss of function of Cla4, Gin4, Nap1, or the septins is lethal in a cln1Δ cln2Δ background, suggesting that these proteins have a role in G1 events (3, 14, 48). Since Elm1 is required for G1 events and Elm1, Cla4, Gin4, Nap1, and the septins all appear to function in the same signaling network, we wanted to determine whether ELM1 shows genetic interactions with CLN1 and CLN2. We crossed an elm1Δ strain with a cln1Δ cln2Δ strain, but we were unable to recover a viable spore containing the triple elmΔ cln1Δ cln2Δ deletion, suggesting that elm1Δ is synthetically lethal with cln1Δ cln2Δ. To further investigate synthetic lethality with the G1 cyclins, we generated a strain harboring the elm1-as allele in a cln1Δ cln2Δ background. We then performed 1NM-PP1 halo assays with wild-type, elm1-as, cln1Δ cln2Δ, and elm1-as cln1Δ cln2Δ strains. We found that 1NM-PP1 blocked the growth of the elm1-as cln1Δ cln2Δ strain but not that of the other strains, demonstrating that inhibition of Elm1 kinase activity in cln1Δ cln2Δ cells is lethal (Fig. 7A).
FIG. 7.
Inhibition of Elm1 kinase activity in elm1-asΔ cln1 cln2 cells is lethal. (A) The indicated strains were grown to saturation, plated onto YEPD, and analyzed for sensitivity to 1NM-PP1 by a halo assay (strains DK186, AS79, AS96, and AS105). WT, wild type. (B) The elm1-as cln1Δ cln2Δ strain (AS105) was nutrient arrested and released onto a YEPD plate containing 40 μM 1NM-PP1 for 24 h. Cells were removed from the plate with a toothpick and photographed with Nomarski optics. (C) Western blots showing the behavior of the Clb2 protein in elm1-as cln1Δ cln2Δ cells (strain AS105) as a function of time after release from nutrient arrest in the presence of 25 μM 1NM-PP1.
We next determined whether the elm1-as cln1Δ cln2Δ cells arrest at a specific point in the cell cycle when Elm1 kinase activity is inhibited. We synchronized the elm1-as cln1Δ cln2Δ strain in G0 by nutrient arrest. We then plated the nutrient-arrested cells onto a YEPD plate containing 40 μM 1NM-PP1 or onto a control plate. Using a micromanipulator needle, we picked 40 individual cells per plate and observed their growth every 3 h for 12 h and then at 24 and 48 h. By 6 h all of the control cells had divided once, and by 24 h a small colony was produced. In contrast, only 21 of 40 of the elm1-as cln1Δ cln2Δ cells underwent bud emergence by 9 h, indicating a long delay in bud emergence. The buds that emerged became highly elongated and had multiple constrictions along their length (Fig. 6B). By 24 h 30 of 40 of the cells had either 1 or 2 highly elongated buds and by 48 h all of the cells on the 1NM-PP1 plate had an elongated bud phenotype, and manipulation of the cells with a microneedle suggested that none of the cells had undergone cytokinesis.
To determine whether the elm1-as cln1Δ cln2Δ cells are able to exit G1 and enter mitosis when Elm1 kinase activity is inhibited, we released the elm1-as cln1Δ cln2Δ cells from a nutrient arrest, placed them into liquid medium containing 25 μM 1NM-PP1, and then took samples every 30 min and used Western blotting to probe for the presence of the Clb2 mitotic cyclin. We found that consistent with a prolonged delay in G1, synthesis of the Clb2 protein was delayed relative to that of a mock-treated control (Fig. 6C). The fact that the Clb2 cyclin was synthesized suggests, however, that the cells were able to proceed into mitosis. To further define the arrest point of the elm1-as cln1Δ cln2Δ cells, we determined whether the cells were able to undergo nuclear division after inhibition of elm1-as kinase activity. We released the elm1-as cln1Δ cln2Δ cells from a nutrient arrest in liquid culture, added 1NM-PP1, and took samples every hour for 6 h. Nuclear division was observed in the mock-treated control by 3 h; however, the cells treated with 1NM-PP1 became arrested with a single nucleus located at the bud neck up to 6 h after release from the nutrient arrest. We attempted to perform tubulin staining to further assess where the elm1-as cln1Δ cln2Δ cells arrest but found that they exhibited highly aberrant microtubule organization with numerous elongated microtubules, and it was therefore not possible to use spindle staining as a cell cycle marker.
DISCUSSION
Elm1 is required for G1 events.
A number of observations have suggested that Elm1 carries out functions during G2 and mitosis. elm1Δ cells undergo a prolonged delay at the short-spindle stage of mitosis (41). The delay is eliminated in swe1Δ cells and Elm1 is required for the full hyperphosphorylation of Swe1 during mitosis, consistent with the idea that Elm1 is required for regulation of Swe1 (18, 41). Elm1 is also required for hyperphosphorylation of Gin4 during mitosis, and elm1Δ shows strong genetic interactions with the mitotic cyclins (41). Finally, elm1Δ cells show defects in cytokinesis that are strongly enhanced by swe1Δ (9, 41).
In this study, however, we used specific inhibition of elm1-as kinase activity to show that Elm1 is also required for G1 events. Inhibition of elm1-as kinase activity early in G1 causes prolonged delays in bud emergence, septin polarization, and Cln2 synthesis. Furthermore, elm1Δ causes synthetic lethality in cln1Δ cln2Δ cells and inhibition of Elm1 kinase activity in cln1Δ cln2Δ cells is lethal. Specific inhibition of Elm1 kinase activity later in the cell cycle reveals a requirement for Elm1 during G2/M, consistent with the phenotype of elm1Δ cells (8, 9, 41).
In previous work we found that elm1Δ cells do not appear to undergo a delay in G1, since the Clb2 cyclin appears with normal timing after release from an α-factor arrest, and the majority of elm1Δ cells have a G2 DNA content (41). G1 progression in elm1Δ cells cannot be determined by the timing of bud emergence due to the highly aberrant morphology of these cells. The apparent lack of a G1 delay in elm1Δ cells may be due to the accumulation of suppressors in elm1Δ cells that have been grown for many generations. Another possibility is that regulatory mechanisms eliminate the G1 delay once cells have developed the highly aberrant morphology characteristic of elm1Δ cells. For example, one might imagine that the G1 delay caused by inhibition of elm1-as kinase activity is due to a defect in cell growth and that the delay is no longer observed once cells have become highly elongated and have undergone defective cytokinesis due to loss of the mitotic functions of Elm1.
Elm1 kinase activity is required for septin organization during G1.
Inhibition of elm1-as kinase activity causes delayed localization of the septins to the site of bud emergence and a failure to form a tight septin ring. The fact that septins localize to the site of bud emergence during G1 is somewhat puzzling, because there are no reports that the septins play a role in bud emergence and analysis of temperature-sensitive septin mutants has suggested that the septins execute their functions later in the cell cycle (19, 21, 28). It therefore seems unlikely that the delay in bud emergence is due to a delay in localization of the septins. It seems more likely that both bud emergence and septin organization during G1 are dependent upon signals from the Elm1 kinase.
Possible functions of Elm1 during G1 and G2/M.
Inhibition of elm1-as kinase activity causes a delay in CLN2 transcription and is lethal in cln1Δ cln2Δ cells. Transcription of CLN2 in wild-type cells is thought to be triggered by CLN3, although the mechanisms by which it does so are poorly understood (13, 17, 42, 44). In the absence of CLN3, transcription of CLN1 and CLN2 is delayed but is eventually triggered by an alternative pathway that appears to work through Bck2 (16, 46). CLN3 is essential for viability in cln1Δ cln2Δ cells (11). Taken together, these observations are consistent with a model in which Elm1 is required for Cln3 function. In this model, Elm1 might be involved either in activating Cln3/Cdc28 complexes or in a pathway that mediates the functions of activated Cln3/Cdc28 complexes.
Elm1 is a member of an intriguing group of proteins that are required both for Swe1 regulation and for viability in cln1Δ cln2Δ cells. Other members of this group include Cla4, Gin4, Bud2, Nap1, and a member of the septin family called Cdc12 (Gin4 is allelic to the Cla6 gene identified in reference 14) (3, 15, 48). The requirement for Elm1, Cla4, Gin4, Nap1, and Cdc12 in cln1Δ cln2Δ cells is poorly understood. The genetic interactions with G1 cyclins suggest that these proteins carry out G1 functions, yet a number of observations suggest that they carry out functions during G2/M. Gin4, for example, undergoes hyperphosphorylation and activation during mitosis, which leads to assembly of a complex that includes Gin4, Nap1, and the septins (1, 33). Within the complex, Gin4 appears to directly phosphorylate the Shs1 septin during mitosis. Furthermore, Nap1 binds to the Clb2 mitotic cyclin and Nap1, Elm1, and Cla4 are required for hyperphosphorylation of Gin4 during mitosis (1, 25, 43). Finally, Elm1, Cla4, Gin4, Nap1, and a member of the septin family show strong genetic interactions with the mitotic cyclins: the phenotype caused by deletion of any of these genes is strongly enhanced in clb1Δ clb3Δ clb4Δ cells, which are dependent upon the CLB2 cyclin for survival (1, 10, 26, 41, 43). In the case of ELM1 and CLA4, loss of function of these genes in clb1Δ clb3Δ clb4Δ cells is nearly lethal.
It has been unclear whether these proteins carry out separable functions at multiple times during the cell cycle or whether they carry out a single function during G1 that is required later in the cell cycle. For example, it is possible that proteins like Elm1 function only once during G1 for completion of an event that is required later in the cell cycle. If successful completion of this event were monitored by Swe1, then failure to complete the event would lead to a Swe1-dependent G2/M delay. In this study, however, the ability to rapidly and specifically inhibit Elm1 kinase activity at different times during the cell cycle allowed us to show that Elm1 is likely to be required early in G1 and again at G2/M. Since loss of function of CLA4, GIN4, and the septins causes phenotypes similar to those of elm1Δ, these proteins may also be required at multiple times during the cell cycle. Recent work has shown that inhibition of an analog-sensitive cla4 allele causes a delay in bud emergence in swe1Δ cells, consistent with the idea that Cla4 also carried out G1 functions (45).
There are a number of models that might account for the functions of Elm1, Nap1, Cla4, Gin4, and the septins. The fact that Elm1 is required for timely bud emergence and that Elm1, Cla4, Gin4, Nap1, and the septins are required for regulation of Swe1 and for timely entry into mitosis suggests the interesting possibility that these proteins are part of a signaling network that decides when passage through key cell cycle stages is to be triggered. For example, proteins like Elm1 might assess external conditions such as nutrient availability, or internal conditions such as cell size, and then send a signal to trigger passage through the cell cycle when conditions are appropriate. At G2/M, these proteins appear to inactivate Swe1 to trigger entry into mitosis, but their targets during G1 are unknown. An alternative possibility is that the proteins control events such as septin organization or cell growth that are required both for entry into mitosis and for bud emergence. These two possibilities are not mutually exclusive. For example, it is possible that there are proteins that both control cell growth and monitor cell size or nutrient availability.
The discovery that Elm1 plays a role in G1 events is of particular interest, because the signaling pathways that control G1 events are poorly understood in all eukaryotic cells. A better understanding of the molecular functions of the Elm1 kinase during G1 and G2/M will require identification of proteins that associate with Elm1 as well as targets of Elm1 kinase activity.
Acknowledgments
We thank Eric Weiss, David Drubin, Jeff Ubersax, and David Morgan for helpful advice and/or sharing unpublished results. We also thank Stephanie Anastasia for help with experiments.
This work was supported by the Pew Biomedical Scholars program and by a grant from the National Institutes of Health.
REFERENCES
- 1.Altman, R., and D. R. Kellogg. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benton, B. K., A. H. Tinkelenberg, D. Jean, S. D. Plump, and F. R. Cross. 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12:5267-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, A., O. Buzko, S. Heyeck-Dumas, I. Jung, B. Kraybill, Y. Liu, K. Shah, S. Ulrich, L. Witucki, F. Yang, C. Zhang, and K. Shokat. 2000. Unnatural ligands for engineered proteins: new tools for chemical genetics. Annu. Rev. Biophys. Biomol. Struct. 29:577-606. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, A. C., C.-Y. Kung, K. Shah, L. Witucki, K. M. Shokat, and Y. Liu. 1999. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. J. Am. Chem. Soc. 121:627-631. [Google Scholar]
- 6.Bishop, A. C., K. Shah, Y. Liu, L. Witucki, C. Kung, and K. M. Shokat. 1998. Design of allele-specific inhibitors to probe protein kinase signaling. Curr. Biol. 8:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. [DOI] [PubMed] [Google Scholar]
- 8.Blacketer, M. J., C. M. Koehler, S. G. Coats, A. M. Myers, and P. Madaule. 1993. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 13:5567-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouquin, N., Y. Barral, R. Courbyrette, M. Blondel, M. Snyder, and C. Mann. 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 113:1435-1445. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, C., R. Altman, D. Schieltz, J. Yates, and D. R. Kellogg. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 143:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross, F. 1990. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signalling pathway. Mol. Cell. Biol. 10:6482-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, F. 1995. Starting the cell cycle: what's the point? Curr. Opin. Cell Biol. 7:790-797. [DOI] [PubMed] [Google Scholar]
- 13.Cross, F. R., and A. H. Tinkelenberg. 1991. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65:875-883. [DOI] [PubMed] [Google Scholar]
- 14.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 15.Cvrckova, F., and K. Nasmyth. 1993. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 12:5277-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Como, C. J., H. Chang, and K. T. Arndt. 1995. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol. Cell. Biol. 15:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgington, N. P., M. J. Blacketer, T. A. Bierwagen, and A. M. Myers. 1999. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford, S. K., and J. R. Pringle. 1991. Cellular morphogenesis in the S. cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12:281-292. [DOI] [PubMed] [Google Scholar]
- 20.Gould, K. L., and P. Nurse. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342:39-45. [DOI] [PubMed] [Google Scholar]
- 21.Haarer, B. K., and J. R. Pringle. 1987. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol. Cell. Biol. 7:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadwiger, J. A., C. Wittenberg, H. E. Richardson, M. de Barros-Lopes, and S. I. Reed. 1989. A novel family of cyclin homologs that control G1 in yeast. Proc. Natl. Acad. Sci. USA 86:6255-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanks, S. K., and A. M. Quinn. 1991. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200:38-62. [DOI] [PubMed] [Google Scholar]
- 24.Harvey, S. L., and D. R. Kellogg. 2003. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 13:264-275. [DOI] [PubMed] [Google Scholar]
- 25.Kellogg, D. R., A. Kikuchi, T. Fujii-Nakata, C. W. Turck, and A. Murray. 1995. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130:661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellogg, D. R., and A. W. Murray. 1995. NAP1 acts with Clb2 to perform mitotic functions and suppress polar bud growth in budding yeast. J. Cell Biol. 130:675-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew, D. J., and S. I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares, C. De Virgilio, and J. R. Pringle. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106-119. [DOI] [PubMed] [Google Scholar]
- 29.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, J. X., Q. Lu, and M. Grunstein. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10:1327-1340. [DOI] [PubMed] [Google Scholar]
- 31.McMillan, J. N., M. S. Longtine, R. A. L. Sia, C. L. Theesfeld, E. S. G. Bardes, J. R. Pringle, and D. J. Lew. 1999. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19:6929-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan, J. N., R. A. L. Sia, and D. J. Lew. 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortensen, E., H. McDonald, J. Yates, and D. R. Kellogg. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13:2091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norbury, C., and P. Nurse. 1992. Animal cell cycles and their control. Annu. Rev. Biochem. 61:441-470. [DOI] [PubMed] [Google Scholar]
- 35.Nurse, P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344:503-508. [DOI] [PubMed] [Google Scholar]
- 36.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 37.Richardson, H. E., C. W. Wittenberg, F. Cross, and S. I. Reed. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59:1127-1133. [DOI] [PubMed] [Google Scholar]
- 38.Russell, P., and P. Nurse. 1987. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49:559-567. [DOI] [PubMed] [Google Scholar]
- 39.Shulewitz, M. J., C. J. Inouye, and J. Thorner. 1999. Hsl7 localizes to a septin ring and serves as an adapter in regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sia, R. A. L., E. S. G. Bardes, and D. J. Lew. 1998. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 17:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreenivasan, A., and D. Kellogg. 1999. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol. Cell. Biol. 19:7983-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuart, D., and C. Wittenberg. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cyclin cells. Genes Dev. 9:2780-2794. [DOI] [PubMed] [Google Scholar]
- 43.Tjandra, H., J. Compton, and D. R. Kellogg. 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr. Biol. 8:991-1000. [DOI] [PubMed] [Google Scholar]
- 44.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss, E. L., A. C. Bishop, K. M. Shokat, and D. G. Drubin. 2000. Chemical genetic analysis of the budding yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 46.Wijnen, H., and B. Futcher. 1999. Genetic analysis of the shared role of CLN3 and BCK2 at the G1-S transition in Saccharomyces cerevisiae. Genetics 153:1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wittenberg, C., K. Sugimoto, and S. I. Reed. 1990. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell 62:225-237. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerman, Z., and D. R. Kellogg. 2001. The Sda1 protein is required for passage through Start. Mol. Biol. Cell 12:201-219. [DOI] [PMC free article] [PubMed] [Google Scholar]