Abstract
Transcriptional activation of the heat shock genes during the heat shock response in Drosophila has been intimately linked to phosphorylation of histone H3 at serine 10, whereas repression of non-heat-shock genes correlates with dephosphorylation of histone H3. It is then possible that specific kinase and/or phosphatase activities may regulate histone phosphorylation and therefore transcription activation and repression, respectively. We find that treatment of cells with strong phosphatase inhibitors interferes with the genome-wide dephosphorylation of histone H3 normally observed at non-heat-shock genes during heat shock. Mutants in protein phosphatase type 2A (PP2A) also display reduced genome-wide H3 dephosphorylation, and sites of H3 phosphorylation that do not contain heat shock genes remain transcriptionally active during heat shock in PP2A mutants. Finally, the SET protein, a potent and highly selective inhibitor of PP2A activity that inhibits PP2A-mediated dephosphorylation of Ser10-phosphorylated H3, is detected at transcriptionally active regions of polytene chromosomes. These results suggest that activation and repression of gene expression during heat shock might be regulated by changes in PP2A activity controlled by the SET protein.
Covalent modifications of the N-terminal tail domains of the core histones within the chromatin fiber have been proposed to act as signals from the genome to the cellular machinery for the regulation of various processes including transcription, mitotic segregation, and gene silencing (19). Phosphorylation of the Ser10 residue of the N-terminal arm of histone H3 is a covalent posttranslational histone modification that is linked to chromosome condensation and segregation during mitosis (14). However, increases in the total amount of phosphorylated H3 during treatment of quiescent cells with growth factors and phorbol esters (7), as well as the association of the phosphorylated histone isoform with immediate-early gene promoter regions during epidermal growth factor-mediated gene activation, have identified Ser10 phosphorylation as a modification possibly associated with the regulation of transcription (42, 43). Results from other experiments have also recognized H3 phosphorylation as a crucial step in the process of GCN5 histone acetyltransferase binding and subsequent acetylation of histones at epidermal growth factor-regulated genes during transcription initiation (9).
The heat shock response of Drosophila melanogaster is an ideal system for studying the processes of transcription activation and repression (6, 35). During heat shock, the transcription and translation of gene products cease, accompanied by the extremely rapid induction of the heat shock genes (27, 30, 41). The change in the transcriptional profile of Drosophila cells during heat shock is also reflected in the distribution of Ser10-phosphorylated histone H3 within the genome, visualized by the staining of salivary gland polytene chromosomes via standard immunochemical methods. After heat shock, H3 phosphorylation appears at sites containing the heat shock genes, whereas H3 becomes dephosphorylated at previously active genes concomitantly with their transcriptional repression. However, it is not known at this time whether histone phosphorylation occurs during the process of transcription initiation or elongation. This change in the distribution of phosphorylated H3 during heat shock is a dynamic process that requires the activity of a functional heat shock transcription factor (HSF) (32), further illustrating an intimate association of Ser10-phosphorylated H3 with active transcription (9, 10, 32, 43). The link between phosphorylated histone H3 and transcription suggests that this process may be regulated by the levels of H3 phosphorylation, which may in turn be determined by the activity of specific protein kinases and/or phosphatases.
Serine/threonine protein phosphatase type 2A (PP2A) is a heterotrimer consisting of catalytic, structural, and regulatory subunits. Although the catalytic and structural subunits are highly conserved and essential for proper enzymatic activity among all human and yeast isoforms of PP2A (18), the regulatory subunit is by contrast highly variable, imparting cellular compartment targeting information and substrate specificity for PP2A catalytic activity (20, 26). Inhibitory factors are known to attenuate PP2A catalytic activity by competing with the regulatory subunit for association with the PP2A trimer. This replacement of the regulatory subunit with other proteins can result in the partial or total loss of PP2A catalytic activity (15, 16, 44). This loss or misregulation of PP2A activity results in a variety of defects, such as increased cellular transformation (44) and abnormal chromosomal segregation (25). While PP2A has numerous roles in various cellular processes, such as cell cycle control (25, 39), and the integration of cellular signaling pathways, such as the mitogen-activated protein kinase pathway (28, 40, 45), a clear link between PP2A activity and the regulation of transcription has yet to be established.
Here we report that PP2A has a role in the process of transcription activation and inactivation. The PP2A enzyme is capable of dephosphorylating the Ser10-phosphorylated histone H3 isoform. Treatment of Drosophila salivary glands with strong phosphatase inhibitors and analysis of polytene chromosomes isolated from PP2A mutants demonstrate that loss of PP2A activity results in disruption of the characteristic redistribution of phosphorylated histone H3 during heat shock, with an increase of the phosphorylated H3 isoform at sites that do not contain heat shock genes. Further, these non-heat-shock gene-containing loci are actively transcribed and contain activated RNA polymerase II complexes. We also find that the SET protein, a strong cellular inhibitor of PP2A, is present at regions of active transcription within polytene chromosomes and can prevent dephosphorylation of Ser10-phosphorylated H3 by PP2A catalysis. These results suggest that regulation of PP2A activity via the SET protein might modulate changes in gene expression in Drosophila.
MATERIALS AND METHODS
Induction of the heat shock response and preparation of polytene chromosomes.
Drosophila stocks were maintained in standard medium at 22°C. Oregon R larvae were used as wild-type controls in all experiments presented. For induction of the heat shock response, wandering third-instar Oregon R and twsP mutant larvae were placed in 1.5-ml microcentrifuge tubes with punctured caps. The microcentrifuge tubes were then incubated for 20 min in a water bath maintained at 37°C. After removal from the water bath, salivary glands were immediately isolated and fixed in formaldehyde-acetic acid fixative to prevent recovery from heat shock (38). Fixed salivary glands were subsequently squashed in 45% acetic acid on subbed slides. The slides were frozen in liquid nitrogen and stored dry at −70°C until used.
Drug treatment of salivary glands and bromouridine triphosphate (BrUTP) labeling.
Salivary glands were isolated from wandering third-instar Oregon R and twsP mutant larvae in complete Grace's medium (Sigma). For treatment, isolated glands were placed in glass culture tubes containing indicated concentrations of okadaic acid (Sigma) diluted in Grace's medium and heat shocked in a water bath at 37°C for 20 min. Following heat shock, the salivary glands were immediately fixed and squashed as above.
For in vivo labeling of nascent transcripts, we followed the method of Westwood and colleagues (8), with the following exceptions: salivary glands were incubated in culture tubes containing 10 mM BrUTP (Sigma) in complete Grace's medium with 400 μM DOTAP (Roche Molecular Biochemicals)/ml for 20 min at 37°C prior to squashing as above.
Immunocytochemistry.
Slides were incubated overnight in antibody dilution buffer (1× phosphate-buffered saline, 1% bovine serum albumin, 0.05% Triton X-100) containing primary antibodies at concentrations of 1:100 anti-Ser10-phosphohistone H3 (kindly provided by David Allis and also purchased from Upstate Biotechnologies), 1:15 anti-SET, and antibromodeoxyuridine (Roche Molecular Biochemicals) or 1:1 anti-hyperphosphorylated RNA polymerase II (H5) (a gift from Joseph Gall). Following incubation, slides were washed three times in antibody dilution buffer and incubated with a 1:250 dilution of either fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G or Texas Red-conjugated goat anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories) for 1 h at 37°C. DNA probes were detected with a 1:100 dilution of FITC-conjugated antidigoxigenin (Roche). Slides were washed three times in antibody dilution buffer, rinsed briefly in phosphate-buffered saline, stained with 0.5 μg of 4′,6-diaminidine-2-phenylindole (DAPI)/ml, and mounted in Vectashield antifade mounting medium (Vector Laboratories) for viewing.
Preparation of Drosophila protein extracts.
Fifty wandering third-instar larvae were homogenized in cold sodium dodecyl sulfate (SDS) sample buffer (20 mM sodium phosphate, 2% SDS, 0.001% bromophenol blue, 0.2 M DTT, 2% glycerol) using five strokes of a Dounce homogenizer. For heat-shocked larval extracts, 50 wandering third-instar larvae were heat shocked as described above and then immediately homogenized. Following homogenization, protein samples were immediately run on SDS-15% polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes for immunodetection. Following transfer to PVDF, the presence of Ser10-phosphorylated histone H3 in larval samples was detected by following standard immunoblot techniques, using anti-Ser10-phosphohistone H3 (kindly provided by David Allis, and also purchased from Upstate Biotechnologies).
Preparation of DNA probes and in situ hybridization.
Digoxigenin-labeled DNA probes were prepared using the Prime-A-Gene random priming kit (Promega). The template used for random priming was a fragment corresponding to nucleotides +219 to +575 of the Drosophila hsp70 transcription unit amplified via PCR. Labeled probes were ethanol precipitated and stored in hybridization buffer (4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 1× Denhardt's containing 0.4 mg of salmon sperm DNA/ml) until ready for use.
Polytene chromosomes were prepared and frozen in liquid N2 as described above. Coverslips were removed, and the slides were stored in ethanol at −70°C until ready for use. Slides were allowed to warm to room temperature, air-dried, and incubated in 2× SSC for 60 min at 65°C. The slides were then dehydrated through an ethanol series, air-dried, and denatured for 3 min in 0.07 M NaOH. Following denaturation, the slides were dehydrated again through an ethanol series and allowed to air dry at room temperature prior to hybridization.
For hybridization, boiled probes were added to the dried, denatured polytene squashes and covered immediately with a coverslip. The slide and coverslip were sealed with rubber cement and incubated at 37°C overnight in a humidified chamber. Following hybridization, coverslips were removed and the slides were washed under high-stringency conditions before proceeding directly to immunocytochemical treatment, as described above.
Preparation of SET antibody and in vitro SET protein expression.
A Drosophila full-length SET cDNA was cloned in-frame into the pET15b vector (InVitrogen), expressed as a poly-His fusion protein, and purified using standard nickel-binding chromatography. Antibody production was carried out as previously described (12). For SET protein expression, a pET 15b vector (InVitrogen) containing the full-length SET cDNA was used as a template in the TnT coupled reticulocyte lysate expression system (Promega). SET protein was detected in the reticulocyte lysate using standard immunodetection protocols and anti-SET antibody.
In vitro PP2A assay.
PP2A activity was measured as follows. Briefly, 5 μg of core histones prepared from Colcemid-treated HeLa cells (Upstate Biotechnology) was used as a substrate in assay buffer (10 mM HEPES [pH 7.0], 1 mM dithiothreitol, 1 mM MnCl2, 10 μg of BSA/ml, 50 μM leupeptin) containing 0.1 U of purified PP2A AC dimer (Upstate Biotechnology). Where indicated, either in vitro-translated SET protein (1 μl of the completed rabbit reticulocyte translation reaction, see above) or 50 nM okadaic acid (Sigma) were included in the assay buffer. Assay reactions were incubated for 60 min at 37°C and stopped by the 1:1 addition of 2× protein sample buffer (0.125 M Tris-Cl, 4% SDS, 20% glycerol, 2% β-mercaptoethanol [pH 6.8]) and boiling at 100°C for 5 min. Assay samples were immediately run on 15% SDS-polyacrylamide gels and transferred to PVDF membranes for immunodetection analysis. Following transfer to PVDF, the presence of Ser10-phosphorylated histone H3 in assay samples was detected by following standard immunoblot techniques, using anti-Ser10-phosphohistone H3.
RESULTS
Inhibition of PP2A prevents widespread dephosphorylation of Ser10-phosphorylated histone H3 during heat shock.
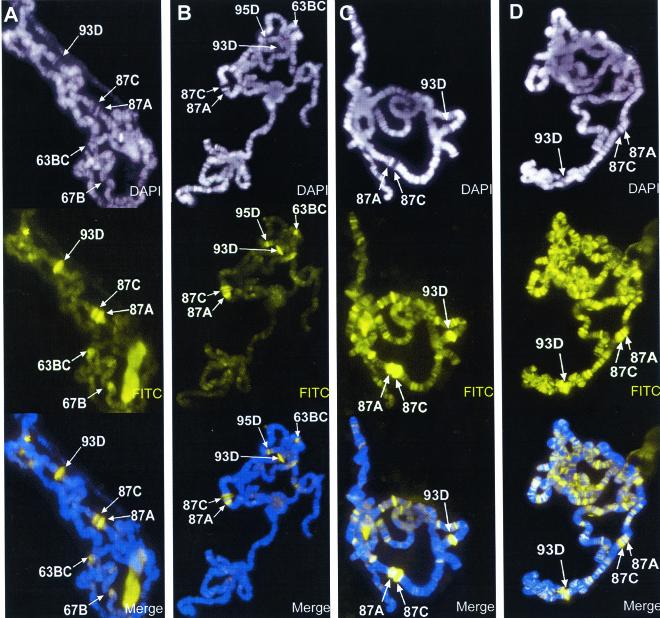
Treatment of quiescent cells with okadaic acid, a potent inhibitor of both protein phosphatase type 1 (PP1) and the catalytic subunit of PP2A (11), is known to induce a rapid increase in the total level of detected phosphorylated histone H3 (7). In order to determine if the inhibition of either PP1 or PP2A catalytic activities could inhibit heat shock-mediated histone H3 dephosphorylation at non-heat-shock gene-containing loci of polytene chromosomes, we incubated Drosophila salivary glands in culture medium containing increasing concentrations of okadaic acid under heat shock conditions and examined the distribution of Ser10-phosphorylated histone H3 in polytene chromosomes. In the absence of okadaic acid, after a 20-min heat shock at 37°C, histone H3 became dephosphorylated in the Ser10 residue at all sites previously containing transcriptionally active genes, whereas H3 is phosphorylated de novo at the heat shock genes concomitantly with their activation (Fig. 1A). When salivary glands were heat shocked in culture medium containing 10 nM okadaic acid, no discernible effect was observed on the presence of Ser10-H3 phosphorylation at non-heat-shock gene loci (Fig. 1B). By contrast, an increase in the concentration of okadaic acid in the culture medium to 50 nM has a dramatic effect on the distribution of Ser10-phosphorylated H3 after heat shock (Fig. 1C). In addition to the heat shock genes, phosphorylated H3 remains present and is detected at many loci that are not known to contain heat shock genes (Fig. 1C). Raising the concentration of okadaic acid to 100 nM results in a further increase in the number of sites containing phosphorylated histone H3 (Fig. 1D). These results suggest that either PP2A or PP1 catalytic activity is involved in the dephosphorylation of histone H3 during the process of transcription inactivation of non-heat-shock genes as a result of the heat shock response. It is well established that okadaic acid can selectively inhibit PP2A without affecting PP1 activity in vitro at concentrations below 100 nM, although it inhibits both phosphatase activities at higher concentrations (11). Although the actual concentration of okadaic acid within the salivary glands cannot be controlled or assayed directly, we do observe an effect on the distribution of histone H3 dephosphorylation when salivary glands are incubated in culture medium containing 50 nM okadaic acid. Since this concentration would represent the absolute maximum intracellular concentration of okadaic acid in our assay, these results suggest that PP2A could be responsible for the observed effects.
FIG. 1.
Effect of okadaic acid on histone H3 phosphorylation during heat shock. Salivary glands isolated from wild-type larvae were incubated in Grace's medium containing 0 nM (A), 10 nM (B), 50 nM (C), and 100 nM (D) okadaic acid. All salivary glands were heat shocked at 37°C for 20 min before squashing, immunostained for Ser10-phosphorylated histone H3 (FITC, yellow), and counterstained with DAPI (white). Merged images are a composite of both channels shown, antibody staining in yellow and DAPI counterstaining in blue. In all photographs, identifiable heat shock loci are labeled; although not all heat shock genes are visible in these photographs, data shown in other figures indicate that all contain Ser10-phosphorylated histone H3. Comparison of treated with untreated chromosomes indicates that okadaic acid prevents the genome-wide dephosphorylation of Ser10-phosphorylated histone H3 isoforms during heat shock.
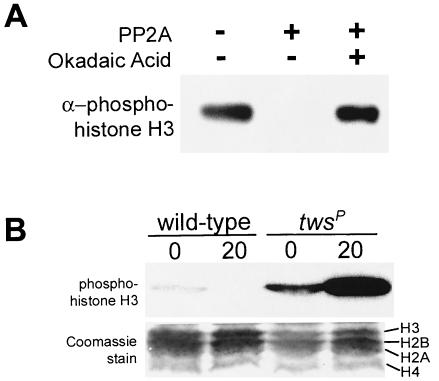
Although PP1 activity is known to dephosphorylate phosphorylated H3 in vivo during mitosis (17), PP2A-mediated dephosphorylation of Ser10-phosphorylated H3 has yet to be described. Given the high identity and conservation of the PP2A enzyme (17) and histone N-terminal domains (18) among eukaryotes, we incubated core HeLa cell histones containing the Ser10-phosphorylated H3 isoform with purified human PP2A in vitro and detected the presence of phosphohistone H3 via Western blotting (Fig. 2A). We found that the PP2A enzyme was indeed capable of dephosphorylating H3 in vitro and that PP2A-mediated H3 dephosphorylation was inhibited in the presence of 50 nM okadaic acid (Fig. 2A). We therefore conclude that the PP2A holoenzyme is capable of dephosphorylating Ser10-phosphorylated histone H3. These results further support our previous conclusion that the effect on the overall distribution of Ser10-phosphorylated histone H3 in polytene chromosomes prepared from okadaic acid-treated salivary glands (see Fig. 1) could be due to the inhibition of PP2A by okadaic acid.
FIG. 2.
Protein phosphatase type 2A dephosphorylates Ser10-phosphorylated histone H3 both in vivo and in vitro. (A) Purified core histones containing Ser10-phosphorylated histone H3 were incubated with purified PP2A in the presence or absence of 50 nM okadaic acid as indicated. The PP2A enzyme strongly dephosphorylates Ser10-phosphorylated histone H3. Catalytic activity of PP2A is strongly inhibited by incubation with 50 nM okadaic acid. (B) Whole-larval extracts were prepared from wild-type and twsP mutant larvae that either were not heat shocked (0) or were heat shocked for 20 min (20), resolved on an SDS-15% polyacrylamide gel, and immunostained for the presence of the Ser10-phosphorylated isoform of histone H3. The twsP mutant extracts contained a greater concentration of Ser10-phosphorylated histone H3 than wild-type controls both before and after heat shock.
Histone H3 dephosphorylation is reduced in hypomorphic PP2A mutants during heat shock.
To further analyze the effect of the loss of in vivo PP2A activity on the phosphorylation status of Ser10 phosphorylated histone H3 during heat shock, we examined flies bearing the twsP hypomorphic PP2A mutation. The Drosophila twsP mutation is caused by the insertion of a P-element into the regulatory subunit of PP2A, resulting in pupal lethality (24, 25). The twsP mutation results in a 50% reduction of endogenous wild-type PP2A catalytic activity and also produces a variety of cell cycle and mitotic defects (25). Based on the results described above, twsP mutant larvae should contain a greater total level of phosphorylated histone H3 than wild-type larvae. To examine this, total protein extracts were prepared from wild-type and twsP mutant larvae and analyzed via Western blotting for the presence of the Ser10-phosphorylated histone H3 isoform both before and after heat shock. Indeed, twsP mutant larvae contained a greater amount of phosphohistone H3 than wild-type larvae both before and during heat shock (Fig. 2B). This difference is possibly attributable to the reduction of total PP2A enzymatic activity described in twsP mutants (25).
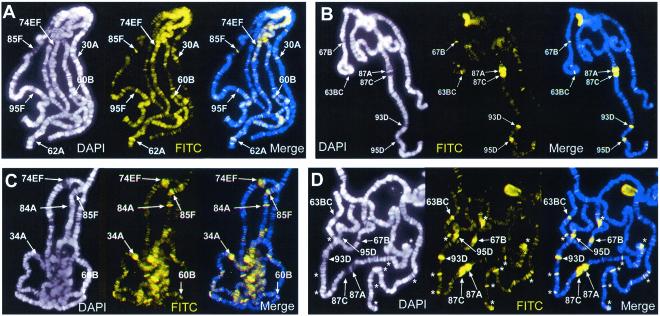
In addition, polytene chromosomes isolated from these mutants should resemble polytene chromosomes isolated from okadaic acid-treated salivary glands in that they should display Ser10-phosphorylated histone H3 at both heat shock and non-heat-shock gene loci during thermal stress. We find that this is indeed the case. Prior to heat shock, the distributions of phosphorylated H3 isoforms in polytene chromosomes prepared from wild-type and twsP mutant larvae are virtually indistinguishable (Fig. 3A and C). After a 20-min heat shock, polytene chromosomes prepared from heat-shocked wild-type larvae show phosphorylated H3 primarily at the heat shock loci. By contrast, chromosomes from twsP mutant larvae show a greater number of sites containing Ser10-phosphorylated H3 at non-heat-shock loci than wild-type controls (Fig. 3B and D). The presence of the Ser10-phosphorylated histone H3 isoform at numerous non-heat-shock gene-containing loci in twsP mutant larvae after heat shock supports our hypothesis that PP2A may have a role in the regulation of transcription during heat shock by controlling histone H3 dephosphorylation.
FIG. 3.
Comparison of histone H3 phosphorylation in wild-type (A and B) versus twsP (C and D) mutant larvae. Wild-type and mutant larvae were heat shocked for 0 min (not heat shocked) (A and C) or 10 min (B and D). Polytene chromosomes were prepared and immunostained for Ser10-phosphorylated histone H3 (FITC, yellow) and counterstained with DAPI (white) to view the DNA. Merged images are a composite of both channels shown, with antibody staining (FITC) in yellow and DAPI counterstaining in blue. Prominent ecdysone gene-containing loci are marked in non-heat-shocked chromosomal spreads (A and C). In heat-shocked samples, visible heat shock loci are labeled, and asterisks indicate regions of strong labeling that do not contain heat shock genes (B and D). After a 10-min heat shock, puffing and phosphorylation are observed only at the heat shock loci in wild-type larvae (B). By contrast, numerous loci that do not contain heat shock genes are phosphorylated in twsP mutants during heat shock (D).
Non-heat-shock genes are transcriptionally active in twsP mutants during heat shock.
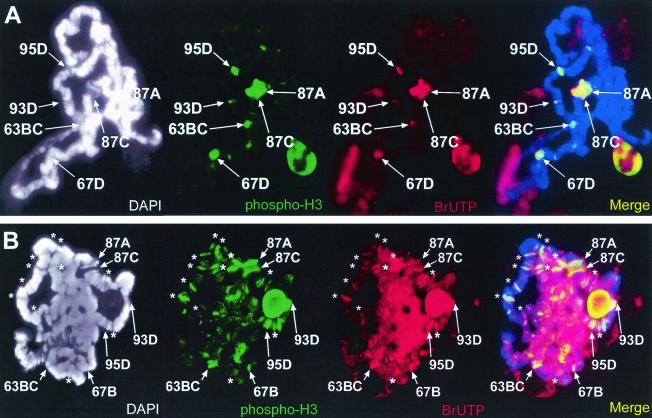
Whereas the puffs of polytene chromosomes are known to be sites of active transcription (41), the presence of the phosphorylated H3 isoform at non-heat-shock genes in okadaic acid-treated and twsP mutant polytene chromosomes may not necessarily be accompanied by active transcription. To address this concern, we incubated salivary glands from both wild-type and twsP mutant larvae at 37°C in tissue culture medium containing 10 μM BrUTP. After incorporation of BrUTP into mRNA, it is possible to visualize the location of nascent mRNA transcripts on polytene chromosomes via immunocytochemistry using antibodies against BrUTP (8). We found that overall, the location of nascent transcripts corresponds almost exclusively with H3 phosphorylation in polytene chromosomes from both wild-type (Fig. 4A) and twsP mutant larvae (Fig. 4B). As expected, nascent transcripts were detected at the heat shock puffs in polytene chromosomes from both wild-type and twsP mutants during heat shock. In twsP mutant larvae, transcripts were also observed at H3-phosphorylated chromosomal loci, including loci that did not contain heat shock genes, indicating that genes at these loci were actively transcribing under heat shock conditions (Fig. 4B).
FIG. 4.
Non-heat-shock genes are transcribed in twsP mutants after heat shock. Salivary glands isolated from wild-type (A) and twsP mutant (B) larvae were heat shocked for 20 min at 37°C in Grace's medium containing bromouridine (BrUTP). Polytene chromosomes were then immunostained for the presence of BrUTP (red) and Ser10-phosphorylated histone H3 (phospho-H3, green). Chromosome spreads were also counterstained with DAPI (white) to view the DNA. DAPI counterstain information is represented as blue in merged images. Visible heat shock loci are indicated in all panels. Strong regions of labeling that do not contain heat shock genes are indicated with an asterisk. Merged images indicate that numerous non-heat-shock gene loci containing the Ser10-phosphorylated histone H3 isoform in twsP mutants are also actively transcribed after heat shock.
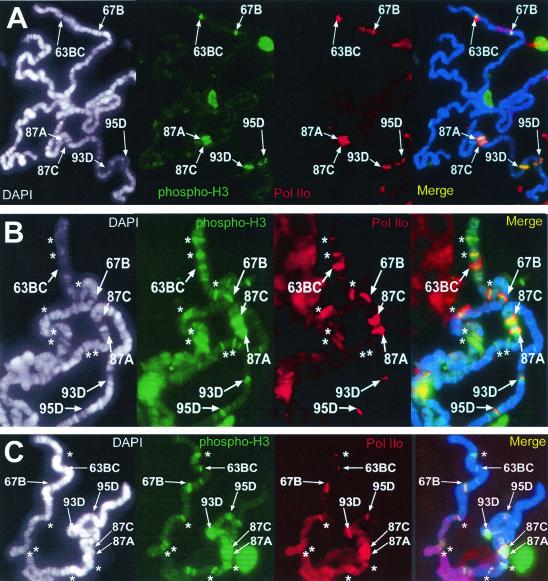
To further demonstrate that the genes at these non-heat-shock gene-containing loci in polytene chromosomes from twsP mutant larvae are actively transcribed during heat shock, we immunostained heat-shocked polytene chromosomes prepared from wild-type and twsP mutant larvae with antibodies specific for C-terminal domain hyperphosphorylated RNA polymerase II (Pol IIo) (34). This phosphorylated isoform of the RNA polymerase complex is the activated form of this enzyme (34) and is detected solely at actively transcribing loci, such as the heat shock puffs of Drosophila polytene chromosomes during heat shock (22). C-terminal hyperphosphorylated RNA polymerase II complexes are detected at all heat shock gene loci containing the Ser10-phosphorylated H3 isoform in both wild-type (Fig. 5A) and twsP mutant (Fig. 5B) polytene chromosomes during heat shock. However, in addition to the heat shock genes, polytene chromosomes isolated from twsP mutant larvae also display Pol IIo staining at H3-phosphorylated chromosomal loci that do not contain heat shock genes (Fig. 5B). The C-terminal phosphorylated isoform of RNA polymerase II is also detected at both heat shock and non-heat-shock gene loci in polytene chromosomes prepared from salivary glands that have been treated with 50 nM okadaic acid (Fig. 5C). The agreement of in vivo BrUTP incorporation data with the RNA Pol IIo results suggests that sites containing Ser10-phosphorylated H3 isoforms in twsP mutants are indeed actively transcribing during heat shock, suggesting that the reduced level of PP2A activity in these mutants results in a reduced inactivation of transcription of non-heat-shock genes due to incomplete dephosphorylation of the Ser10-phosphorylated H3 isoform at these chromosomal loci.
FIG. 5.
Sites that contain Ser10-phosphorylated histone H3 isoform also contain active RNA polymerase II during heat shock. Polytene chromosomes were prepared from wild-type (A) and twsP mutant (B) larvae. In addition, salivary glands from wild-type larvae were treated with 50 nM okadaic acid (C). All samples were heat shocked for 20 min at 37°C, immunostained with antibodies against hyperphosphorylated RNA Pol II (Pol IIo, red) and the Ser10-phosphorylated histone H3 isoform (phospho-H3, green), and counterstained with DAPI (white) to view the DNA. DAPI counterstain information is represented as blue in merged images. Visible heat shock gene-containing loci are labeled in all panels. The active form of RNA polymerase II was observed at all sites containing the Ser10-phosphorylated H3 isoform, including the heat shock gene loci (A,B, and C). Additionally, prominent Pol IIo labeling was detected at regions that do not contain heat shock genes (indicated with asterisks) in twsP mutant (B) and okadaic acid-treated (C) chromosomes.
The SET oncoprotein inhibits PP2A-mediated H3 dephosphorylation and is present at actively transcribed regions.
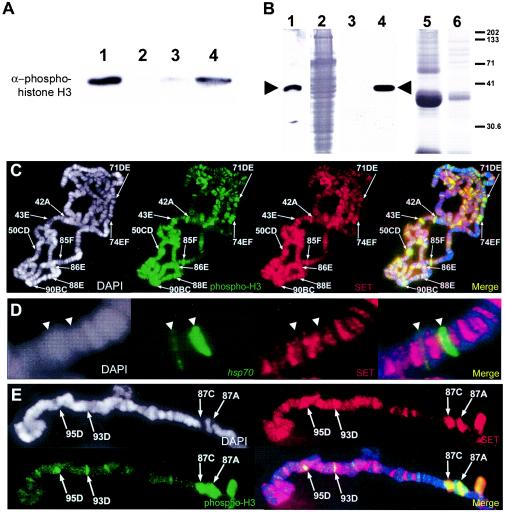
If PP2A activity is responsible for the inactivation of transcription via the dephosphorylation of histone H3, then a reasonable hypothesis is that PP2A activity might be partially or completely inhibited at the sites of actively transcribing genes, which contain Ser10-phosphorylated histone H3. One candidate inhibitor of PP2A activity is the I-2/SET oncoprotein, a potent and highly specific PP2A inhibitor that is localized primarily to the cell nucleus (21, 29). Aberrant SET activity results in misregulation of the catalytic activity of PP2A, producing abnormal patterns of gene expression and cell cycle defects that lead to acute undifferentiated leukemogenesis (29). It should be noted that the SET oncoprotein does not contain a SET domain, a protein module responsible for lysine 9 methylation of histone H3 and subsequent gene silencing via the formation of heterochromatin (37). Given the described roles of SET as an in vivo inhibitor of PP2A activity (21, 29, 33), we were particularly interested to determine if SET can inhibit PP2A-mediated dephosphorylation of Ser10-phosphorylated histone H3 in our previously described in vitro assay. We find that Drosophila SET protein prepared via expression and translation in an in vitro reticulocyte lysate system can indeed prevent dephosphorylation of Ser10-phosphorylated H3. In vitro reticulocyte lysate translation reactions that did not contain SET protein (i.e., uninduced lysate) showed minimal inhibition of PP2A activity (Fig. 6A).
FIG. 6.
The SET oncoprotein inhibits PP2A-mediated dephosphorylation of phosphohistone H3 and is present at transcriptionally active loci. (A) Purified human core histones containing Ser10-phosphorylated histone H3 (lane 1) were incubated with purified human PP2A (lane 2). The presence of 1 μl of in vitro reticulocyte lysate-translated SET is capable of inhibiting PP2A-mediated dephosphorylation of histone H3 (lane 4). Translation reactions that did not contain SET protein showed minimal inhibition of PP2A activity (lane 3). (B) Antibodies to the SET gene product were generated and used to detect SET in whole Drosophila pupa extracts (lane 1) and the in vitro-translated SET protein expressed via a reticulocyte lysate system and utilized in a PP2A activity assay (lanes 3 and 4). Lane 3 contains 5 μl of reticulocyte lysate that does not contain the expressed SET protein. Lane 4 contains 5 μl of reticulocyte lysate containing expressed SET. Lane 2 shows the Coomassie staining of lane 1. Lanes 5 and 6 show Coomassie staining of lanes 3 and 4, respectively. Molecular mass size standards (in kilodaltons) are indicated at right. (C,D, and E) Genomic distribution of SET protein in polytene chromosomes prepared from wild-type larvae before (C) and after (E) heat shock and immunostained for the presence of SET (SET, red) and the Ser10-phosphorylated histone H3 isoform (phospho-H3, green). Chromosome spreads were also counterstained with DAPI (white) to view the DNA. DAPI counterstain information is represented as blue in merged images. Before heat shock, the SET protein is detected at puffed and H3-phosphorylated (transcriptionally active) loci, including the ecdysone-regulated genes, which are labeled (C). In situ hybridization with a fragment of the hsp70 transcription unit (hsp70, green) reveals that prior to heat shock, the SET protein (SET, red) does not colocalize with the hsp70 genes (D). During heat shock, strong SET staining is also detected at the actively transcribing heat shock puffs (E).
If SET is indeed involved in the regulation of PP2A activity during transcription activation or repression, then a reasonable prediction would be that SET is associated with actively transcribing regions of polytene chromosomes that contain the Ser10-phosphorylated histone H3 isoform. To test this hypothesis, we prepared antibodies against the full-length Drosophila SET gene product. Anti-SET antibodies detect a single band in extracts prepared from Drosophila pupae (Fig. 6B, lane 1). Further, anti-SET antibodies also detect in vitro reticulocyte lysate-translated SET protein used in PP2A activity assays (Fig. 6B, lane 4). We then used anti-SET antibodies to stain polytene chromosomes prepared from wild-type larvae (Fig. 6C). As expected, SET was detected at all sites containing the Ser10-phosphorylated histone H3 isoform before heat shock, including the strongly puffed regions that contain ecdysone-inducible genes (Fig. 6C).
To determine if SET was present at a particular heat shock gene under non-heat-shock conditions, we used immunocytochemistry and in situ hybridization with a portion of the hsp70 transcription unit as a probe to examine the distribution of SET protein on polytene chromosomes relative to the position of the hsp70 genes prior to heat shock. We found that before temperature elevation, the SET protein is absent or present at very low levels in the chromosome regions containing hsp70 genes (Fig. 6D). Following heat shock, SET staining increases dramatically at the heat shock gene loci, which also contain Ser10-phosphorylated H3 (Fig. 6E). This suggests that SET might be recruited to an actively transcribed gene during activation of transcription. Nevertheless, the SET protein is still present after heat shock at many loci that do not contain phosphorylated H3, suggesting that additional mechanisms must be in place to rapidly inhibit SET activity without requiring its disappearance from genes that are transcriptionally inactivated after temperature elevation.
DISCUSSION
The close association of the Ser10-phosphorylated histone H3 isoform with actively transcribing genes suggests that the regulation of H3 phosphorylation and the regulation of transcription may also be linked through the activities of specific protein kinases and/or phosphatases (9, 10, 32, 43). Mitotic dephosphorylation of the phosphorylated histone H3 isoform by the actions of protein phosphatases has been previously described (31, 36). Further, Drosophila mutations in the PP2A catalytic subunit arrest during early embryogenesis, display hypercondensed chromosomes, and lack segregational fidelity during mitosis (39). This strongly implies that PP2A is capable of acting on phosphohistone H3 as a substrate, given the fact that H3 phosphorylation is essential for proper mitotic chromosomal condensation (5, 13).
Our data suggest a role for PP2A in the process of transcriptional control, possibly through the dephosphorylation of the Ser10-phosphorylated H3 isoform. This conclusion rests on the following experimental observations. Incubation of salivary glands with 50 nM okadaic acid results in the maintenance of Ser10-phosphorylated H3 at sites of non-heat-shock genes in polytene chromosomes after heat shock. Although okadaic acid can inhibit both PP2A and PP1, it is known that its effect is specific for PP2A at this concentration when tested with in vitro assays. PP2A is capable of dephosphorylating Ser10-phosphorylated histone H3 in an in vitro assay. In addition, hypomorphic mutations in a regulatory subunit of PP2A also result in the perdurance of phosphorylated H3 at non-heat-shock genes after thermal stress. The significance of this result is highlighted by the fact that the twsP allele used in these studies only decreases PP2A activity to 50% of normal levels. Finally, SET, an endogenous inhibitor of PP2A catalytic activity, prevents PP2A-mediated dephosphorylation of the Ser10-phosphorylated H3 isoform in an in vitro assay and is detected at chromosomal loci containing actively transcribing genes upon their induction. Taken together, these observations support the conclusion that inhibition or attenuation of the catalytic activity of PP2A results in a lack of dephosphorylation of H3 associated with non-heat-shock genes, ultimately preventing the transcriptional repression of these genes.
The inference of a causal relationship between histone H3 phosphorylation and transcriptional activation is based on cytological observations of polytene chromosome staining using specific antibodies. The resolution of these measurements does not allow firm conclusions on whether H3 phosphorylation is associated with specific gene regions. Nevertheless, the perfect overlap in the patterns of the signals elicited by antibodies against the phosphorylated tail of histone H3 and activated RNA polymerase II suggests that indeed phosphorylated histone H3 is associated with active genes. Additional experiments involving chromatin immunoprecipitation analysis would be required to accurately map the specific gene regions associated with this modification and determine whether H3 phosphorylation is involved in the release of the paused RNA polymerase holoenzyme, transcriptional elongation, or other aspects of transcription. The nature of the protein kinase responsible for phosphorylation of histone H3 in actively transcribed genes is not known; but, in principle, this kinase could be pTEF-b. pTEF-b kinase activity is essential for the C-terminal phosphorylation, and subsequent activation, of the RNA polymerase II holoenzyme during transcription (23). We find that the phosphorylation of histone H3 is unaffected at the heat shock gene loci in polytene chromosomes prepared from salivary glands treated with the pTEF-b kinase inhibitor DRB (data not shown), suggesting that serine-10 phosphorylation of histone H3 would occur independently of RNA polymerase II phosphorylation and by a mechanism involving a different protein kinase. Since a link between PP2A activity and mitotic Ser10 H3 phosphorylation is suggested by the phenotype of mts mutants (39), PP2A activity may therefore be focused on Ser10-phosphorylated histone H3 rather than the C-terminal domain of RNA polymerase II during repression of transcription.
Since the processes of transcription activation and repression are highly regulated, control of PP2A activity in the nucleus must be an important step in this process. We find that the SET protein, a highly selective and potent cellular inhibitor of PP2A (21, 29), inhibits PP2A-mediated dephosphorylation of Ser10-phosphorylated H3 and colocalizes with regions of active transcription. In the context of the hsp70 gene, prior to gene induction, SET is not detected or is present at very low levels at the 87A and 87C chromosomal subdivisions where the hsp70 genes are located. Following activation of transcription, the amount of SET protein present at these two sites, as well as at other heat shock gene loci, increases dramatically, and this increase might result in inhibition of PP2A activity and subsequent increase in H3 phosphorylation. Attenuation of PP2A activity in vivo involves the replacement of the regulatory subunit of the PP2A heterotrimer with another competing protein, resulting in unregulated or nonspecific PP2A activity (18). This strategy of PP2A inhibition has been observed in several cases, including the action of the simian virus 40 small tumor antigen (44). It remains a strong possibility that the inhibition of PP2A catalytic activity by the SET protein at actively transcribing loci might occur via this method of competition with the regulatory subunit for association with the PP2A heterotrimer.
The detection of SET at some chromosomal loci that do not contain Ser10-phosphoryated H3 during heat shock suggests that the SET protein may itself be regulated as a consequence of the Drosophila heat shock response. This regulation might be accomplished by the presence of additional proteins or by covalent modification of the SET protein. Covalent posttranslational modification of the SET protein may be important for the PP2A inhibitory activity of the SET protein, since we find that recombinant SET protein expressed and purified from bacterial cells is not capable of inhibiting PP2A-mediated dephosphorylation of Ser10-phosphorylated histone H3 (data not shown), but SET protein produced in a eukaryotic translation system is perfectly capable of inhibiting PP2A activity (Fig. 6A). A possible modification of the SET protein that could subsequently regulate its activity could be phosphorylation. SET does possess a consensus site for protein kinase C (1) and is known to exist as a phosphoprotein (33), suggesting a regulatory mechanism whereby the inhibition of PP2A by SET might be modulated through the phosphorylation or dephosphorylation of SET. In keeping with the observed kinetics of the heat shock response (27, 30, 41), dephosphorylation of the phospho-SET protein could serve as an extremely rapid means of inactivating SET at non-heat-shock genes. It is interesting that SET-mediated inhibition of PP2A can depend upon other cofactors, such as the SEB and HRX proteins, which regulate SET-PP2A interactions and subsequent PP2A inhibition during leukemogenesis (2, 29). Determining the precise effect of the heat shock response on the SET protein would therefore shed light on the role of SET during transcription.
The precise role of Ser10-phosphorylation of histone H3 tails in transcription is not known. One possibility is that this modification alters chromatin structure in a manner required for the assembly of the transcription complex, or it might serve as a signal for the recruitment of specific proteins at the promoter or for further covalent modifications of histone H3. An interesting alternative is suggested by recent results of Ahmad and Henikoff (9), who have found that transcriptional activation of rDNA arrays in Drosophila correlates with the replacement of histone H3 for the H3.3 variant. Since the antibodies used in our studies do not distinguish between H3 and H3.3, it is possible that the de novo phosphorylation we observe at sites of active transcription takes place on H3.3 molecules as they are incorporated into chromatin. Alternatively, phosphorylation of H3 might be a prerequisite for subsequent exchange with H3.3. This explanation would account for the lack of observations implicating H3 phosphorylation as a general mechanism for transcriptional activation of yeast genes. Since the only form of histone H3 found in yeast corresponds to the H3.3 variant (3, 4), an exchange between H3 and H3.3, and therefore H3 phosphorylation, might not be required for activation of transcription in this organism.
Acknowledgments
We thank Mariano Labrador and Fabien Mongelard for discussions and critical comments on the manuscript and Tatiana Gerasimova for advice with cytological analysis. We also thank Jean-Paul Paraiso and members of the Westwood lab for technical advice with BrUTP labeling. y w; twsP/TM6B flies were kindly provided by Adelaide Carpenter and David Glover. Anti-Ser10-phosphorylated H3 antibodies were a gift from David Allis. RNA polymerase IIo (H5) antibodies were a gift from Joseph Gall.
This work was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
REFERENCES
- 1.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 340:231-235. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H. T., F. S. Nallaseth, G. Walter, and D. C. Tkachuk. 1997. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J. Biol. Chem. 272:28407-28414. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, K., and S. Henikoff. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad, K., and S. Henikoff. 2002. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 5.Ajiro, K., H. Yasuda, and H. Tsuji. 1996. Vanadate triggers the transition from chromosome condensation to decondensation in a mitotic mutant (tsTM13) inactivation of p34cdc2/H1 kinase and dephosphorylation of mitosis-specific histone H3. Eur. J. Biochem. 241:923-930. [DOI] [PubMed] [Google Scholar]
- 6.Ashburner, M., and J. J. Bonner. 1979. The induction of gene activity in Drosophilia by heat shock. Cell 17:241-254. [DOI] [PubMed] [Google Scholar]
- 7.Barratt, M. J., C. A. Hazzalin, E. Cano, and L. C. Mahadevan. 1994. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc. Natl. Acad. Sci. USA 91:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, W. Y., N. A. Winegarden, J. P. Paraiso, M. L. Stevens, and J. T. Westwood. 2000. Visualization of nascent transcripts on Drosophila polytene chromosomes using BrUTP incorporation. BioTechniques 29:934-936. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 10.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson, J. F., and C. F. Holmes. 1999. Molecular mechanisms underlying inhibition of protein phosphatases by marine toxins. Front. Biosci. 4:D646-D658. [DOI] [PubMed] [Google Scholar]
- 12.Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. Simonova, and V. G. Corces. 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82:587-597. [DOI] [PubMed] [Google Scholar]
- 13.Guo, X. W., J. P. Th'ng, R. A. Swank, H. J. Anderson, C. Tudan, E. M. Bradbury, and M. Roberge. 1995. Chromosome condensation induced by fostriecin does not require p34cdc2 kinase activity and histone H1 hyperphosphorylation, but is associated with enhanced histone H2A and H3 phosphorylation. EMBO J. 14:976-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 15.Hong, Y., E. J. Lubert, D. W. Rodgers, and K. D. Sarge. 2000. Molecular basis of competition between HSF2 and catalytic subunit for binding to the PR65/A subunit of PP2A. Biochem. Biophys. Res. Commun. 272:84-89. [DOI] [PubMed] [Google Scholar]
- 16.Hong, Y., and K. D. Sarge. 1999. Regulation of protein phosphatase 2A activity by heat shock transcription factor 2. J. Biol. Chem. 274:12967-12970. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 18.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kamibayashi, C., R. Estes, R. L. Lickteig, S. I. Yang, C. Craft, and M. C. Mumby. 1994. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J. Biol. Chem. 269:20139-20148. [PubMed] [Google Scholar]
- 21.Li, M., A. Makkinje, and Z. Damuni. 1996. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 271:11059-11062. [DOI] [PubMed] [Google Scholar]
- 22.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 24.Mayer-Jaekel, R. E., H. Ohkura, P. Ferrigno, N. Andjelkovic, K. Shiomi, T. Uemura, D. M. Glover, and B. A. Hemmings. 1994. Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J. Cell Sci. 107:2609-2616. [DOI] [PubMed] [Google Scholar]
- 25.Mayer-Jaekel, R. E., H. Ohkura, R. Gomes, C. E. Sunkel, S. Baumgartner, B. A. Hemmings, and D. M. Glover. 1993. The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell 72:621-633. [DOI] [PubMed] [Google Scholar]
- 26.McCright, B., A. M. Rivers, S. Audlin, and D. M. Virshup. 1996. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271:22081-22089. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie, S. L., S. Henikoff, and M. Meselson. 1975. Localization of RNA from heat-induced polysomes at puff sites in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 72:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millward, T. A., S. Zolnierowicz, and B. A. Hemmings. 1999. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24:186-191. [DOI] [PubMed] [Google Scholar]
- 29.Minakuchi, M., N. Kakazu, M. J. Gorrin-Rivas, T. Abe, T. D. Copeland, K. Ueda, and Y. Adachi. 2001. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur. J. Biochem. 268:1340-1351. [DOI] [PubMed] [Google Scholar]
- 30.Mirault, M. E., M. Goldschmidt-Clermont, L. Moran, A. P. Arrigo, and A. Tissieres. 1978. The effect of heat shock on gene expression in Drosophila melanogaster. Cold Spring Harb. Symp. Quant. Biol. 42:819-827. [DOI] [PubMed] [Google Scholar]
- 31.Murnion, M. E., R. R. Adams, D. M. Callister, C. D. Allis, W. C. Earnshaw, and J. R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656-26665. [DOI] [PubMed] [Google Scholar]
- 32.Nowak, S. J., and V. G. Corces. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey, A. V., S. H. Mellon, and W. L. Miller. 2003. Protein phosphatase 2A and phosphoprotein SET regulate androgen production by P450c17. J. Biol. Chem. 278:2837-2844. [DOI] [PubMed] [Google Scholar]
- 34.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 35.Pauli, D., A. P. Arrigo, and A. Tissieres. 1992. Heat shock response in Drosophila. Experientia 48:623-629. [DOI] [PubMed] [Google Scholar]
- 36.Preuss, U., G. Landsberg, and K. H. Scheidtmann. 2003. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz, D. C., K. Ayyanathan, D. Negorev, G. G. Maul, and F. J. Rauscher III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shopland, L. S., and J. T. Lis. 1996. HSF recruitment and loss at most Drosophila heat shock loci is coordinated and depends on proximal promoter sequences. Chromosoma 105:158-171. [DOI] [PubMed] [Google Scholar]
- 39.Snaith, H. A., C. G. Armstrong, Y. Guo, K. Kaiser, and P. T. Cohen. 1996. Deficiency of protein phosphatase 2A uncouples the nuclear and centrosome cycles and prevents attachment of microtubules to the kinetochore in Drosophila microtubule star (mts) embryos. J. Cell Sci. 109:3001-3012. [DOI] [PubMed] [Google Scholar]
- 40.Sontag, E. 2001. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal. 13:7-16. [DOI] [PubMed] [Google Scholar]
- 41.Spradling, A., S. Penman, and M. L. Pardue. 1975. Analysis of drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell 4:395-404. [DOI] [PubMed] [Google Scholar]
- 42.Strelkov, I. S., and J. R. Davie. 2002. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 62:75-78. [PubMed] [Google Scholar]
- 43.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 8:1231-1241. [DOI] [PubMed] [Google Scholar]
- 44.Yang, S. I., R. L. Lickteig, R. Estes, K. Rundell, G. Walter, and M. C. Mumby. 1991. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol. Cell. Biol. 11:1988-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zolnierowicz, S. 2000. Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem. Pharmacol. 60:1225-1235. [DOI] [PubMed] [Google Scholar]