Abstract
Coeliac disease, the most common intestinal disorder of western populations, is an autoimmune enteropathy caused by an abnormal immune response to dietary gluten peptides that occurs in genetically susceptible individuals carrying the HLA-DQ2 or -DQ8 haplotype. Despite the recent progresses in understanding the molecular mechanisms of mucosal lesions, it remains unknown how increased amounts of gluten peptides can enter the intestinal mucosa to initiate the inflammatory cascade. Current knowledge indicates that different gluten peptides are involved in the disease process in a different manner, some fragments being ‘toxic’ and others ‘immunogenic’. Those defined as ‘toxic’ are able to induce mucosal damage either when added in culture to duodenal endoscopic biopsy or when administered in vivo, while those defined as ‘immunogenic’ are able to specifically stimulate HLA-DQ2- or DQ8-restricted T cell clones isolated from jejunal mucosa or peripheral blood of coeliac patients. These peptides are able to trigger two immunological pathways: one is thought to be a rapid effect on the epithelium that involves the innate immune response and the other represents the adaptive immune response involving CD4+ T cells in the lamina propria that recognize gluten epitopes processed and presented by antigen presenting cells. These findings are the subject of the present review.
Keywords: coeliac disease, gluten, innate immunity, adaptive immunity
Definition and clinical description of coeliac disease
Coeliac disease (CD) may be considered the most common chronic inflammatory condition since the estimated prevalence in Western Countries is near to one per cent [1,2]. CD is an autoimmune enteropathy caused by an abnormal immune response to dietary gluten that occurs in genetically susceptible individuals [3]. The unique well established genetic factor is the HLA-DQ region at 6p21.3 [4] which, however, contributes no more than 40% of the risk, the non-HLA genes being the stronger determinant of CD susceptibility [5]. The broad spectrum of gluten-sensitive intestinal mucosal changes that are characteristic, albeit not pathognomonic of this condition, ranges from a complete disruption of the mucosal architecture with villous flattening and crypt hyperplasia, to a slight increase of inflammatory infiltrate in both the epithelium and lamina propria [6]. Furthermore, from in vivo and ex vivo challenges it has been shown that all the lesions comprise a dynamically interrelated series of events that, however, do not necessarily occur in the same patient [7–9]. The wide spectrum of clinical manifestations comprises the classical features of intestinal malabsorption to atypical or asyntomatic cases that now are believed to represent the majority of patients [10]. The only treatment currently available is a lifelong strict adherence to a gluten-free diet that is followed by an amelioration or a normalization of the histological lesions [3,6].
As far as pathogenesis is concerned, CD represents a unique and privileged model since both an external trigger, the gluten peptides [11], and the autoantigen, the ubiquitous enzyme tissue transglutaminase (tTG) [12], have been identified. However, despite the great advances of the last decade in understanding the molecular mechanisms of mucosal lesions, our knowledge about the immune recognition of gluten and the consequent immune response is still far from being complete. Two pathways have been hypothesized to be triggered by these peptides: one is the direct effect on the epithelium that involves the innate immune response [13], the other represents the adaptive immune response involving CD4+ T cells in the lamina propria that recognize processed gluten epitopes [14].
The external trigger
For the vast majority of human beings, cereals represent an important source of nutrients, whereas for CD patients certain cereal products represent poisons that not only destroy small intestinal mucosa [6], but also predispose to gastrointestinal malignancy [15,16]. These products constitute the storage protein fraction of the endosperm of the grains which, in the wheat, is classified into four classes depending on their solubility, the albumins being soluble in water, the globulins soluble in salt solution, the gliadins soluble in alcohol solution and the glutenins insoluble in neutral aqueous or saline solution and ethanol. Gluten, which is provided by both gliadins and glutenins, is the product of a ball of wheat flour dough that has been exhaustively washed in tap water, and the baking qualities of the wheat depends on its ability to trap carbon dioxide in dough. No nutritional value has been attributed to gluten. When collectively considered, the alcohol soluble fractions of cereals are designated as prolamins, a term reflecting the particular amino acid composition that is a high content of proline and glutamine and, depending on the cereal, they have been termed secalin for rye, hordein for barley, avenin for oats other than gliadin for wheat [17,18]. The gluten peptides and the related prolamins are responsible for triggering mucosal lesions in CD patients [18]. Among them, only gliadin has been investigated in great detail. Gliadin is an extremely heterogeneous mixture of proteins that contains at least 40 components which can be assigned, on the basis of their electrophoretic mobility at acidic pH, to four major groups (i.e. α-, β-, γ-, and ω-gliadins) [19] or, more modernly, into three major types according to their N-terminal amino acid sequence, designated as α-, γ- and ω-types [20,21]. Even if the correspondence between the old and the new nomenclature is not complete, it may be assumed that the electrophoretically separated α- and β-gliadins mainly constitute the α-type gliadins, while γ- and ω- gliadins are comprised into the γ- and ω-types, respectively [11].
Current understanding indicates that different gluten peptides are involved in the disease process in a different manner, some fragments being ‘toxic’ and others ‘immunogenic’. Specifically, a fragment is defined ‘toxic’ if it is able to induce mucosal damage either when added in culture to duodenal mucosal biopsy [22], or when administered in vivo on proximal and distal intestine [6,23,24], whereas a fragment is defined ‘immunogenic’ if it is able to specifically stimulate HLA-DQ2- [25–28] or DQ8- [29–31] restricted T cell lines and T cell clones derived from jejunal mucosa or peripheral blood of coeliac patients. More in detail, as concerns ‘toxicity’, using in vitro jejunal biopsy organ culture, it was demonstrated that α-, β-, γ-, and ω-gliadins have a decreasing toxicity and the presence of similar N-terminal aminoacid sequence in rye and barley prolamins suggests that a certain amino acid sequence may constitute the toxic determinant [32,33]. After the full 266 amino acid sequencing of A-gliadin [34], a fraction of α-gliadin, a panel of gliadin peptides was synthesized and tested to evaluate their toxicity [22,35–40] as listed in Table 1, while only in the last few years, by combining reverse phase high performance liquid chromatography and mass spectrometry, a very high number of gliadin peptides have been purified and tested for their immunogenicity. The immunogenic peptides (or epitopes) are listed in Table 2[25–31,41–46], and it is remarkable that some of them are immunodominant (those with activity designed as + + +), which is the ability of such epitopes to elicit a strong T cell response. In addition, this term refers to another two factors, which may be considered as the consequences of the previous feature, that are: first, an immunodominant epitope elicits a specific T cell response from intestinal biopsies of nearly all patients, whereas those which are simply immunogenic stimulate T cells only in some patients, and second, if dietary gluten is administered to a patients who has been on a gluten-free diet, T cells responsive toward the immunodominant epitopes are detectable in peripheral blood significantly earlier than T cells that respond to the immunogenic epitopes [47]. Furthermore, some peptides identified as inactive or simply immunogenic in their native form become very potent stimulators of T cells after treatment with tTG (see below) [48], a process that can lead to epitope focusing during the course of the disease [49].
Table 1.
Toxic gliadin peptides.
| Amino acid sequences | Position | Toxicity | References |
|---|---|---|---|
| VPVPQLQPQNPSQQQPQEQ | α 3–21 | – | [22,36,37] |
| PGQQQPFPPQQPY | α 31–43 | + | [39] |
| PGQQQPFPPQQPYPQPQPF | α 31–49 | + | [22,36,37] |
| PGQQQPFPPQQPYPQPQPFPSQQPY | α 31–55 | + | [36,39] |
| PQPQPFPSQQPY | α 44–55 | + | [39] |
| SQQPYLQLQPFPQPQLPY | α 51–70 | + | [40] |
| LQLQPFPQPQLPYPQPQLPY | α 56–75 | + | [57] |
| QQYPLGQGSFRPSQQNPQA | α 202–220 | – | [37] |
| LGQGSFRPSQQN | α 206–217 | + | [38] |
Table 2.
Immunogenic gliadin peptides.
| Amino acid sequences | Position | Immunogenicity | References |
|---|---|---|---|
| VRVPVPQLQPQNPSQQQPQ | α-gliadin: 1–19 | + | [41] |
| QNPSQQQPQEQVPLVQQQ | α-gliadin: 11–28 | + | [41] |
| QVPLVQQQQFPGQQQPFPPQ | α-gliadin: 21–40 | + | [41] |
| PGQQQPFPPQQPYPQPQPF | α-gliadin: 31–49 | + | [41] |
| FPGQQQPFPPQQPYPQPQPF | α-gliadin: 30–49 | + | [41] |
| QPYPQPQPFPSQQPYLQL | α-gliadin: 41–58 | + | [41] |
| PQPFPSQQPYLQLQPFPQ | α-gliadin: 46–63 | + | [42] |
| PQPQLPYPQPQLPY | α-gliadin: 62–75/(a) | +/++ + | [41,43] |
| QLQPFPQPQLPY | α-gliadin: 57–68 (a) | +/++ + | [25] |
| QLQPFPQ | α-gliadin: 57–63 (a) | + + + | [26] |
| LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | α-gliadin: 57–89/(a) | +/++ + | [44] |
| QLQPFPQPQLPY | α-gliadin: 58–69/(a) | +/++ + | [44] |
| PQPQLPYPQPQLPY | α-gliadin: 63–76/(a) | +/++ + | [44] |
| PFRPQQPYPQPQPQ | α-gliadin: 93–106 (a) | + | [45] |
| LIFCMDVVLQ | α-gliadin: 123–132 | + | [46] |
| QQPLQQYPLGQGSFRPSQQNPQAQG | α-gliadin: 198–222 | + | [27] |
| QYPLGQGSFRPSQQNPQA | α-gliadin: 203–220/(a) | +/+ | [31] |
| PSGQGSFQPS | α-gliadin: 205–214 | – | [27] |
| PSGQGSFQPSQQ | α-gliadin: 205–216/(a) | +/++ + | [27,29] |
| SGQGSFQPSQQN | α-gliadin: 206–217/(a) | +/++ + | [27,29] |
| QGSFQPSQQN | α-gliadin: 208–217/(a) | –/++ + | [27,29] |
| LQPQQPFPQQPQQPYPQQPQ | γ-gliadin: 60–79 | + | [43] |
| FPQQPQQPYPQQPQ | γ-gliadin: 66–78 | + | [43] |
| FSQPQQQFPQPQ | γ-gliadin: 102–113/(a) | –/+ | [43] |
| OQPQQSFPEQQ | γ-gliadin: 134–153/(a) | +/++ + | [28] |
| VQGQGIIQPQQPAQL | γ-gliadin: 222–236/(a) | +/+ | [45] |
| QQQQPPFSQQQQSPFSQQQQ | glutenin: 40–59/(a) | –/+ | [45] |
| QQPPFSQQQQPLPQ | glutenin: 46–60/(a) | –/+ | [45] |
| SGQGQRPGQWLQPGQGQQGYYPTSPQQSGQGQQLGQ | glutenin: 707–742/(a) | +/+ | [30] |
| PGQGQQGYYPTSPQQSGQ | glutenin: 719–736 | + | [30] |
| GYYPTSPQQSGQGQQLGQ | glutenin: 725–742 | + | [30] |
| GYYPTSPQQSG | glutenin: 725–735 | + | [30] |
| QGYYPTSPQQS | glutenin: 724–734/(a) | + – | [30] |
| QQGYYPTSPQQSG | glutenin: 723–735 | + | [30] |
| GQQGYYPTSPQQSG | glutenin: 722–735 | + | [30] |
| GQQGYYPTSPQQS | glutenin: 722–734 | + | [30] |
(a): deamidated.
As mentioned, a common feature among gluten epitopes is the presence of multiple proline and glutamine residues, which make them exceptionally impregnable by gastric, pancreatic and intestinal digestive proteases [50]. In addition, the repetitive presence of these residues makes the peptides a preferred substrate of tTG, whose main function is to catalyse the covalent and irreversible crosslinking of a glutamine residue in glutamine-donor proteins with a lysine residue in glutamine-acceptor proteins which results in the formation of an ɛ-(γ-glutamyl)-lysine (isopeptidyl) bond [51]. However, apart from crosslinking its substrates, tTG can also hydrolyse peptide-bound glutamine to glutamic acid either at a lower pH or when no acceptor proteins are available [52], a process leading to an enhanced immunogenicity of gluten peptides [49]. Furthermore, these peptides naturally adopt a structural configuration characterized by a left-handed polyproline II helical conformation, which is the one preferred by all bound HLA class II ligands [53].
Glutenins are also involved in T cell response, and specifically the native peptide HLA-DQ8-restricted glt04 707–742, whose minimal stimulatory sequence (residues 724–734) is repetitively present in the glutenin molecules [30], and the high molecular weight glutenin proteins in a HLA-DQ2-restricted and gliadin epitope-cross reactivity fashion [54]. It is remarkable that tTG specific deamidation does not influence the T cell recognition of the former peptide, whereas it is determinant in the latter.
More recently, using algorithms based on the spacing between glutamine and proline residues and on the presence or absence of other amino acids within the peptide, Vader et al. [32,55] predicted there are as many as 50 or more immunogenic peptides in gluten, hordeins, and secalins, but almost none in avenins. However, even though previous studies have also found the absence of oats toxicity in both paediatric and adult CD patients [56–58], Sollid and co-workers [59] recently provided evidence of stimulatory capacity of such peptides overall after deamidation by tTG. They clearly demonstrated that even if some of the avenin-reactive T cells were originally primed to gluten and responded to avenin because of cross-reactivity, there is also a specific avenin-driven immune response.
Finally, even though discrepancies exist between toxicity and immunogenicity, since a peptide defined as toxic does not necessarily activate intestinal T cells from coeliac patients and, conversely, some of the immunostimulant peptides may not prove toxic to the intestinal mucosa of CD patients, it is not excluded that the same peptide may have both capacities simultaneously [40,60]. Further studies using a combination of epitope identification by T cell clones and epitope confirmation by in vitro biopsy challenge will identify a valuable map of the gliadin fragments and other gluten peptides involved in disease pathogenesis.
Gluten and the innate immune response
Although a number of studies have been published concerning the role of T cells in inducing intestinal damage [61–63], little is known about the early stages in which gluten starts the whole process. The primary event of a gluten-induced immune response requires epitope-bearing oligopeptides to have access to lamina propria inside the relatively impermeable surface of the intestinal epithelial layer. Ordinarily, such oligopeptides are efficiently hydrolysed into amino acids, di- or tri-peptides by peptidases located in the brush border membrane of the differentiated enterocytes before they can be transported across the epithelium. The importance of digestive processing for gluten antigenicity is supported not only by the observed proximal to distal gradient of the coeliac lesions in the small intestine, but mainly by the lack of an immune response following the prolyl endopeptidase digestion of the 33-mer immunodominat peptide [44]. An increased transport throughout the intestinal epithelium of both toxic and immunogenic gliadin fragments has recently been demonstrated only in active CD [64], pleading against a constitutive defect and suggesting that the immaturity of enterocytes and/or inflammation are responsible for this event. On the same line are the results previously obtained by Zimmer et al. [65] who demonstrated that gliadin is translocated into vacuoles positive for HLA class II antigens in enterocytes of untreated CD patients, but not in those of healthy controls. It is well known that under inflammatory and/or stress stimuli, enterocytes express not only HLA class II molecules, but also MHC class I chain-related gene (MIC) and HLA E molecules that are recognized by the natural killer receptors NKG2D and CD94 present on intraepithelial lymphocytes (IELs) [66,67]. The recent discoveries that interleukin (IL)-15, a key regulatory cytokine which supports the homeostasis between innate and adaptive immunity [68], may be produced by intestinal epithelial cells [69] and is a potent stimulant of IELs [70], and that gliadin causes an overexpression of MIC-A in the intestinal mucosa of CD patients [71] have focused the attention on IL-15 as a key molecule in the pathogenesis of CD [67,72–74]. Maiuri et al. [75] demonstrated that the toxic gliadin peptide 31–43 is able to induce an innate immune response by up-regulating the expression of IL-15, cyclo-oxygenase-2, and the activation markers CD25 and CD83 in lamina propria cells having the characteristics of macrophages, monocytes, and dendritic cells, without binding to HLA-DQ2 or -DQ8 and without stimulating CD4+ T cells. This study clearly shows that certain gluten peptides elicit an innate response, while others drive the adaptive immunity. Interestingly, the innate response to peptide 31–43 is only found in HLA-DQ2 positive CD patients, but not in HLA-DQ2 positive noncoeliac controls. This peculiar innate response to gluten may explain why only some of DQ-2 positive individuals develop CD, the prevalence of DQ2-positivity in western populations ranging between 25% and 35%[4]. Recently, another gluten peptide that activates the innate immune system has emerged [76], and the existence of more such fragments can be expected.
However, it remains unknown how very small amounts of gluten peptides can enter the intestinal mucosa of genetically susceptible subjects to initiate the inflammatory cascade. In rat intestine, gliadin administration leads to increased permeability of tight junctions, mediated by zonulin [77], and in human coeliac mucosa an increased zonulin expression has been demonstrated [78]. It can thus be hypothesized that in CD a gliadin-dependent or -independent breaking of the mucosal barrier facilitates the delivery of gluten peptides to the lamina propria via the paracellular route.
Another unsolved problem is how gluten peptides can cross the basement membrane to directly interact with immune cells present in the lamina propria. In this regard, intestinal epithelial cells have been shown to release exosome-like vesicles morphologically similar to those secreted by professional antigen presenting cells (APC) in fact, very recently, Van Niel et al. [79] clearly demonstrated that exosomes derived from intestinal epithelial cells are antigen presenting vesicles bearing HLA class II/peptide complexes and that they are up-regulated by interferon-γ treatment. Gluten fragments may therefore interact with the underlying immune system through exosomes released from the basolateral side of enterocytes. Once present in the lamina propria, peptides undergo capturing and processing by professional APC. Up to now, no direct information is available concerning the mechanisms of antigen processing and presentation of gluten peptides by subepithelial dendritic cells either in normal condition or in CD, even though an interesting paper has recently been published concerning the influence of digested wheat gluten on bone marrow-derived dendritic cells of BALB/c mice [80]. The authors demonstrated that gluten peptides are able to induce both maturation and expression of costimulatory molecules by dendritic cells, and the secretion of chemokines responsible for leucocyte activation.
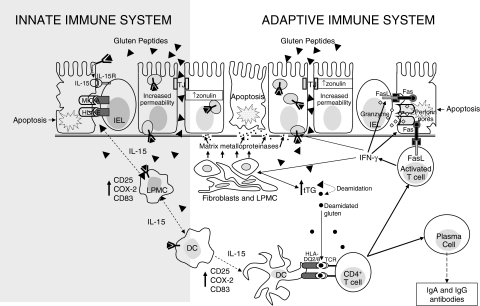
The described alterations in transport and processing of gluten peptides by intestinal mucosa of CD patients likely participate in the pathogenesis of the disease and the combined presence of toxic and immunogenic sequences may stimulate synergistically both the innate and adapive immune system (Fig. 1).
Fig. 1.
Epithelial transport and immune recognition of gluten in coeliac disease. In coeliac disease gluten peptides may cross the intestinal epithelium either via the paracellular route or via the transcellular route. In the first case this occurs as a consequence of an increased permeability due to an up-regulation of zonulin (which might be gliadin-dependent or gliadin-independent), in the second case it occurs by using enterocytic vesicles carrying the HLA class II-peptides complex. These vesicles are able to cross the basal membrane allowing intact gluten peptides to have access to the lamina propria. Here, they can activate both dendritic cells and lamina propria mononuclear cells to produce interleukin 15 which in turn causes an up-regulation of stress protein by enterocytes that are recognized by the natural killer receptors present on intraepithelial lymphocytes. On the other hand, native and tTG-deamidated gluten peptides are presented by mature dendritic cells to T cell receptors by using the heterodimer encoded by HLA-DQ2 or DQ8 genes A and B. This bond strongly activates CD4+ T cells which, by secreting pro-inflammatory cytokines, stimulate cytotoxic T cells and fibroblasts to produce a particular matrix metalloproteinase pattern which is responsible for degradation of both extracellular matrix and basement membrane. Activated T cells also become able to trigger enterocyte apoptosis by producing molecules like Fas ligand and granzyme, which are responsible for cytotoxicity, leading to the characteristic mucosal lesions. Stimulated CD4+ T cells are also able to induce lymphocyte B differentiation into plasma cells producing specific antigladin and anti-tissue transglutaminase antibodies. DC, dendritic cell; FasL, Fas ligand; HLA, human leucocyte antigen; IL, interleukin; IEL, intraepithelial lymphocyte; LPMC, lamina propria mononuclear cell; TCR, T cell receptor; TJ, tight junction; tTG, tissue transglutaminase.
Gluten and the adaptive immune response
The presence of an increased amount of gluten peptides in the lamina propria together with a genetic predisposition greatly increases the risk of breaking the oral tolerance toward these peptides. As concerns the genetic predisposition, a condition necessary but not sufficient for the development of the disease process, two highly polymorphic genes called DQA1 and DQB1, encoding the α/β transmembrane heterodimer expressed on the surface of APC [81] have been well characterized. The vast majority of patients, approximately 95%, have the DQ2 haplotype (DQA1*0501/DQB1*0201, termed HLA-DQ2.5 hereafter) and most of them are DR3-DQ2 homozygotes (DRB1*0301-DQA1*0501-DQB1*0201) or are DR5-DQ7 and DR7-DQ2 heterozygotes (that is DRB1*11/12-DQA1*0505-DQB1*0301 and DRB1*07-DQA1*0201-DQB1*0202, respectively) [82]. A second HLA-DQ2 molecule exists (DQA1*0201/DQB1*0202, termed HLA-DQ2.2 hereafter) with peptide-binding properties that are virtually identical to those of HLA-DQ2.5 [83]. However, HLA-DQ2.2 does not predispose for CD unless it is expressed together with HLA-DQ2.5 [84]. Specifically, individuals homozygous for HLA-DQ2.5 genotype or HLA-DQ2.2/2.5 heterozygous have a higher risk of developing CD, while those HLA-DQ2.5/non DQ2.2 heterozygous have only a slightly increased risk [82]. These findings have recently been confirmed by an in vitro study aimed at evaluating the ability of gluten peptides to stimulate T cell clones in the presence of an HLA-DQ2 positive EBV-transformed lymphoblastoid cell line used as APC [84]. The authors concluded that the risk associated with the HLA-DQ2 haplotype depends on the impact of the number and quality of the HLA-DQ molecules on gluten peptide presentation to T cells. The remainder of DQ2 negative coeliac patients (about 5%) mostly have a DQ8 molecule encoded by DQB1*0302 and DQA1*0301 on a DR4 haplotype [85].
In CD, the HLA-DQ2 and -DQ8 molecules bind protein fragments in their peptide grooves, and these complexes are expressed on the cellular surface of APC to be recognized by a specific population of CD4+ T cells. A very important finding is that optimal binding of some gluten peptides to HLA-DQ2/DQ8 requires their prior modification by tTG [86,87]. As previously mentioned [49,52], this enzyme has a high avidity for prolamins and possesses, at low pH, a deamidation activity which transforms neutral glutamine residues into negatively charged glutamic acid [88]. The introduction of negative charges into gluten peptides at specific positions favours their interaction with basic amino acids located in the anchor positions of HLA-DQ2/DQ8 molecules, and subsequently enhances peptide binding and T cell stimulation [86,87]. Whether immunogenic gluten peptides are recognized by mucosal T cells in an isolated form and/or in a complexed form, since the presence of gluten-tTG complexes has recently been demonstrated in coeliac mucosa [89], remains to be established.
It is of interest to note that the expression levels of HLA-DQ are under the influence of cytokines and particularly of interferon-γ[90]. Local production of interferon-γ, e.g. as the result of an infection, would thus lead to elevated HLA-DQ expression by professional and nonprofessional APC like enterocytes and, consequently, gluten peptide presentation, which can contribute to the development and/or progression of a gluten-specific T cell response. Furthermore, the X-ray crystal structure of the soluble domain of HLA-DQ2 bound to a deamidated epitope of αI-gliadin has recently been reported, and demonstrates the presence of an intricate hydrogen-bonding network between the two molecules [53]; it was demonstrated that the deamidated αI-gliadin has a 25-fold higher affinity compared with the nondeamidated counterpart probably due to having a more potent hydrogen-bond acceptor that can form additional protein-to protein hydrogen bonds, resulting in a more stable gliadin-DQ2 complex.
This properly presented epitope engages the T cell receptor on CD4+ T lymphocytes which then become able to activate cytotoxic T lymphocytes, macrophages, stromal cells and B lymphocytes by secreting a peculiar pattern of cytokines and matrix metalloproteinase [91], which in turn cause tissue damage and antibody production (Fig. 1).
Conclusions
It is hoped that the knowledge of the exact nature of the gluten peptides and of the mechanisms of their intestinal handling in CD patients will make it feasible to inhibit or prevent the disease by blocking the different steps of gluten recognition. It has been demonstrated that the substitution of a glutamine residue by a proline residue in both glutenin and gliadin epitopes lacked T cell recognition and then immunostimulatory activity [32], and it is also demonstrated that by a single amino acid substitution, the 31–49 peptide looses its toxic property [92]. Theoretically, the specific substitution at DNA level may allow the detoxification of gluten peptides without affecting the baking properties, but this procedure have some intrinsic problems since several nucleotide mutations might be necessary to eliminate T cell reactivity, gluten peptides are encoded by many different genes on distinct chromosomes and the final viability of such genetically modified wheat is unknown. Further difficulties are due to the presence of different epitopes by age, children being sensitive to an enormous number of epitopes in contrast to adult patients [45,49], and to the possibility that the removal of known epitopes could reveal alternative epitopes like what happens for avenin [59]. The alternative might be to detoxify gluten by the action of bacterial prolyl endopeptidase [44], such as vaccinotherapy by intranasal administration of whole gliadin or its epitopes able to induce systemic T-cell anergy [93,94], or the use of analogue peptides able to bind HLA class II molecules [53]. The future availability of an animal model of CD may make it possible to evaluate all the new therapeutical strategies for this pathological condition [95,96].
References
- 1.Fasano A, Berti I, Gerarduzzi T, et al. Prevalence of celiac disease in at-risk and not-risk groups in the United States: a large multicenter study. Arch Intern Med. 2003;163:286–92. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- 2.Maki M, Mustalahti K, Kokkonen J, et al. Prevalence of celiac disease among children in Finland. N Engl J Med. 2003;348:2517–24. doi: 10.1056/NEJMoa021687. [DOI] [PubMed] [Google Scholar]
- 3.Sollid LM. Celiac disease: dissecting a complex inflammatory disorder. Nature Rev Immunol. 2002;2:647–55. doi: 10.1038/nri885. [DOI] [PubMed] [Google Scholar]
- 4.Sollid LM, Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910–22. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- 5.Bevan S, Popat S, Braegger CP, et al. Contribution of the MHC region to the familial risk of coeliac disease. J Med Genet. 1999;36:687–90. [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh MN. Gluten, major histocompatibility complex, and the small intestine. Gastroenterology. 1992;102:330–54. [PubMed] [Google Scholar]
- 7.Marsh MN, Crowe PT. Morphology of the mucosal lesion in gluten sensitivity. Baillieres Clin Gastroenterol. 1995;9:273–93. doi: 10.1016/0950-3528(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 8.Marsh MN, Loft DE, Garner V, et al. The temporal/dose–response of coeliac mucosa to graded oral challenges with Frazer's fraction III. Eur J Gastroenterol Hepatol. 1992;4:667–73. [Google Scholar]
- 9.Freedman AR, Macartney JC, Nelufer JM, Ciclitira PJ. Timing of infiltration of T lymphocytes induced by gluten into the small intestine in coeliac disease. J Clin Pathol. 1987;40:741–5. doi: 10.1136/jcp.40.7.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biagi F, Corazza GR. Clinical features of coeliac disease. Dig Liver Dis. 2002;34:225–8. doi: 10.1016/s1590-8658(02)80197-9. [DOI] [PubMed] [Google Scholar]
- 11.Wieser H. The precipitating factor in coeliac disease. Baillieres Clin Gastroenterol. 1995;9:191–207. doi: 10.1016/0950-3528(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 12.Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, Schuppan D. Identification of tissue transglutaminase as the autoantigen in celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 13.Schuppan D, Esslinger B, Dieterich W. Innate immunity and coeliac disease. Lancet. 2003;362:3–4. doi: 10.1016/S0140-6736(03)13843-3. [DOI] [PubMed] [Google Scholar]
- 14.Scuppan D. Corrent concepts of celiac disease pathogenesis. Gastroenterology. 2000;119:234–42. doi: 10.1053/gast.2000.8521. [DOI] [PubMed] [Google Scholar]
- 15.Corrao G, Corazza GR, Bagnardi V, et al. Mortality in patients with coeliac disease and their relatives: a cohort study. Lancet. 2001;358:356–61. doi: 10.1016/s0140-6736(01)05554-4. [DOI] [PubMed] [Google Scholar]
- 16.Howdle PD, Jalal PK, Holmes GK, Houlston RS. Primary small-bowel malignancy in the UK and its association with coeliac disease. QJM. 2003;96:345–53. doi: 10.1093/qjmed/hcg058. [DOI] [PubMed] [Google Scholar]
- 17.Kreis M, Forde BG, Rahman S, Miflin BJ, Shewry PR. Molecular evolution of the seed storage proteins of barley, rye and wheat. J Mol Biol. 1985;183:499–502. doi: 10.1016/0022-2836(85)90017-8. [DOI] [PubMed] [Google Scholar]
- 18.Sturgess RP, Ellis HJ, Ciclitira PJ. Cereal chemistry, molecular biology, and toxicity in coeliac disease. Gut. 1991;32:1055–60. doi: 10.1136/gut.32.9.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones RW, Taylor NW, Senti FR. Electrophoresis and fractionation of wheat gluten. Arch Biochem Biophys. 1959;84:363–76. doi: 10.1016/0003-9861(59)90599-5. [DOI] [PubMed] [Google Scholar]
- 20.Wieser H, Môdl A, Seilmeir W, Belitz HD. High performance liquid chromatography of gliadins from different wheat varieties. amino acid composition and N-terminal amino acid sequence of components. Z Lebensm Unters Forsch. 1987;185:371–8. doi: 10.1007/BF01042814. [DOI] [PubMed] [Google Scholar]
- 21.Shewry PR, Tatham AS, Forde J, et al. The classification and nomenclature of wheat gluten proteins: a reassessment. J Cer Sci. 1986;4:97–106. [Google Scholar]
- 22.Howdle PD, Corazza GR, Bullen AW, Losowsky MS. Gluten sensitivity of small intestinal mucosa in vitro: quantitative assessment of histologic change. Gastroenterology. 1981;80:442–50. [PubMed] [Google Scholar]
- 23.Ellis HJ, Ciclitira PJ. In vivo gluten challenge in celiac disease. Can J Gastroenterol. 2001;15:243–7. doi: 10.1155/2001/127241. [DOI] [PubMed] [Google Scholar]
- 24.Lahteenoja H, Maki M, Viander M, Raiha I, Vilja P, Rantala I, Toivanen A, Syrjanen S. Local challenge on oral mucosa with an alpha-gliadin related synthetic peptide in patients with celiac disease. Am J Gastroenterol. 2000;95:2880–7. doi: 10.1111/j.1572-0241.2000.02257.x. [DOI] [PubMed] [Google Scholar]
- 25.Arentz-Hansen H, Korner R, Molberg Ø, et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. 2000;191:603–12. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. 2000;6:337–42. doi: 10.1038/73200. [DOI] [PubMed] [Google Scholar]
- 27.Van de Wal Y, Kooy YMC, van Veelen PA, et al. Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc Natl Acad Sci USA. 1998;95:10050–4. doi: 10.1073/pnas.95.17.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjostrom H, Lundin KEA, Molberg Ø, et al. Identification of a gliadin T-cell epitope in coeliac disease: general importance of gliadin deamidation for intestinal T-cell recognition. Scand J Immunol. 1998;48:111–5. doi: 10.1046/j.1365-3083.1998.00397.x. [DOI] [PubMed] [Google Scholar]
- 29.van de Wal Y, Kooy YMC, van Veelen P, Pena S, Mearin L, Papadopoulos G, Koning F. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol. 1998;161:1585–8. [PubMed] [Google Scholar]
- 30.van de Wal Y, Kooy YMC, van Veelen P, Vader W, August SA, Drijfhout JW, Pena SA, Koning F. Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol. 1999;29:3133–9. doi: 10.1002/(SICI)1521-4141(199910)29:10<3133::AID-IMMU3133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 31.Mazzarella G, Maglio M, Paparo F, et al. An immunodominant DQ8 restricted gliadin peptide activates small intestinal immune response in in vitro cultured mucosa from HLA-DQ8 positive but not HLA-DQ8 negative coeliac patients. Gut. 2003;52:57–62. doi: 10.1136/gut.52.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vader LW, Stepniak DT, Bunnik EM, Kooy YM, de Haan W, Drijfhout JW, Van Veelen PA, Koning F. Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology. 2003;125:1105–13. doi: 10.1016/s0016-5085(03)01204-6. [DOI] [PubMed] [Google Scholar]
- 33.Wieser H, Springer G, Belitz HD, Ashkenazi A, Idar D. Toxicity of different wheat gliadins in coeliac disease. Z Lebensm Unters Forsch. 1982;175:321–6. doi: 10.1007/BF01136247. [DOI] [PubMed] [Google Scholar]
- 34.Kasarda DD, Okita TW, Bernardin JE, Baecker PA, Nimmo CC, Lew EJ, Dietler MD, Greene FC. Nucleic acid (cDNA) and amino acid sequences of alpha-type gliadins from wheat (Triticum aestivum) Proc Natl Acad Sci USA. 1984;81:4712–6. doi: 10.1073/pnas.81.15.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson A, MacDonald TT, McClure JP, Holden RJ. Cell-mediated immunity to gliadin within the small-intestinal mucosa in coeliac disease. Lancet. 1975;1:895–7. doi: 10.1016/s0140-6736(75)91689-x. [DOI] [PubMed] [Google Scholar]
- 36.de Ritis G, Auricchio S, Jones HW, Lew EJ, Bernardin JE, Kasarda DD. In vitro (organ culture) studies of the toxicity of specific A-gliadin peptides in celiac disease. Gastroenterology. 1988;94:41–9. doi: 10.1016/0016-5085(88)90607-5. [DOI] [PubMed] [Google Scholar]
- 37.Sturgess R, Day P, Ellis HJ, Lundin KE, Gjertsen HA, Kontakou M, Ciclitira PJ. Wheat peptide challenge in coeliac disease. Lancet. 1994;343:758–61. doi: 10.1016/s0140-6736(94)91837-6. [DOI] [PubMed] [Google Scholar]
- 38.Mantzaris G, Jewell DP. In vivo toxicity of a synthetic dodecapeptide from A gliadin in patients with coeliac disease. Scand J Gastroenterol. 1991;26:392–8. doi: 10.3109/00365529108996500. [DOI] [PubMed] [Google Scholar]
- 39.Maiuri L, Troncone R, Mayer M, Coletta S, Picarelli A, De Vincenzi M, Pavone V, Auricchio S. In vitro activities of A-gliadin-related synthetic peptides: damaging effect on the atrophic coeliac mucosa and activation of mucosal immune response in the treated coeliac mucosa. Scand J Gastroenterol. 1996;31:247–53. doi: 10.3109/00365529609004874. [DOI] [PubMed] [Google Scholar]
- 40.Martucci S, Fraser JS, Biagi F, Corazza GR, Ciclitira PJ, Ellis HJ. Characterizing one of the DQ2 candidate epitopes in coeliac disease: A-gliadin 51–70 toxicity assessed using an organ culture system. Eur J Gastroenterol Hepatol. 2003;15:1293–8. doi: 10.1097/00042737-200312000-00007. [DOI] [PubMed] [Google Scholar]
- 41.Lundin KEA, Scott H, Hansen T, Paulsen G, Halstensen TS, Fausa O, Thorsby E, Sollid LM. Gliadin-specific, HLA-DQ (α1*0501,β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J Exp Med. 1993;178:187–96. doi: 10.1084/jem.178.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Wal Y, Kooy YM, Drijfhout JW, Amons R, Koning F. Peptide binding characteristics of the coeliac disease-associated DQ (alpha1*0501, beta1*0201) molecule. Immunogenetics. 1996;44:246–53. doi: 10.1007/BF02602553. [DOI] [PubMed] [Google Scholar]
- 43.Arentz-Hansen H, McAdam SN, Molberg Ø, et al. Celiac lesion T cells recognize epitopes that cluster in regions of gliadin rich in proline residues. Gastroenterology. 2002;123:803–9. doi: 10.1053/gast.2002.35381. [DOI] [PubMed] [Google Scholar]
- 44.Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–9. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 45.Vader W, Kooy Y, van Veelen P, et al. The gluten response in children with in celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology. 2002;122:1729–37. doi: 10.1053/gast.2002.33606. [DOI] [PubMed] [Google Scholar]
- 46.Gianfrani C, Troncone R, Mugione P, et al. Celiac disease association with CD8+ T cell responses: identification of a novel gliadin-derived HLA-DQ2-restricted epitope. J Immunol. 2003;170:2719–26. doi: 10.4049/jimmunol.170.5.2719. [DOI] [PubMed] [Google Scholar]
- 47.Stern M, Ciclitira PJ, van Eckert R, et al. Analysis and clinical effects of gluten in celiac disease. Eur J Gastroenterol Hepatol. 2001;13:741–7. doi: 10.1097/00042737-200106000-00023. [DOI] [PubMed] [Google Scholar]
- 48.Molberg Ø, McAdam SN, Korner R, et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med. 1998;4:713–7. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- 49.Martucci S, Corazza GR. Spreading and focusing of gluten epitopes in celiac disease. Gastroenterology. 2002;122:2072–5. doi: 10.1053/gast.2002.34102. [DOI] [PubMed] [Google Scholar]
- 50.Hausch F, Shan L, Santiago NA, Gray GM, Khosla C. Intestinal digestive resistance of immunodominant gliadin peptides. Am J Physiol Gastrointest Liver Physiol. 2002;283:G996–1003. doi: 10.1152/ajpgi.00136.2002. [DOI] [PubMed] [Google Scholar]
- 51.Folk JE. Mechanism and basis for specificity of transglutaminase-catalyzed ɛ-(γ-glutamyl) lysine bond formation. Adv Enzymol Relat Areas Mol Biol. 1983;54:1–56. doi: 10.1002/9780470122990.ch1. [DOI] [PubMed] [Google Scholar]
- 52.Folk JE, Finlayson JS. The ɛ-(γ-glutamyl) lysine crosslink and the catalytic role of transglutaminases. Adv Protein Chem. 1977;31:1–133. doi: 10.1016/s0065-3233(08)60217-x. [DOI] [PubMed] [Google Scholar]
- 53.Kim C-H, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci USA. 2004;101:4175–9. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molberg Ø, Solheim Flæte NS, Jensen T, Lundin KE, Arentz-Hansen H, Anderson OD, Kjersti Uhlen A, Sollid LM. Intestinal T-cell responses to high-molecular-weight glutenins in celiac disease. Gastroenterology. 2003;125:337–44. doi: 10.1016/s0016-5085(03)00890-4. [DOI] [PubMed] [Google Scholar]
- 55.Vader LW, de Ru A, van der Wal Y, et al. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med. 2002;195:643–9. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffenberg EJ, Haas J, Drescher A, Barnhurst R, Osberg I, Bao F, Eisenbarth G. A trial of oats in children with newly diagnosed celiac disease. J Pediatr. 2000;137:361–6. doi: 10.1067/mpd.2000.109003. [DOI] [PubMed] [Google Scholar]
- 57.Janatuinen EK, Kemppainen TA, Julkunen RJK, Kosma VM, Maki M, Heikkinen M, Uusitupa MI. No harm from five year ingestion of oats in coeliac disease. Gut. 2002;50:332–5. doi: 10.1136/gut.50.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kilmartin C, Lynch S, Abuzakouk M, Wieser H, Feighery C. Avenin fails to induce a Th1 response in coeliac tissue following in vitro culture. Gut. 2003;52:47–52. doi: 10.1136/gut.52.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arentz-Hansen H, Fleckenstein B, Molberg Ø, et al. The molecular basis for oat intolerance in patients with celiac disease. PLOS Med. 2004;1:84–92. doi: 10.1371/journal.pmed.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraser JS, Engel W, Ellis HJ, Moodie SJ, Pollock EL, Wieser H, Ciclitira PJ. Coeliac disease: in vivo toxicity of the putative immunodominant epitope. Gut. 2003;52:1698–702. doi: 10.1136/gut.52.12.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDonald TT, Spencer J. Evidence that activated mucosal T cells play a role in the pathogenesis of enteropathy in human small intestine. J Exp Med. 1988;16:1341–9. doi: 10.1084/jem.167.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciccocioppo R, Di Sabatino A, Parroni R, et al. Increased enterocyte apoptosis and Fas/Fas Ligand system in celiac disease. Am J Clin Pathol. 2001;115:494–503. doi: 10.1309/UV54-BHP3-A66B-0QUD. [DOI] [PubMed] [Google Scholar]
- 63.Di Sabatino A, Ciccocioppo R, D’Alò Parroni R, Millimaggi D, Cifone MG, Corazza GR. Intraepithelial and lamina propria lymphocytes show distinct pattern of apoptosis whereas both populations are active in Fas based cytotoxicity in coeliac disease. Gut. 2001;49:380–6. doi: 10.1136/gut.49.3.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matysiak-Budnik T, Candalh C, Dugave C, Namane A, Cellier C, Cerf-Bensussan N, Heyman M. Alterations of the intestinal transport and processing of gliadin peptides in celiac disease. Gastroenterology. 2003;125:696–707. doi: 10.1016/s0016-5085(03)01049-7. [DOI] [PubMed] [Google Scholar]
- 65.Zimmer KP, Naim H, Weber P, Ellis HJ, Ciclitira PJ. Targeting of gliadin peptides, CD8, alpha/beta-TCR and gamma/delta-TCR to Golgi complexes and vacuoles within celiac disease enterocytes. FASEB J. 1998;12:1349–57. doi: 10.1096/fasebj.12.13.1349. [DOI] [PubMed] [Google Scholar]
- 66.Braud VM, Allan DS, O'Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 67.Jabri B, Patey-Mariaud De Serre N, Cellier C, et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118:867–79. doi: 10.1016/S0016-5085(00)70173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tagaya Y, Bamford RN, DeFilippis AP, Waldmann TA. IL-15: a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity. 1996;4:329–36. doi: 10.1016/s1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- 69.Reinecker H, MacDermott R, Mirau S, Dignass A, Podolsky DK. Intestinal epithelial cells both express and respond to interleukin-15. Gastroenterology. 1996;111:1706–13. doi: 10.1016/s0016-5085(96)70036-7. [DOI] [PubMed] [Google Scholar]
- 70.Ebert EC. Interleukin 15 is a potent stimulant of intraepithelial lymphocytes. Gastroenterology. 1998;115:1439–45. doi: 10.1016/s0016-5085(98)70022-8. [DOI] [PubMed] [Google Scholar]
- 71.Martin-Pagola A, Perez-Nanclares G, Ortiz L, et al. MICA response to gliadin in intestinal mucosa from celiac patients. Immunogenetics. 2004;56:549–54. doi: 10.1007/s00251-004-0724-8. [DOI] [PubMed] [Google Scholar]
- 72.Maiuri L, Ciacci C, Auricchio S, Brown V, Quarantino S, Londei M. Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology. 2000;119:996–1006. doi: 10.1053/gast.2000.18149. [DOI] [PubMed] [Google Scholar]
- 73.Maiuri L, Ciacci C, Vacca L, Ricciardelli I, Auricchio S, Quarantino S, Londei M. IL-15 drives the specific migration of CD94+ and TCR-γδ+ intraepithelial lymphocytes in organ cultures of treated celiac patients. Am J Gastroenterol. 2001;96:150–6. doi: 10.1111/j.1572-0241.2001.03437.x. [DOI] [PubMed] [Google Scholar]
- 74.Hue S, Mention JJ, Monteiro RC, et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21:367–77. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 75.Maiuri L, Ciacci C, Ricciardelli I, et al. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet. 2003;362:30–7. doi: 10.1016/S0140-6736(03)13803-2. [DOI] [PubMed] [Google Scholar]
- 76.Tuckova L, Novotna J, Novak P, et al. Activation of macrophages by gliadin fragments: isolation and characterization of active peptide. J Leuk Biol. 2002;71:625–31. [PubMed] [Google Scholar]
- 77.Clemente MG, De Virgiliis S, Kang JS, et al. Early effects of gliadin on enterocyte intracellular signalling involved in intestinal barrier function. Gut. 2003;52:218–23. doi: 10.1136/gut.52.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fasano A, Not T, Wang W, Uzzau S, Berti I, Tomassini A, Goldblum SE. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. 2000;355:1518–9. doi: 10.1016/S0140-6736(00)02169-3. [DOI] [PubMed] [Google Scholar]
- 79.Van Niel G, Mallegol J, Bevilacqua C, et al. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52:1690–7. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nikulina M, Habich C, Flohé SB, Scott FW, Kolb HI. Wheat gluten causes dendritic cell maturation and chemokine secretion. J Immunol. 2004;173:1925–33. doi: 10.4049/jimmunol.173.3.1925. [DOI] [PubMed] [Google Scholar]
- 81.Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ α/β heterodimer. J Exp Med. 1989;169:345–50. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Louka AS, Nilsson S, Olsson M, et al. HLA in coeliac disease families: a novel test of risk modification by the ‘other’ haplotype when at least one DQA1*05-DQB1*02 haplotype is carried. Tissue Antigens. 2002;60:147–54. doi: 10.1034/j.1399-0039.2002.600205.x. [DOI] [PubMed] [Google Scholar]
- 83.van de Wal Y, Kooy YM, Drijfhout JW, Amons R, Papadopoulos GK, Koning F. Unique peptide binding characteristics of the disease-associated DQ (alpha 1*0501, beta 1*0201) vs the non-disease-associated DQ (alpha 1*0201, beta 1*0202) molecule. Immunogenetics. 1997;46:484–92. doi: 10.1007/s002510050309. [DOI] [PubMed] [Google Scholar]
- 84.Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, Spaenij L, Koning F. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA. 2003;100:12390–5. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sidney J, del Guercio MF, Southwood S, Sette A. The HLA molecules DQA1*0501/B1*0201 and DQA1*0301/B1*0302 share an extensive overlap in peptide binding specificity. J Immunol. 2002;169:5098–108. doi: 10.4049/jimmunol.169.9.5098. [DOI] [PubMed] [Google Scholar]
- 86.Quarsten H, Molberg Ø, Fugger L, McAdam SM, Sollid LM. HLA-binding and T cell recognition of a tissue transglutaminase-modified gliadin epitope. Eur J Immunol. 1999;29:2506–14. doi: 10.1002/(SICI)1521-4141(199908)29:08<2506::AID-IMMU2506>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 87.Molberg Ø, McAdam SN, Lundin KEA, Kristiansen C, Arentz-Hansen H, Kett K, Sollid LM. T cells from celiac disease lesions recognize gliadin epitopes deamidated in situ by endogenous tissue transglutaminase. Eur J Immunol. 2001;31:1317–23. doi: 10.1002/1521-4141(200105)31:5<1317::AID-IMMU1317>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 88.Fleckenstein B, Molberg Ø, Qiao SW, Schmid DG, von der Mulbe F, Elgstoen K, Jung G, Sollid LM. Gliadin T cell epitope selection by tissue transglutaminase in celiac disease. role of enzyme specificity and pH influence on the transamidation versus deamidation reactions. J Biol Chem. 2002;277:34109–16. doi: 10.1074/jbc.M204521200. [DOI] [PubMed] [Google Scholar]
- 89.Ciccocioppo R, Di Sabatino A, Ara C, Biagi F, Perilli M, Amicosante G, Cifone MG, Corazza GR. Gliadin and tissue transglutaminase complexes in normal and coeliac duodenal mucosa. Clin Exp Immunol. 2003;134:516–24. doi: 10.1111/j.1365-2249.2003.02326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 91.Ciccocioppo R, Di Sabatino A, Bauer M, et al. Matrix metalloproteinase pattern in celiac duodenal mucosa. Laboratory Invest. 2005;85:397–407. doi: 10.1038/labinvest.3700225. [DOI] [PubMed] [Google Scholar]
- 92.Biagi F, Ellis HJ, Parnell ND, Shidrawi RG, Thomas PD, O'Reilly N, Corazza GR, Ciclitira PJ. A non-toxic analogue of a celiac-activating gliadin peptide: basis for immunomodulation? Aliment Pharmacol Ther. 1999;13:945–50. doi: 10.1046/j.1365-2036.1999.00512.x. [DOI] [PubMed] [Google Scholar]
- 93.Maurano F, Siciliano RA, De Giulio B, Luongo D, Mazzeo MF, Troncone R, Auricchio S, Rossi M. Intranasal administration of one alpha gliadin can downregulates the immune response to whole gliadin in mice. Scand J Immunol. 2001;53:290–5. doi: 10.1046/j.1365-3083.2001.00869.x. [DOI] [PubMed] [Google Scholar]
- 94.Senger S, Luongo D, Maurano F, et al. Intranasal administration of a recombinant alpha-gliadin down-regulates the immune response to wheat gliadin in DQ8 transgenic mice. Immunol Lett. 2003;88:127–34. doi: 10.1016/s0165-2478(03)00069-5. [DOI] [PubMed] [Google Scholar]
- 95.Black KE, Murray JA, David CS. HLA-DQ determines the response to exogenous wheat proteins: a model of gluten sensitivity in transgenic knockout mice. J Immunol. 2002;169:5595–600. doi: 10.4049/jimmunol.169.10.5595. [DOI] [PubMed] [Google Scholar]
- 96.Marietta E, Black K, Camilleri M, Krause P, Rogers RS, 3rd, David C, Pittelkow MR, Murray JA. A new model for dermatitis herpetiformis that uses HLA-DQ8 transgenic NOD mice. J Clin Invest. 2004;114:1090–7. doi: 10.1172/JCI21055. [DOI] [PMC free article] [PubMed] [Google Scholar]