Abstract
Hyper-immunoglobulin E (IgE) syndrome (HIES) is one of the primary immunodeficiency syndromes. Although the cytokine dysregulation is suggested to play a role in its pathophysiology, the causative gene has not yet been identified. To investigate the pathophysiology and candidate genes involved in this disease, we performed microarray analysis of unstimulated peripheral CD4+ T cells and CD14+ cells, as well as peripheral blood mononuclear cells (PBMNC) stimulated with Staphylococcus aureus isolated from HIES patients and healthy controls. By microarray analysis, 38 genes showed over 2-fold differences between the HIES patients and healthy controls in purified CD14+ cells, although only small differences in the gene expression profiles were observed between the two groups in purified CD4+ T cells. RGC32 expression levels showed the greatest difference between the two groups, and were significantly elevated in HIES compared with those in severe atopic dermatitis or healthy controls using real-time PCR. A significantly larger number of lysosome-related genes were up-regulated, and significantly larger number of genes related to cell growth and maintenance were down-regulated in HIES. After the stimulation of PBMNC with Staphylococcus aureus, 51 genes showed over 3-fold differences between HIES patients and healthy controls. A significantly large number of immunoglobulin-related genes were up-regulated in HIES. The distinct patterns of gene expression profiles and RGC32 expression levels will be useful for understanding the pathophysiology and for diagnosis of HIES, respectively.
Keywords: hyper-immunoglobulin E syndrome, microarray/ genomics/ proteomics, immunodeficiency–primary
Introduction
Hyper-immunoglobulin E (IgE) syndrome (HIES) is a rare primary immunodeficiency, characterized by severe recurrent staphylococcal infections, eczema, and markedly elevated levels of IgE [1,2]. Pneumatoceles, dental and skeletal abnormalities and neutrophil defects are sometimes observed in patients with HIES [3,4]. Since the causative gene of HIES has not been identified, the diagnosis of HIES is sometimes difficult because of the similarity of symptoms to severe atopic dermatitis, particularly during early infancy.
Previous studies suggested that there might be a Th1/Th2 imbalance, especially a defect in Th1 induction [5–9], which may account for the high serum levels of IgE and eosinophilia in HIES. In other reports, mitogen-induced secretion of IL-4 and IFN-γ by PBMNC was not different between patients with HIES and healthy controls [10,11]. Susceptibilities to bacterial and fungal infections in HIES indicate possible defects in phagocytes and T cells, while bone and dental abnormalities suggest certain defects in monocyte-lineage cells such as osteoclasts and osteoblasts.
Thus, to assess the pathogenesis and pathophysiology of the HIES, we purified CD4+ T cells and CD14+ cells from HIES patients and analysed the whole gene expression with microarray. HIES showed characteristic gene expression patterns compared with controls. Analysis of distinct gene expression patterns would be useful in the understanding of the pathophysiology as well as in the diagnosis of HIES.
Methods
Patients
Eight patients (median age 16·5 years, range 5–34 years; 3 males and 5 females) with HIES were enrolled in this study. The patients’ characteristics are shown in Table 1. The samples obtained from patients 4 and 7 (Table 1) were used in the microarray analysis. As the controls, peripheral blood was obtained from 17 age-matched patients with severe atopic dermatitis (median age 8 years, range 1·0–34 years; 11 males and 6 females) and 15 healthy donors (median age 11 years, range 3–20 years, 8 males and 7 females). All the atopic patients had typical skin manifestation with increased serum levels of IgE (range 183–19504 IU/ml, median 3438 IU/ml) with or without eosinophilia (range 308–2018/µl, median 860/µl), and did not have the recurrent infection, lung cyst, abnormal facies, or bone abnormalities. The median values (ranges) of points of HIES diagnosis scoring system [12] were 25 (18–31) in HIES patients, and 16 (4–25) in atopic dermatitis patients (P < 0·01, by Mann–Whitney's test). Informed consent was obtained from all patients and healthy donors. This study was approved by Regional Committee of Ethics for Human research at Faculty of Medicine of Kyushu University.
Table 1.
Clinical characteristics of the patients with HIES.
| Patient | Age | Gender | Eczematous rash | Skin abscesses | Lymphadenitis | Pneumonia | Lung cyst | Characteristic facies | Retained primary teeth | Fractures with minor trauma | Articular over-extension | Scoliosis | Serum IgE level (IU/ml) | St. aureus-specific IgE (U/ml)* | Eosinophil count (cells/µl) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5y | Female | + | + | + | + | – | + | – | – | + | – | 40100 | 2260 | 2088 |
| 2 | 9y | Male | + | + | + | + | – | + | – | + | – | + | 27180 | 1675 | 9520 |
| 3 | 11y | Male | + | + | + | + | – | – | – | – | – | – | 17420 | 1331 | 1884 |
| 4 | 16y | Female | + | + | + | + | + | + | + | + | – | – | 37780 | 2219 | 1198 |
| 5 | 17y | Female | + | + | – | + | – | + | – | – | – | + | 16000 | 460 | 3663 |
| 6 | 18y | Female | + | + | + | + | – | + | – | – | – | – | 24807 | 2305 | 1064 |
| 7 | 18y | Female | + | + | + | + | + | + | + | – | – | + | 18300 | 716 | 18300 |
| 8 | 34y | Male | + | + | + | + | – | – | + | + | + | – | 28100 | 14530 | 1480 |
Normal range: < 400 U/ml.
Isolation of CD4+ T cells and CD14+ cells
PBMNC were separated immediately using density-gradient centrifugation using LSM (Cappel-ICN Immunobiologicals, Costa Mesa, CA, USA). CD4+ T cells were purified using cell sorter (EPICS ALTRA; Beckman Coulter, Hialeah, FL, USA) after staining with phycoerythrin (PE)-conjugated anti-CD4 antibody (Beckman Coulter). CD14+ cells were isolated using CD14 MicroBeads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). The purity of CD4+ T cells or CD14+ cells was higher than 97%.
PBMNC stimulation with Staphylococcus aureus
PBMNC were incubated in 48 well-culture plates at the concentration of 1 × 106 cells/ml. Heat killed Staphylococcus aureus (frozen at the concentration of 1 × 109 CFU/ml) was added at the concentration of 1 × 107 CFU/ml. After 6 h, the cells were harvested, washed twice with phosphate buffered saline, and used for total RNA extraction.
RNA extraction, amplification and labelling
Total RNA was extracted using Isogen (Nippon Gene, Tokyo, Japan). For the microarray analysis of purified CD4+ T cells or CD14+ cells, linear amplification of RNA was carried out using an Amino Allyl Message Amp aRNA Kit (Ambion, Austin, TX, USA). Briefly, double-stranded complementary DNA (cDNA) was synthesized from total RNA using oligo dT primer with a T7 RNA polymerase promoter site added to the 3′ end. Then, in vitro transcription was performed in the presence of amino allyl UTP to produce multiple copies of amino allyl labelled complementary RNA (cRNA). Amino allyl labelled cRNA was purified, and then 15 µg of cRNA was reacted with N-hydroxy succinimide esters of Cy3 (Amersham Pharmacia Biotech, Piscataway, NJ, USA) for cRNA from the purified cells, and Cy5 (Amersham Pharmacia Biotech) for that from 5 control PBMNC, according to the protocol of Hitachi Software Engineering (Yokohama, Japan).
For the gene expression analysis of PBMNC before and after the stimulation with Staphylococcus aureus, cDNA microarray was applied. After the linear amplification using RiboAmp RNA Amplification Kit (Arcturus Bioscience Mountain View, CA, USA), FluoroLink Cy3-dUTP- or FluoroLink Cy5-dUTP-labelled cDNA was synthesized using RNA Fluorescence Labeling Core Kit (M-MLV version) ver.2·0 (Takara Bio, Tokyo Japan), according to the manufacturer's instructions.
Microarray analysis
Microarray analysis for the purified CD4+ T cells and CD14+ cells was performed using an AceGene Human Oligo Chip 30K (Hitachi Software Engineering) that contains approximately 30 000 genes. Each gene expression level of CD4+ T cells and CD14+ cells from the patients or controls was determined by comparison with that from the standard sample. The arrays were scanned by FLA-8000 (Fuji Photo Film, Tokyo, Japan), and changed to the numerical values by ArrayVision (Amersham Biosciences). The numerical data were normalized using LOWESS method. In the microarray analysis of CD4+ T cells, data from 2 HIES patients and those from 2 healthy controls were compared. In the analysis of CD14+ cells, data from 2 HIES patients were compared with those from 2 healthy controls and one with severe atopic dermatitis to find out genes that were expressed specifically in HIES. Genes that were consistently up-regulated or consistently down-regulated in both HIES patients compared with all controls, and that showed over 2·0-fold differences by the comparison between the two groups in the mean expression levels were selected.
For the microarray analysis of PBMNC before and after the stimulation with Staphylococcus aureus, Agilent Human 1 cDNA Microarray (Agilent Technology, Palo Alto, CA, USA) was used, which contains 13 767 genes. The changes of the gene expression were analysed by direct comparison of the samples before and after the stimulation with Staphylococcus aureus in two patients with HIES, 2 healthy controls. The data were scanned by FLA-8000 (Fuji Photo Film), and changed to the numerical values by ArrayVision (Amersham Biosciences). The ratios of each gene expression level before and after the stimulation were obtained and normalized by being divided by the median value in each plate. Genes that were expressed consistently higher or consistently lower in both HIES patients compared with all controls, and that showed over 3·0-fold differences by the comparison between the two groups in the mean expression levels were selected.
The data with low signal-to-noise ratios (S/N < 3) were not used for further analysis. The data were analysed using Gene Spring software (Silicon Genetics, Redwood City, CA, USA).
Real-time PCR
An Assay-on-Demand Gene Expression Product Hs.00204129_m1 (Applied Biosystems) was used for primers and probes for response gene to complement 32 (RGC32). A Pre-Developed TaqMan Assay Reagent Human ACTB (β-actin) (Applied Biosystems) was used for the internal control. TaqMan assay was performed according to the manufacturer's instructions. In brief, PCR primer set and TaqMan probe for the target gene were added in the TaqMan Universal PCR Master Mix (Applied Biosystems) at the final volume of 25 µl. PCR condition was as follows: 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of amplification at 94°C for 15 s and 60°C for 1 min. PCR amplification and the detection of the amounts of PCR products were performed by ABI PRISM 7700 Sequence Detector (Applied Biosystems). The gene expression levels were described by the relative ratios of the levels of target genes to those of the internal control (β-actin gene).
Statistical analyses
Chi-square test was used to analyse the deviated distribution of genes in each Gene Ontology into the two groups. One-way anova test was used for the comparison of gene expression levels in three groups, and comparison of the two groups among 3 was performed with Scheffe's test. The differences were considered to be significant when the P-value < 0·05.
Results
To evaluate the gene expression profiles of circulating CD4+ T cells and CD14+ cells (monocytes) from patients with HIES, we extracted mRNAs from purified cells and performed microarray analysis, together with those from non-HIES controls. Six up-regulated and 4 down-regulated genes in HIES showed over 2-fold differences compared with controls in CD4+ T cells (Table 2), while 33 up-regulated and 5 down-regulated genes in HIES showed over 2-fold differences compared with controls in CD14+ cells (Table 3). The expression levels of major Th1- or Th2-related genes in CD4+ T cells and CD14+ cells did not show more than 2-fold differences between HIES patients and controls (Tables 2 and 3). In addition, there were only small differences in the expression levels of most cytokine-related genes between HIES patients and controls (data, not shown). Among genes with over 2-fold differences in the expression between HIES patients and controls, there was no gene with a functional relevance to the abnormalities of bone and neutrophils, and increased serum levels of IgE, which might directly relate to the pathogenesis of HIES.
Table 2.
Microarray analysis of purified CD4+ T cells between HIES and healthy controls.
| Gene name | Synonyms | GeneBank | *Fold difference |
|---|---|---|---|
| HIES > Control | |||
| Haemoglobin, delta | HBD | NM_000519 | 2·4 (3·0, 2·0) |
| Chromosome 6 open reading frame 111 | C6orf111 | AF161424 | 2·3 (2·5, 2·1) |
| Haemoglobin, alpha 2 | HBA2 | AF281258 | 2·3 (2·5, 2·1) |
| No name | XM_031045 | 2·2 (2·2, 2·2) | |
| Dynein, cytoplasmic, light intermediate polypeptide 2 | DNCLI2 | NM_006141 | 2·2 (2·8, 1·7) |
| Topoisomerase-related function protein 4–2 | AF089897 | 2·0 (2·2, 1·9) | |
| HIES < Control | |||
| Polymerase (RNA) II (DNA directed) polypeptide D | POLR2D | BC010427 | 1/3·7 (1/3·8, 1/3·6) |
| No name | AC026785·3.13728·33112·2 | 1/2·3 (1/2·7, 1/1·9) | |
| Sarcoglycan, gamma (35 kDa dystrophin-associated glycoprotein) | SGCG | NM_000231 | 1/2·3 (1/2·7, 1/1·9) |
| No name | U82695·2.1·167460·8 | 1/2·0 (1/2·6, 1/1·6) | |
The difference of mean gene expression levels between 2 HIES patients and 2 healthy controls in microarray analysis is given. Values in parentheses are the fold differences of the gene expression in each HIES patient compared with the mean expression value in healthy controls. Genes that showed more than 2 fold expressional differences between HIES and healthy controls were selected.
Table 3.
Microarray analysis of purified CD14+ cells between HIES and healthy controls.
| Gene name | Synonyms | GeneBank | *Fold difference |
|---|---|---|---|
| HIES > Control | |||
| Response gene to complement 32 | RGC32 | NM_014059 | 3·8 (4·2, 3·5) |
| Homeo box A1 | HOXA1 | NM_005522 | 3·8 (3·9, 3·7) |
| Cathepsin L | CTSL | NM_001912 | 3·2 (2·1, 4·8) |
| C-type lectin, superfamily member 2 (activation-induced) | CLECSF2 | NM_005127 | 3·1 (1·8, 5·3) |
| Epstein-Barr virus induced gene 2 | EBI2 | NM_004951 | 2·9 (2·8, 2·9) |
| Fatty acid binding protein 5 (psoriasis-associated) | FABP5 | NM_001444 | 2·8 (1·6, 5·0) |
| Myristoylated alanine-rich protein kinase C substrate | MARCKS | XM_047795 | 2·6 (2·2, 3·2) |
| Interleukin 1 receptor, type II | IL1R2 | NM_004633 | 2·6 (3·4, 1·9) |
| Adaptor-related protein complex 3, sigma 1 subunit | AP3S | XM_003926 | 2·4 (2·6, 2·3) |
| CD3D antigen, delta polypeptide (TiT3 complex) | CD3D | M12727 | 2·4 (1·6, 3·7) |
| Carboxypeptidase, vitellogenic-like | CPVL | AF106704 | 2·4 (3·2, 1·8) |
| Receptor-interacting serine-threonine kinase 2 | RIPK2 | NM_003821 | 2·3 (3·0, 1·8) |
| Hypothetical protein FLJ20186 | FLJ20186 | NM_017702 | 2·3 (1·5, 3·7) |
| Protein tyrosine phosphatase, receptor type, E | PTPRE | NM_006504 | 2·3 (2·5, 2·1) |
| X Kell blood group precursor-related, Y-linked | XKRY | NM_004677 | 2·3 (3·1, 1·6) |
| Toll-like receptor 1 | TLR1 | NM_003263 | 2·3 (1·8, 2·8) |
| Spermidine/spermine N1-acetyltransferase | SAT | BC002503 | 2·2 (2·7, 1·9) |
| KIAA0431 protein | KIAA0431 | NM_015251 | 2·2 (2·0, 2·4) |
| Vimentin | VIM | NM_003380 | 2·2 (2·8, 1·8) |
| Pleckstrin homology, Sec7 and coiled-coil domains, binding protein | PSCDBP | NM_004288 | 2·2 (2·3, 2·0) |
| Recombining binding protein suppressor of hairless (Drosophila) | RBPSUH | NM_015874 | 2·2 (1·6, 2·9) |
| Proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | PSMD12 | AB003103 | 2·2 (2·4, 1·9) |
| Multiple ankyrin repeats, single KH-domain (MASK) homolog | MASK | NM_024668 | 2·2 (2·5, 1·9) |
| Ubiquitin-conjugating enzyme E2B (RAD6 homolog) | UBE2B | NM_003337 | 2·2 (1·6, 2·8) |
| Syndecan 2 | SDC2 | AK025488 | 2·2 (1·6, 2·8) |
| Sialyltransferase 10 (alpha-2,3-sialyltransferase VI) | SIAT10 | XM_044802 | 2·1 (1·8, 2·6) |
| Rearranged l-myc fusion sequence | RLF | NM_012421 | 2·1 (2·2, 2·0) |
| RAB6 interacting protein 1 | RAB6IP1 | AK025499 | 2·1 (2·5, 1·8) |
| No name | XM_008071 | 2·1 (2·0, 2·2) | |
| CDC-like kinase 1 | CLK1 | NM_004071 | 2·1 (2·5, 1·7) |
| Hypothetical protein FLJ22965 | FLJ22965 | NM_022101 | 2·1 (2·2, 1·9) |
| Hypothetical protein CL25022 | CL25022 | AF131802 | 2·0 (1·8, 2·3) |
| Opioid growth factor receptor-like 1 | OGFRL1 | NM_024576 | 2·0 (2·2, 1·8) |
| HIES < Control | |||
| Dynein, cytoplasmic, light polypeptide 1 | DNCL1 | NM_003746 | 1/2·9 (1/1·9, 1/4·4) |
| Calmodulin regulated spectrin-associated protein 1 | CAMSAP1 | NM_018627 | 1/2·4 (1/1·7. 1/3·2) |
| Placenta-specific 8 | PLAC8 | NM_016619 | 1/2·1 (1/1·6, 1/2·8) |
| Vacuolar protein sorting 35 (yeast) | VPS35 | NM_018206 | 1/2·0 (1/1·6, 1/2·6) |
| Butyrophilin, subfamily 3, member A1 | BTN3A1 | NM_007048 | 1/2·0 (1/1·5, 1/2·8) |
The difference of mean gene expression levels between 2 HIES patients and 3 controls (2 healthy donors and a atopy patient) in microarray analysis is given. Values in parentheses are the fold differences of the gene expression in each HIES patient compared with the mean expression value in controls. Genes that showed more than 2 fold expressional differences between HIES and healthy controls were selected.
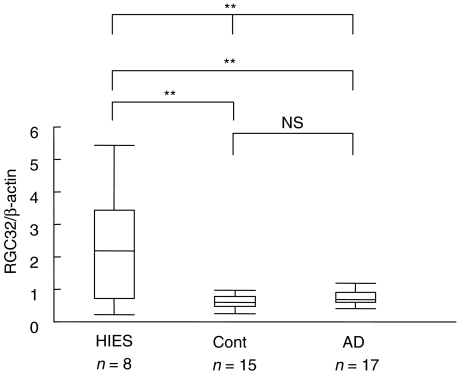
The expression levels of most up-regulated or down-regulated genes in CD14+ cells that showed over 2-fold differences between HIES patients and controls were confirmed by real-time quantitative PCR. The expression levels of RGC32, with the highest expression among 33 up-regulated genes in HIES in CD14+ cells by microarray analysis (Table 3), were significantly increased in PBMNC of HIES patients, compared with those in patients with atopic dermatitis or controls (Fig. 1). We were not able to analyse the expression levels of DNCL1 (dynein, cytoplasmic, light polypeptide 1) gene, with the lowest expression among 5 down-regulated genes in HIES in CD14+ cells by microarray analysis (Table 3), because the primers and probe were unable to be set to obtain precise and reproducible results due to the presence of many cDNA-type pseudogenes.
Fig. 1.
Comparison of RGC32 expression in patients with HIES, severe atopic dermatitis, and healthy controls. RGC32 expression in PBMNC was analysed by real-time PCR. HIES: patients with HIES, Cont: healthy controls, AD: patients with severe atopic dermatitis, NS: not significant. Whiskers indicate the ranges, boxes the 25th to 75th percentiles and horizontal bars inside boxes the median values. Three groups were compared using One-way anova test and each two groups were compared using Scheffe's test. **P < 0·01.
We compared the genes expressed in CD4+ T cells or CD14+ cells between HIES patients and controls, according to Gene Ontology Classifications from Gene Ontology Consortium to assess the differences in functionally related gene groups. At first, the genes that showed consistent expressional differences between HIES patients and controls in the two individual experiments were selected (HIES > normal: 1507 genes, HIES < control: 1116 genes). Among them, statistical analysis was performed in each gene group by the χ2 test (Table 4). In unstimulated CD4+ T cells, there were no gene groups which showed significant expressional differences between HIES patients and controls (data not shown). In unstimulated CD14+ cells, a significantly larger number of lysosome-related genes was up-regulated (26 (1·73%) of 1507 total up-regulated genes, P < 0·05), and a significantly larger number of genes related to the cell growth and maintenance was down-regulated (246 (22·0%) of 1116 total down-regulated genes, P < 0·05) in HIES (Table 4).
Table 4.
Comparison of gene expression according to the Gene Ontology groups.
| HIES > Control | HIES < Control | P-value* | |
|---|---|---|---|
| CD14+ cells | (1507 genes) | (1116 genes) | |
| Lysosome related genes (total 137 genes) | 26 (1·73%) | 8 (0·72%) | < 0·05 |
| Genes related to the cell growth and maintenance (total 2572 genes) | 254 (16·9%) | 246 (22·0%) | < 0·01 |
| MNC stimulated with Staphylococcus aureus | (1194 genes) | (1331 genes) | |
| Immunoglobulin-related genes (total 54 genes) | 29 (2·43%) | 4 (0·30%) | < 0·01 |
χ2 test.
Recurrent staphylococcal infection is one of the characteristics of patients with HIES. We stimulated PBMNC with heat-killed Staphylococcus aureus, and analysed the alteration of mRNA expression levels in HIES patients and healthy controls. Table 5 shows the genes with over 3·0-fold differences in the expression between HIES patients and controls before and after Staphylococcus aureus stimulation. Among cytokine-related genes, the relative expression levels of IL-17 and small inducible cytokine subfamily B (Cys-X-Cys), member 11 (CXCL11) genes after stimulation were decreased to 1/5·5 and 1/3·8, respectively, in HIES (Table 5). Analysis of the differences in functionally related gene groups in stimulated PBMNC from HIES patients and controls according to Gene Ontology Classifications revealed that a significantly larger number of immunoglobulin-related genes were up-regulated in HIES (29 of 1194 total up-regulated genes: 2·43%, P < 0·05 χ2 test, Table 4).
Table 5.
Microarray analysis of the change of gene expression after the stimulation with Staphylococcus aureus.
| Fold increase of gene expression* | |||||
|---|---|---|---|---|---|
| Gene name | Synonyms | Gene bank | HIES | Control | Fold difference** HIES/Control |
| HIES > Control | |||||
| Ubinuclein 1 | UBN1 | AF108460 | 3·5 | 1/1·4 | 4·9 (5·0, 4·9) |
| Rho guanine nucleotide exchange factor (GEF) 1 | ARHGEF1 | Y09160 | 2·7 | 1/1·7 | 4·7 (2·7, 8·3) |
| Hydroxymethylbilane synthase | HMBS | X04808 | 3·4 | 1/1·3 | 4·3 (1·4, 13) |
| Bromodomain containing 3 | BRD3 | D26362 | 2·5 | 1/1·6 | 4·0 (3·7, 4·4) |
| Incyte EST | 2·9 | 1/1·3 | 3·9 (2·8, 5·2) | ||
| KIAA0582 protein | KIAA0582 | AK000856 | 1·3 | 1/3·0 | 3·8 (3·4, 4·2) |
| A kinase (PRKA) anchor protein (yotiao) 9 | AKAP9 | NM_005751 | 2·1 | 1/1·8 | 3·7 (4·6, 2·9) |
| Haloacid dehalogenase-like hydrolase domain | HDHD1A | M86934 | 2·6 | 1/1·4 | 3·7 (1·3, 11) |
| KIAA0555 gene product | AL137976 | 5·1 | 1·4 | 3·7 (2·3, 5·8) | |
| Ral GEF with PH domain and SH3 binding motif 2 | RALGPS2 | AK001106 | 1·6 | 1/2·2 | 3·6 (3·2, 3·9) |
| Tripartite motif-containing 2 | TRIM2 | AB011089 | 1·5 | 1/2·4 | 3·5 (2·6, 4·7) |
| Serine (or cysteine) proteinase inhibitor, member 1 | SERPINI1 | NM_005025 | 2·1 | 1/1·6 | 3·5 (7·9, 1·5) |
| Cysteine conjugate-beta lyase; cytoplasmic | CCBL1 | X82224 | 1·4 | 1/2·4 | 3·4 (3·3, 3·5) |
| Homeo box A7 | HOXA7 | AJ005814 | 3·2 | 1·0 | 3·3 (3·7, 3·0) |
| Scaffold attachment factor B2 | SAFB2 | D50928 | 3·3 | 1·0 | 3·3 (4·5, 2·4) |
| 5′-nucleotidase, ecto (CD73) | NT5E | X55740 | 1·4 | 1/2·3 | 3·3 (3·7, 2·9) |
| Mad4 homolog | AL040187 | 1·4 | 1/2·3 | 3·3 (3·3, 3·3) | |
| Aldo-keto reductase family 1, member C1 | AKR1C1 | M86609 | 5·8 | 1·8 | 3·3 (2·5, 4·3) |
| Hypothetical protein FLJ13213 | FLJ13213 | AK000867 | 2·6 | 1/1·3 | 3·3 (4·5, 2·4) |
| Human FLI1 gene for ERGB transcription fuctor | AB012624 | 2·0 | 1/1·6 | 3·3 (3·1, 3·4) | |
| Myosin XVB, pseudogene | MYO15B | AK026339 | 1·3 | 1/2·5 | 3·2 (3·3, 3·2) |
| Adducin 2 (beta) | ADD2 | S81079 | 1·6 | 1/2·0 | 3·2 (2·5, 4·2) |
| Zinc finger protein 236 | ZNF236 | AF085244 | 1·7 | 1/1·9 | 3·2 (4·1, 2·5) |
| Yippee-like 1 (Drosophila) | YPEL1 | AW006162 | 3·1 | 1·0 | 3·2 (2·0, 5·1) |
| Incyte EST | 1·1 | 1/3·0 | 3·2 (3·3, 3·0) | ||
| NIMA (never in mitosis gene a)-related kinase 1 | NEK1 | AL050385 | 3·3 | 1·1 | 3·1 (5·0, 2·0) |
| Protocadherin 9 | PCDH9 | AF169692 | 1·7 | 1/1·8 | 3·1 (4·2, 2·3) |
| Poly (ADP-ribose) polymerase family, member 2 | ADPRTL2 | AK001980 | 2·1 | 1/1·4 | 3·0 (3·2, 2·8) |
| Calcium/calmodulin-dependent protein kinase (CaM kinase) II delta | CAMK2D | AF071569 | 1·5 | 1/2·0 | 3·0 (1·7, 5·3) |
| HIES < Control | |||||
| Solute carrier organic anion transporter family, member 1A2 | SLCO1A2 | U21943 | 1·8 | 24 | 1/14 (1/29, 1/6·7) |
| Normal mucosa of oesophagus specific 1 | NMES1 | AK026298 | 4·6 | 37 | 1/8·1 (1/10, 1/6·3) |
| Syndecan 2 | SDC2 | J04621 | 4·1 | 23 | 1/5·6 (1/12, 1/2·7) |
| Interleukin 17 | IL17 | U32659 | 1/1·3 | 4·1 | 1/5·5 (1/8·0,1/3·7) |
| TBC1 domain family, member 15 | TBC1D15 | AK022147 | 1/2·0 | 2·2 | 1/4·5 (1/6·9, 1/2·9) |
| Syndecan 4 (amphiglycan, ryudocan) | SDC4 | D13292 | 1·0 | 4·6 | 1/4·4 (1/5·2, 1/3·7) |
| Recombining binding protein suppressor of hairless (Drosophila) | RBPSUH | AW968632 | 1/2·3 | 1·7 | 1/4·0 (1/4·8, 1/3·3) |
| Humour necrosis factor, alpha-induced protein 6 | TNFAIP6 | M31165 | 5·5 | 22 | 1/3·9 (1/3·0, 1/5·2) |
| Hypothetical protein FLJ10199 | FLJ10199 | AI216532 | 1·0 | 3·8 | 1/3·9 (1/6·2, 1/2·4) |
| Small inducible cytokine subfamily B (Cys-X-Cys), member 11 | CXCL11 | AF030514 | 1·5 | 5·6 | 1/3·8 (1/2·7, 1/5·4) |
| Cathepsin L | CTSL | AI041851 | 5·5 | 21 | 1/3·8 (1/7·7, 1/1·9) |
| Enhancer of rudimentary homolog (Drosophila) | ERH | U66871 | 1/1·9 | 1·9 | 1/3·6 (1/3·0, 1/4·3) |
| Tryptophanyl-tRNA synthetase | WARS | X59892 | 1/1·7 | 2·1 | 1/3·6 (1/3·8, 1/3·4) |
| Epithelial membrane protein 1 | EMP1 | Y07909 | 1·2 | 4·4 | 1/3·6 (1/5·6, 1/2·3) |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13 kDa | NDUFA5 | U53468 | 1/3·7 | 1/1·1 | 1/3·4 (1/4·1, 1/2·9) |
| Hect domain and RLD 5 | HERC5 | AB027289 | 7·5 | 25 | 1/3·3 (1/1·2, 1/8·7) |
| Feline leukaemia virus subgroup C cellular receptor | FLVCR | AK001419 | 1/2·8 | 1·2 | 1/3·3 (1/5·2, 1/2·0) |
| FK506 binding protein 5 | FKBP5 | U42031 | 1/2·3 | 1·3 | 1/3·1 (1/4·5, 1/2·2) |
| Deleted in lymphocytic leukaemia, 2 | DLEU2 | AW978447 | 1/2·8 | 1·1 | 1/3·1 (1/2·1, 1/4·6) |
| Cathepsin L2 | CTSL2 | AB001928 | 4·2 | 13 | 1/3·0 (1/5·7, 1/1·6) |
| Methionine adenosyltransferase II, alpha | MAT2A | F07456 | 1/2·4 | 1·3 | 1/3·0 (1/1·8, 1/5·1) |
| v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | MAF | AF055376 | 1·0 | 2·9 | 1/3·0 (1/3·7, 1/2·5) |
Fold increase of the gene expression after stimulation with Staphylococcus aureus.
Fold difference of the change of the gene expression levels after stimulation with Staphylococcus aureus between two HIS patients and two healthy controls. Values in parentheses are the fold differences of the gene expression in each HIS patient compared with the mean expression value in healthy controls. Genes that showed more than 3 fold expressional differences between HIES and healthy controls were selected.
These results showed unique patterns of the gene expression in unstimulated and stimulated conditions in peripheral blood cells from HIES patients.
Discussion
HIES is a multisystem disorder with the manifestations of recurrent infections especially with Staphylococcus aureus, characteristic facies, hyperextensibility of joints, multiple bone fractures, scoliosis, and delayed shedding of the primary teeth [4]. Therefore, it is likely that HIES is caused by a molecule which affects various biological functions rather than only by a Th1/Th2–related molecule. Chehimi et al. [8] analysed 375 cytokine and chemokine gene expression by microarray, and found that the expression of some chemokine genes was decreased in PBMNC of HIES after the stimulation with PHA, which may account for the decreased responsiveness against microorganisms. Because of the complex multisystem nature of this disease, we thought more comprehensive analysis of gene expression profiles in purified populations without and with the stimulation of Staphylococcus aureus would be necessary.
It is worthy of note that there were no remarkable differences in the whole gene expression levels of CD4+ T cells containing Th1/Th2 cells between HIES patients and controls in an unstimulated condition. On the other hand, in unstimulated CD14+ cells, 33 genes were up-regulated in HIES and unique gene expression patterns were observed in HIES (Tables 3 and 4). By quantitative PCR, the expression levels of RGC32 that was the most up-regulated gene in microarray analysis, were significantly elevated in HIES compared with healthy controls and atopic dermatitis (Fig. 1). Although RGC32 appears to be a cell cycle regulatory factor that mediates cell proliferation [13], it was not clear why the expression of RGC32 was most up-regulated in CD14+ cells in HIES and how it was related to the pathogenesis or pathophysiology of HIES. Nevertheless, the analysis of expression levels of RGC32 would be helpful in the diagnosis of HIES. The down-regulation of a significantly large number of genes related to the cell growth and maintenance (Table 4) might contribute to the pathophysiology of HIES such as a decreased responsiveness against microorganisms.
After the stimulation with Staphylococcus aureus, decreased expression levels of IL-17 and CXCL11 genes were observed in PBMNC from patients with HIES (Table 5). IL-17, produced by T cells, is a proinflammatory cytokine that induces production of IL-1β and TNF-α by macrophages [14], and IL-6, IL-8 and GM-CSF by stromal cells [15]. It induces migration and functional maturation of neutrophils [16–18] and proinflammatory cytokine production by keratinocytes [19]. In addition, it is involved in the osteoclastogenesis [20]. CXCL11 gene is located on chromosome 4q21, where HIES is linked. The chemokine, CXCL11 is produced by monocytes and keratinocytes after the stimulation with IFN-γ, and is also involved in the inflammatory response by inducing a chemotaxis of monocytes and activated T cells [21]. Osteoblasts express CXC chemokine receptor 3, the receptor of CXCL11 [22]. It is possible that a decreased expression of IL-17 and CXCL11 genes, of which translated products have pleiotropic biological activities, may play a role in the pathogenesis or pathophysiology of HIES.
One of the remarkable findings was up-regulation of immunoglobulin-related genes after the stimulation with Staphylococcus aureus (Table 4). It is reported that an increase of IgE levels was observed even in the cord blood of HIES [23], suggesting the primary dysregulation of IgE production. Our result suggested that the dysregulation was more generalized in immunoglobulin-related genes, and more prominent with the staphylococcal stimulation.
The number of patients used for the microarray analysis was low in this study, and it is possible that other genes or gene groups have more important meanings than those we picked up in the pathophysiology of HIES. Further genetic analysis based on the unique gene expression profile would clarify the pathophysiology and causative genetic abnormality of HIES, and help to develop a new diagnostic method in the near future.
Acknowledgments
We thank the following doctors for the cooperation in this study: Takefumi Matsuda, Department of Paediatrics, Japanese Red Cross Akita Hospital; Takeshi Ikegaya, Department of Paediatrics, Fujieda Municipal General Hospital; Kimio Minagawa, Department of Paediatrics, Hokkaido Children's Hospital and Medical Centre; Norio Onodera, Department of Paediatrics, Iwate Prefectural Hospital; Yoshihisa Nagatoshi, Department of Paediatrics, National Kyushu Cancer Centre; Satoru Yamaguchi, Department of Paediatrics, Fukuoka University; Koji Shinozaki, Department of Paediatrics, Sinshu University.
References
- 1.Davis SD, Schaller J, Wedgwood RJ. Job's syndrome: recurrent, ‘cold’, staphylococcal abscesses. Lancet. 1966;1:1013–5. doi: 10.1016/s0140-6736(66)90119-x. [DOI] [PubMed] [Google Scholar]
- 2.Buckley RH, Wray BB, Belmarker EZ. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics. 1972;49:59–70. [PubMed] [Google Scholar]
- 3.Hill HR, Ochs HD, Quie PG, Clark RA, Pabst HF, Klebanoff SJ, Wedgwood RJ. Defect in neutrophil granulocyte chemotaxis in Job's syndrome of recurrent cold staphylococcal abscesses. Lancet. 1974;14:617–9. doi: 10.1016/s0140-6736(74)91942-4. [DOI] [PubMed] [Google Scholar]
- 4.Grimbacher B, Holland SM, Gallin JI, et al. Hyper–IgE syndrome with recurrent infections-an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 5.Del Prete G, Tiri A, Maggi E, et al. Defective in vitro production of γ-interferon and tumor necrosis factor-alpha by circulating T cells from patients with the hyper-immunoglobulin E syndrome. J Clin Invest. 1989;84:1830–5. doi: 10.1172/JCI114368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokota S, Mitsuda T, Shimizu H, Ibe M, Ikezawa Z. Hyper IgE syndrome-a disease of imbalanced activation of helper T-cell subsets? Arerugi. 1990;39:442–51. [PubMed] [Google Scholar]
- 7.Borges WG, Augustine NH, Hill HR. Defective interleukin-12/interferon-γ pathway in patients with hyperimmunoglobulinemia E syndrome. J Pediatr. 2000;136:176–80. doi: 10.1016/s0022-3476(00)70098-9. [DOI] [PubMed] [Google Scholar]
- 8.Chehimi J, Elder M, Greene J, Noroski L, Stiehm ER, Winkelstein JA, Sullivan KE. Cytokine and chemokine dysregulation in hyper–IgE syndrome. Clin Immunol. 2001;100:49–56. doi: 10.1006/clim.2001.5039. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson KO, Sigurjonsson OE, Gudmundsson S, Goldblatt D, Weemaes MR, Haraldsson A. Increased expression of interleukin-13 but not interleukin-4 in CD4+ cells from patients with the hyper–IgE syndrome. Clin Exp Immunol. 2002;128:532–7. doi: 10.1046/j.1365-2249.2002.01870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vercelli D, Jabara HH, Cunningham-Rundles C, Abrams JS, Lewis DB, Meyer J, Schneider LC, Leung DY, Geha RS. Regulation of immunoglobulin (Ig) E synthesis in the hyper–IgE syndrome. J Clin Invest. 1990;85:1666–71. doi: 10.1172/JCI114618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez MF, Patino PJ, Montoya F, Montoya CJ, Sorensen RU. deOlarte DG. Interleukin 4 and interferon-γ secretion by antigen and mitogen stimulated mononuclear cells in the hyper–IgE syndrome: No TH-2 cytokine pattern. Ann Allergy Asthma Immunol. 1998;81:443–7. doi: 10.1016/s1081-1206(10)63143-2. [DOI] [PubMed] [Google Scholar]
- 12.Grimbacher B, Schaffer AA, Holland SM, et al. Genetic linkage of hyper–IgE syndrome to chromosome 4. Am J Hum Genet. 1999;65:735–44. doi: 10.1086/302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badea TC, Niculescu FI, Soane L, Shin ML, Rus H. Molecular cloning and characterization of RGC-32, a novel gene induced by complement activation in oligodendrocytes. J Biol Chem. 1998;41:26977–81. doi: 10.1074/jbc.273.41.26977. [DOI] [PubMed] [Google Scholar]
- 14.Jovanovic DV, Di Battista JA, Martel-Pelletier J, Jolicoeur FC, He Y, Zhang M. IL-17 stimulates the production and expression of proinflammatory cytokines, IL-1β and TNF-α, by human macrophages. J Immunol. 1998;160:3513–21. [PubMed] [Google Scholar]
- 15.Fossiez F, Djossou O, Chomarat P, et al. T-cell IL-17 induces stromal cells to produces proinflammatory and hematopoietic cytokines. J Exp Med. 1996;183:2593–603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laan M, Cui ZH, Hoshino H, Lotvall J, Sjostrand M, Gruenert DC, Skoogh BE, Linden A. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J Immunol. 1999;162:2347–52. [PubMed] [Google Scholar]
- 17.Ye P, Rodriguez FH, Kanaly S, et al. Recruitment of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defence. J Exp Med. 2001;194:519–27. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino H, Laan M, Sjostrand M, Lotvall J, Skoogh BE, Linden A. Increased elastase and myeloperoxidase activity associated with neutrophil recruitment by IL-17 in airways in vivo. J Allergy Clin Immunol. 2000;105:143–9. doi: 10.1016/s0091-6749(00)90189-1. [DOI] [PubMed] [Google Scholar]
- 19.Teunissen MB, Koomen CW, de Waal Malefyt R, Wierenga EA, Bos JD. Interleukin-17 and interferon-γ synergize in the enhancement of proinflammatory cytokine production by human keratinocytes. J Invest Dermatol. 1998;111:645–9. doi: 10.1046/j.1523-1747.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 20.Kotake S, Udagawa N, Takahashi N, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345–52. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole KE, Strick CA, Paradis TJ, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–21. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisignoli G, Toneguzzi S, Piacentini A, et al. Human osteoblasts express functional CXC chemokine receptors 3 and 5: Activation by their ligands, CXCL10 and CXCL13, significantly induces alkaline phosphatase and beta-N-acetylhexosaminidase release. J Cell Physiol. 2003;194:71–9. doi: 10.1002/jcp.10188. [DOI] [PubMed] [Google Scholar]
- 23.Dreskin SC, Gallin JI. Evolution of the hyperimmunoglobulin E and recurrent infection (HIE, Job’s) syndrome in a young girl. J Allergy Clin Immunol. 1987;80:746–51. doi: 10.1016/0091-6749(87)90297-1. [DOI] [PubMed] [Google Scholar]