Abstract
Guanylic nucleotides are essential cellular players, and the critical enzyme in their tightly regulated synthesis in Saccharomyces cerevisiae is encoded by the IMD2 gene. The transcription of IMD2 is subject to general repression by nutrient limitation through the cis nutrient-sensing element. It is also subject to specific feedback regulation by the end products of the guanylic nucleotide synthesis pathway. The critical cis element for this latter mechanism is the guanine response element (GRE), a TATAATA sequence which is located 202 nucleotides upstream of the transcription initiation site and which functions as the IMD2 TATA box. We show that the GRE functions in conjunction with a 52-nucleotide stretch near the transcription start site. This very unusual promoter structure ensures low, basal expression of IMD2 and the recruitment of TFIID to the GRE in response to guanylic nucleotide limitation.
IMP dehydrogenase (IMPDH) catalyzes the first committed step of the de novo biosynthesis of guanylic nucleotides. The regulation of GMP synthesis plays a crucial role in cell proliferation, since increased activity of IMPDH has been observed in rapidly dividing mammalian cells, including human leukemic cell lines, solid tumor tissues, and B and T lymphocytes. In turn, antagonists of IMPDH activity, such as mycophenolic acid (MPA), serve as immunosuppressive agents. The importance of IMDPH as a target for medical treatments has led to the generation of much interest in the study of the molecular mechanisms governing the regulation of genes encoding IMPDH (see references 1 and 34 for recent reviews).
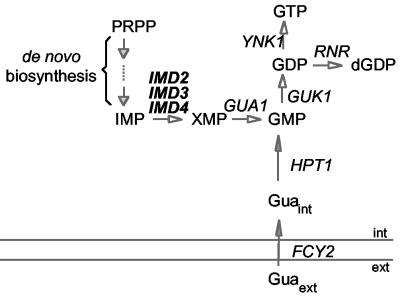
The yeast Saccharomyces cerevisiae has a family of four very closely related genes, IMD1 to IMD4, encoding proteins 80 to 96% identical to each other and also highly homologous (60% amino acid sequence identity) to mammalian IMPDH. It has been demonstrated that the levels of IMD transcripts decrease when cell growth ceases in response to nutrient limitation (8, 12, 13, 30). Additionally, in exponentially growing cells, IMD transcripts are feedback regulated by pathway end products (10, 29). Indeed, the expression of the IMD genes is downregulated when cells are provided with guanine, a GMP precursor that can be directly converted into GMP, allowing bypass of the de novo pathway (Fig. 1). In contrast, when cells are treated with MPA or 6-azauracil (6-AU), which inhibits IMPDH activity and therefore lowers guanylic nucleotide levels, the expression of the IMD genes is upregulated.
FIG. 1.
Schematic representation of the guanylic nucleotide synthesis pathway. Solid lines represent the plasma membrane. ext, extracellular medium; Gua, guanine; int, intracellular medium; PRPP, 5-phosphoribosyl-1-pyrophosphate. Genes are shown in italic type and encode the following enzymatic activities: FCY2, purine cytosine permease; GUA1, guanosine-5′-monophosphate synthetase; GUK1, guanosine-5′-monophosphate kinase; HPT1, hypoxanthine-guanine phosphoribosyltransferase; IMD2, IMD3, and IMD4, IMPDH; RNR, ribonucleotide reductase; and YNK1, nucleoside-5′-diphosphate kinase. IMD1 is not indicated here because it is not expressed (3, 12) and is thought to be a pseudogene.
Various investigators have studied the regulation of the IMD2 gene in more detail (10, 11, 29, 30). On the one hand, studies of fusions of IMD promoters to lacZ demonstrated tighter repression by guanine of the IMD2-lacZ fusion than of other IMD-lacZ fusions (10). On the other hand, strains lacking one or the other IMD gene were tested for resistance to MPA, and only IMD2 was apparently critical (30), while the overexpression of IMD2 led to increased resistance of cells to MPA (9, 30). Escobar-Henriques and Daignan-Fornier previously mapped the promoter of IMD2 and identified an octanucleotide cis element, called the guanine response element (GRE), thought to mediate regulation by both MPA and guanine (10). Those findings were in agreement with the findings of studies by Shaw et al. (30), who mapped the response to 6-AU to a DNA fragment including the GRE. In the latter studies, an additional fragment responsible for reduced IMD2 expression was found close to the transcription start site.
Recently, the regulation of the IMD2 gene has sparked additional interest, because several transcription elongation mutant strains were found to be sensitive to MPA and/or 6-AU, and in some cases this sensitivity could be suppressed by the overexpression of IMD2 (references 26 and 30 and references therein). These findings have led to the use of sensitivity to MPA or 6-AU as a typical assay to characterize factors that may be involved in transcription elongation. However, a recent systematic study of the yeast “disruptome” for mutations affecting resistance to MPA led to the identification of a large number of genes (approximately 100) that are required for cells to be resistant to MPA as well as transcription elongation mutants (9).
In this work, we investigated the regulation of IMD2 in more detail by further characterizing the role of the previously mapped GRE, a TATA box-like sequence required for regulation by MPA and guanine. We made the surprising finding that the GRE is the IMD2 TATA box, considering that it is located 202 nucleotides from the transcription start site. This long distance from the transcription start site was essential to keep IMD2 expression levels low; surprisingly, other TATA box-like sequences present in this promoter and closer to the initiation site could not replace the GRE. Our results are consistent with a model in which the binding of TFIID to the TATA box of the IMD2 promoter would directly respond to the levels of guanylic nucleotides.
MATERIALS AND METHODS
Yeast strains and media.
SD medium is 2% glucose, 0.17% nitrogen base, and 0.5% ammonium sulfate. SDcasa medium is SD medium supplemented with 0.2% Casamino Acids (Difco). Uracil at a final concentration of 200 μg/ml was optionally added. Guanine was optionally added to a final concentration of 0.13 mM (stock solution of 1.3 mM in 0.05 N HCl). MPA or 6-AU (stock solutions of 10 mg/ml in 0.1 M NaOH) were optionally added to various final concentrations.
Saccharomyces cerevisiae wild-type strain Y350 (MATa ura3-52 leu2-3,112 lys2Δ201) was used for all experiments except when otherwise indicated. For some experiments (see Fig. 3B), wild-type strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and ppr2-disrupted strain Y14411 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ygl043w::kanMX4) (both purchased from Euroscarf) were used. Strains Y1648, Y1649, Y1651, and Y1652 (see Fig. 4) and strains Y1653 and Y1655 (see Fig. 6) were constructed as follows. Strain Y350 was used to mutate at the endogenous IMD2 gene the GRE and/or TATA2 boxes, thus generating strains Y1648 (with wild-type GRE and TATA2 boxes), Y1649 (with the GRE box mutated to TACCCATA and with the wild-type TATA2 box), Y1651 (with the wild-type GRE box and with the TATA2 box mutated to TGCAGATTTT), and Y1652 (with the GRE box mutated to TACCCATA and with the TATA2 box mutated to TGCAGATTTT) (mutations are indicated in boldface). These strains were obtained by transforming into strain Y350 plasmids pMAC463B, pMAC464B, pMAC468B, and pMAC469B (URA3 integration derivatives, constructed as described below) digested with ClaI. The URA3 plasmid DNA was cured from the transformants by streaking on 5-fluoro-orotic acid, and the ura3 recombinant strains were analyzed by PCR for the presence of the mutated sequences. A BamHI site (for Y1648 and Y1649) or an EcoRI site (for Y1651 and Y1652) was created at −311 bp upstream of the IMD2 ATG in the mutated sequences, and an additional BamHI site was placed −108 bp upstream of the IMD2 ATG in strains Y1651 and Y1652. Strains Y1653 (with the wild-type GRE box) and Y1655 (with the GRE box mutated to TGTAAATA) were generated in the same manner as strains Y1648 and Y1649 from strain Y643 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG) with plasmids pMAC463B and pMAC466B, respectively. For some experiments (see Fig. 7), isogenic strains GY5 (MATa ura3-52 leu2 trp1Δ1 leu2::PET56 gal2 gcn4Δ taf1::LEU2 pTAF1-HIS3) and GY7 (MATa ura3-52 leu2 trp1Δ1 leu2::PET56 gal2 gcn4Δ taf1::LEU2 ptaf1-4-HIS3) (7) were used.
FIG. 3.
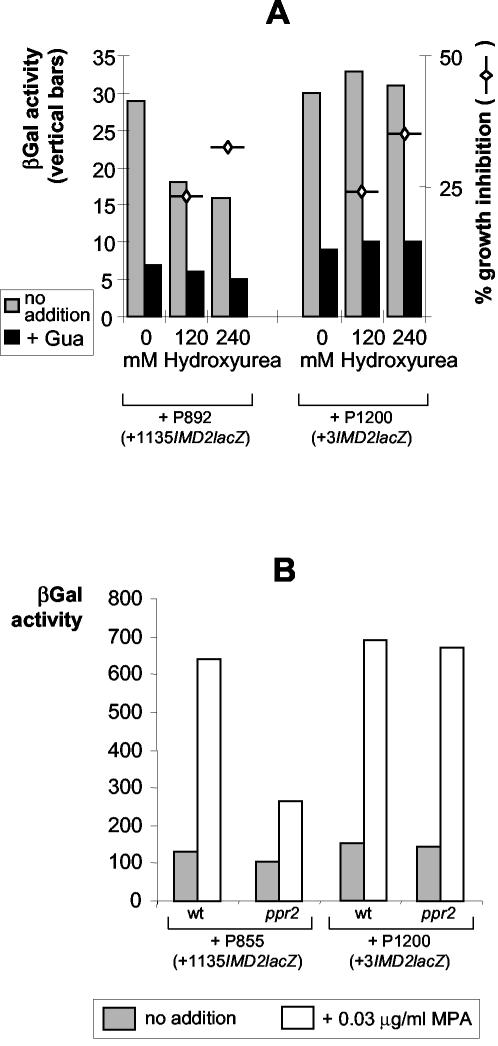
The impaired MPA induction of IMD2 in a ppr2 strain does not involve the GRE. (A) Wild-type cells were transformed with plasmids carrying IMD2-lacZ fusions containing (P892) or not containing (P1200) the coding sequences of IMD2. Expression from these reporter plasmids was measured for wild-type cells grown to exponential phase and then treated for 12 h with hydroxurea, an inhibitor of ribonucleotide reductase that affects cellular proliferation. Gua, guanine; βGal, β-galactosidase. (B) Strains BY4742 (wild type [wt]) and Y14411 (PPR2 disruption [ppr2]) were transformed with the indicated plasmids (Fig. 2) and grown from exponential precultures for 6 h in the presence or absence of MPA. Cells were harvested and assayed for βGal activity.
FIG.4.
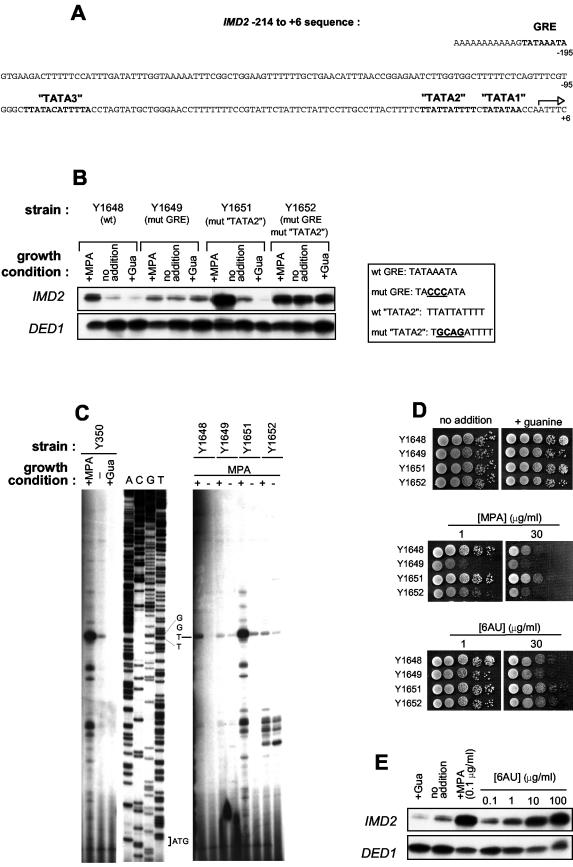
GRE and TATA2 mutations affect endogenous IMD2 transcriptional regulation and resistance of cells to MPA. (A) Sequence of the IMD2 promoter region, with the GRE and AT-rich sequences shown in bold type. (B) Yeast strains carrying the indicated IMD2 sequences were treated or not treated with guanine (Gua) or MPA. Total cellular RNA was extracted, and the levels of transcripts of IMD2 and DED1 (as an internal control) were measured by S1 nuclease analysis. wt, wild type; mut, mutant. (C) The 5′ end of the IMD2 transcripts was characterized by 5′ extension with the indicated yeast strains under various growth conditions. The oligonucleotide used in the extension reactions was also used for sequencing to identify the position of the start site. (D) The indicated yeast strains were grown to exponential phase and plated in serial dilutions on plates containing or not containing Gua, MPA, or 6-AU. Plates were incubated at 30°C for 3 days. (E) S1 nuclease analysis of induction of IMD2 transcripts by MPA and 6-AU.
FIG. 6.
The distance of the GRE from the initiation site is responsible for the low basal expression of IMD2. Wild-type cells were transformed with the indicated plasmids, and transformants were grown to exponential phase and then treated or not treated with guanine (Gua). β-Galactosidase (βGal) activity of the lacZ fusions expressed from the indicated plasmids was measured. Similar effects were observed when mRNA from the fusions was analyzed (data not shown).
FIG. 7.
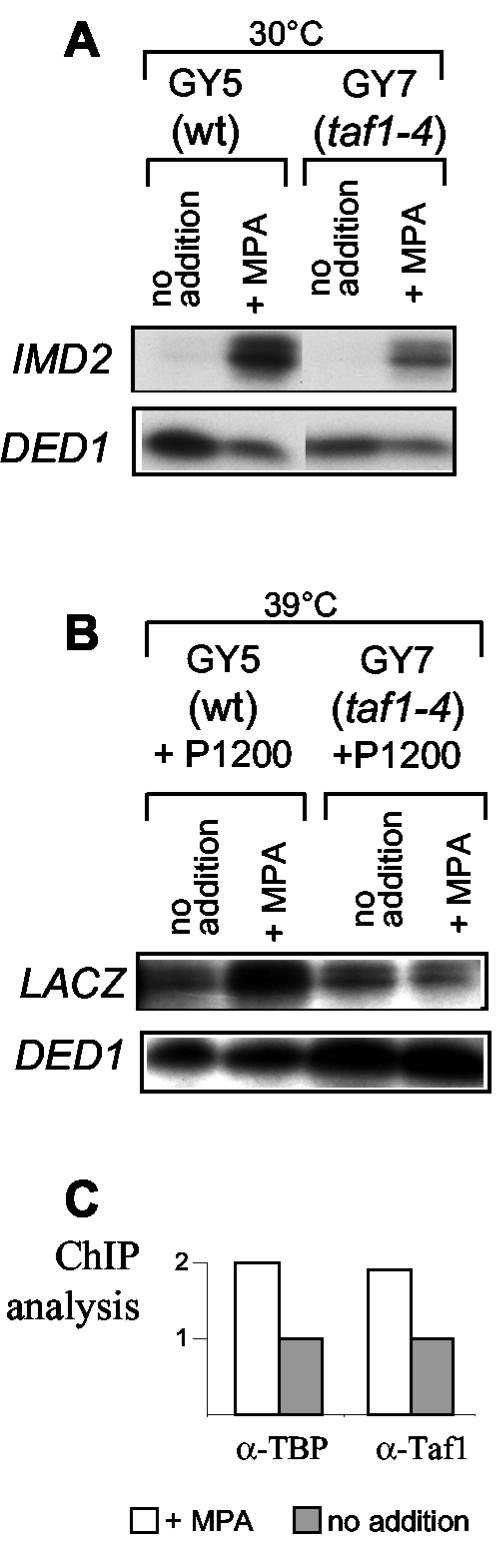
IMD2 expression is not inducible in the absence of a functional TFIID. (A) Total cellular RNA was extracted from isogenic strains carrying wild-type TAF1 (wt) or the temperature-sensitive taf1-4 allele and treated or not treated with MPA. Cells were grown at 30°C, and the levels of transcripts of IMD2 and DED1 (as an internal control) were measured by S1 nuclease analysis. (B) Cells were transformed with a plasmid carrying an IMD2-lacZ fusion that does not contain the coding sequences of the IMD2 gene beyond the ATG (P1200). Transformants were grown to exponential phase and then shifted to 39°C. After 10 min, MPA was added for 1 h. Total cellular RNA was extracted, and lacZ and DED1 (as an internal control) mRNA levels were measured by S1 nuclease analysis. (C) TBP and TAF1 occupancy of the IMD2 promoter was measured by ChIP analysis with wild-type strain Y350 treated or not treated with MPA. The amount of IMD2 promoter chromatin in the control or TBP immunoprecipitates was normalized to that of input DNA, and the indicated values are the relative levels in TBP versus control immunoprecipitates. An analysis of internal controls, showing no change in TBP occupancy, was also performed by PCR amplification of both the ACT1 TATA box region and a non-TATA box-containing region of the IMD2 gene corresponding to its coding sequence (data not shown).
Plasmids.
Plasmid P892 or P913, containing the IMD2-lacZ fusions and 311 bp upstream of the IMD2 ATG, corresponding to 205 bp of the IMD2 promoter, and either a wild-type (TATAAATA) or a mutated (TACCCATA) GRE box, respectively, was described elsewhere (10). The IMD2-lacZ plasmid P855 or P1200, with 355 bp upstream of the IMD2 ATG and either 1,135 bp or only the ATG of the coding region, respectively, was described elsewhere (10, 12). The lacZ fusions were constructed in the URA3 vectors described by Myers and coworkers (25). The IMD2-lacZ fusion plasmid P1881, with the GRE replaced by a canonical TATA box (see Fig. 5A), resulted from cloning of the BamHI-HindIII fragment of the PCR product obtained with oligonucleotides 311TATAB and IMP22 into the BamHI-HindIII sites of vector Yep356. Various other plasmids (see Fig. 6) resulted from cloning of the EcoRI-BamHI fragment of the desired PCR product into the EcoRI-BamHI sites of plasmid P860 (10). The desired PCR product was obtained by using the promoter-coding strand oligonucleotide Eco311 and the appropriate noncoding strand oligonucleotide. Therefore, to create plasmids P1627, P1625, P1623, P1621, P1619, and P1617, the noncoding strand oligonucleotides Bam160, Bam171, Bam180, Bam188, Bam198, and Bam207, respectively, were used.
FIG. 5.
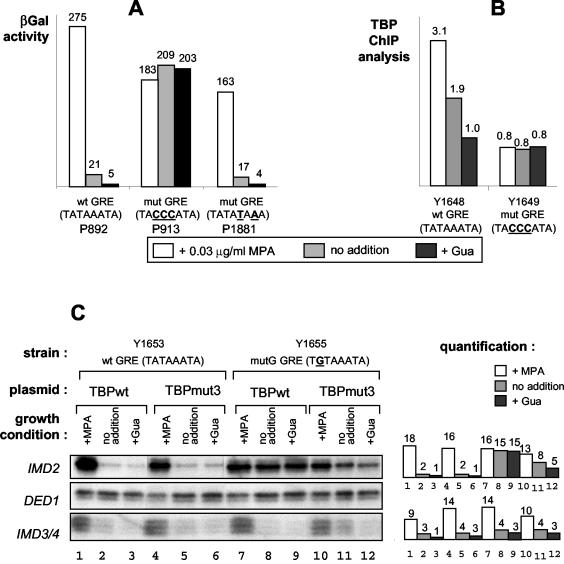
The GRE is the IMD2 TATA box. (A) Plasmids carrying IMD2-lacZ fusions with the indicated GRE sequences (wt, wild type; mut, mutant) were transformed into wild-type Y350 cells that were treated or not treated with guanine (Gua) or MPA. β-Galactosidase (βGal) activity was then measured. (B) The TBP occupancy of the IMD2 promoter was measured by ChIP analysis with isogenic strains carrying either the wild-type or the mutant GRE and treated or not treated with Gua or MPA. The amount of IMD2 promoter chromatin in the control or TBP immunoprecipitates was normalized to that of input DNA, and the indicated values are the relative levels in TBP versus control immunoprecipitates. An analysis of internal controls, showing no change in TBP occupancy, was also performed by PCR amplification of both the ACT1 TATA box region and a non-TATA box-containing region of the IMD2 gene corresponding to its coding sequence (data not shown). (C) A mutant TBP recognizing a TGTAAATA mutant TATA box (mutation in boldface) is able to restore MPA and Gua regulation to a strain with a TGTAAATA mutant GRE box at the IMD2 locus. The indicated strains were transformed with a multicopy plasmid expressing wild-type TBP (TBPwt) or an altered-specificity mutant of TBP (TBPmut3) that can bind the TGTAAATA sequence. Total cellular RNA was extracted from cells grown in the presence or absence of Gua and MPA, and the levels of transcripts of IMD2 and DED1 (as an internal control) were measured by S1 nuclease analysis. A portion of the IMD2 probe can hybridize to the IMD3 and/or IMD4 transcripts, and this hybridization is revealed by a very small labeled DNA fragment protected from S1 nuclease digestion. We included this hybridization product in the figure, since it actually demonstrates that activation by MPA was similar in the different strains. Quantification of the blots (arbitrary units) is shown on the right.
The plasmids used to create the endogenous IMD2 mutant strains were obtained by cloning different fragments into the EcoRI-HindIII sites of the URA3 integration vector YIplac211 as follows. For plasmids pMAC463B and pMAC464B, respectively, used to create strains Y1648 and Y1649, we cloned the PCR product amplified with oligonucleotides 5′ IMD2-1730 and 3′ IMD2-312Bam digested with EcoRI-BamHI together with the EcoRI-HindIII fragments from plasmids P892 and P913, respectively. For plasmids pMAC468B and pMAC469B, respectively, used to create strains Y1651 and Y1652, we cloned the PCR product amplified with oligonucleotides 5′ IMD2-1730 and 3′ IMD2-312Eco digested with EcoRI together with the EcoRI-HindIII fragments from plasmids P2108 and P2110, respectively. P2108 (with a mutated TATA2 box) and P2110 (with mutated GRE and TATA2 boxes) resulted from cloning of the EcoRI-BamHI fragments of the PCR product obtained with the oligonucleotide pairs Eco311-BamTATA2 and Eco311C303-BamTATA2, respectively, into the EcoRI-BamHI sites of plasmid P890 (10). To create strains Y1653 and Y1655, we used pMAC463B and pMAC466B, respectively. pMAC463B construction was described above. To construct pMAC466B, we cloned the PCR product amplified with oligonucleotides 5′ IMD2-1730 and 3′ IMD2-312Bam digested with EcoRI-BamHI together with the BamHI-HindIII fragment from plasmid P1691. P1691 (with the GRE replaced by TGTAAATA) resulted from cloning of the BamHI-HindIII fragment of the PCR product obtained with oligonucleotides IMD2G2B and IMP22 into the BamHI-HindIII sites of vector Yep356.
The oligonucleotides used to PCR amplify the IMD2 fragments from the S288C genomic DNA template were as follows: coding strand—311TATAB, 5′ CGG GAT CCA AGT ATA TAA AGT GAA GAC TTT TTC CA 3′; Eco311, 5′ GGA ATT CAA GTA TAA ATA GTG AAG ACT T 3′; Eco311C303, 5′ CGG AAT TCA AGT ACC CAT AGT GAA GAC TT 3′; 5′IMD2-1730, 5′ CCG GAA TTC AAC ATA TCC TTG CAA 3′; and IMD2G2B, 5′ CGG GAT CCA AGT GTA AAT AGT GAA GTC TTT T 3′; noncoding strand—IMP22, 5′ TCC CCC GGG CCC AAA TAC CGT A 3′; Bam160, 5′ CGG GAT CCA AAA AAA GGT TCC CAG CA 3′; Bam171, 5′ CGG GAT CCC AGC ATA CTA GGT AAA 3′; Bam180, 5′ CGG GAT CCT AGG TAA AAT GTA TAA GC 3′; Bam188, 5′ CGG GAT CCA TGT ATA AGC CCA CGA AA 3′; Bam198, 5′ CGG GAT CCC ACG AAA CTG AGA AAA 3′; Bam207, 5′ CGG GAT CCT GAG AAA AAG CCA CCA A 3′; BamTATA2, 5′ CGG GAT CCT TAT ATA GAA AAT CTG CAG AAA AGT AA 3′; 3′IMD2-312Bam, 5′ CGG GAT CCT TTT TTT TTT TTA TTT TTT CGT TTT 3′; and 3′IMD2-312Eco, 5′ CCG GAA TTC TTT TTT TTT TTT ATT TTT TCG TTT T 3′.
Plasmids expressing either wild-type or altered-specificity mutant TATA box binding protein (TBP) (TBPmut3, TRP1, and 2μm) were kind gifts from Kevin Struhl.
S1 nuclease assays.
Fifty micrograms of total cellular RNA was prepared from cells grown to the desired conditions and analyzed for the presence of specific transcripts as previously described (5). The oligonucleotide used for IMD2 was IMD2S1avantNSE (5′ CCA TCC GGT CTT GGT AGG CTC TTG GTA AAG TCT AGT GCG GTC TTG TAG TCT CTA ATG GCC G 3′); that used for ACT1 was ACT1(S1) (5′ CGA GCA ATT GGG ACC GTG CAA TTC TTC TTA CAG TTA AAT GGG ATG GTG CAA GCG CGC C 3′); and that used for DED1 has already been described (5). Hybridization was performed at 63°C.
β-Galactosidase assays.
Cells were grown overnight in medium selective for the reporter plasmids and used to inoculate fresh SDcasa medium supplemented or not supplemented with 0.03 μg of MPA or guanine/ml. After growth for 6 h (or 12 h [see Fig. 3B]), the cells were collected to measure expression from the IMD2-lacZ fusions as described previously (10). β-Galactosidase activity was expressed in units of optical density at 420 nm × 1,000/(optical density at 600 nm × time [in minutes] × volume [in milliliters]).
Growth plate tests.
Strains Y1648, Y1649, Y1651, and Y1652 transformed with P193 (2μm URA3) and grown to exponential phase were resuspended in water to an optical density at 600 nm of 1. Ten-microliter drops and four serial 1:10 dilutions of this suspension were spotted on SDcasa medium supplemented with guanine, MPA, or 6-AU and incubated for 3 days at 30°C.
IMD2 mRNA 5′ mapping.
A 0.5-pmol quantity of the labeled IMD2 probe (IMD2 prim ext, 5′ GGC TCT TGG TAA AGT CTA GT 3′) and 20 μg of total cellular RNA were used to perform 5′ extension analysis with hybridization and primer extension reaction steps according to standard protocols, 200 U of Moloney murine leukemia virus reverse transcriptase (GIBCO BRL), but no actinomycin D. The extension product was ethanol precipitated and loaded onto a 6% sequencing gel.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (7) with exponentially growing Y1648, Y1649, and Y350 cultures treated for 1 h with guanine or 0.03 μg of MPA/ml. One of the IMD2 real-time PCR amplifications (see Fig. 5B) was performed with oligonucleotides 5′GREbis (5′ CAA AAT TAT TGG TTT TCG TAA CC 3′) and 3′GREbis (5′ GCC ACC AAG ATT CTC CGG T 3′). Another IMD2 real-time PCR amplification (see Fig. 7C) was performed with oligonucleotides 5′GREtris (5′ AAA AAA AAG TAT AAA TAG TGA AGA CTT 3′) and 3′GRE (5′ CAG CAA AAA ACT TCC AGC CG 3′).
RESULTS
Repression of IMD2 by nucleotide feedback and repression by nutrient limitation occur through separate cis elements.
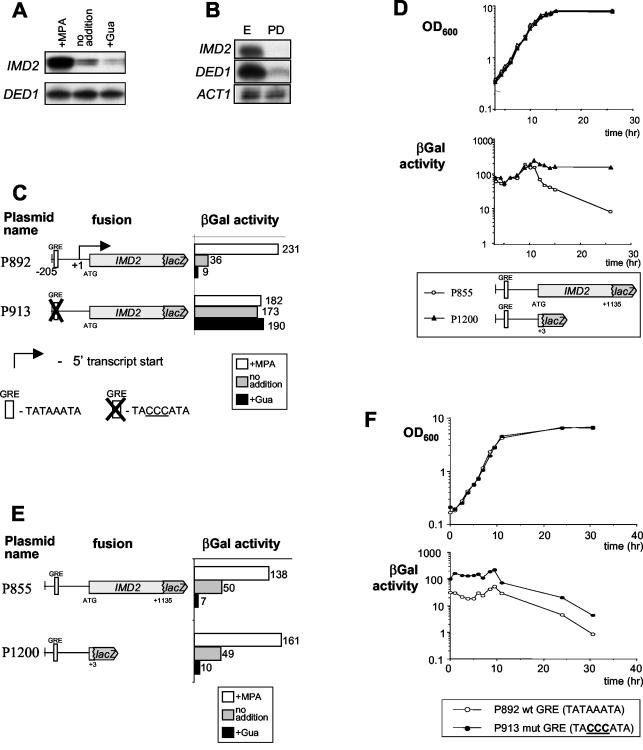
Previous experiments indicated that IMD2 gene expression is induced by MPA and repressed by guanine. However, these results were obtained either with lacZ fusion constructs, which are subject to artifacts, or by Northen blot analysis, which did not allow discrimination among the very similar IMD2 to IMD4 genes. Therefore, S1 nuclease analysis with probes specific to IMD2 was used to measure IMD2 mRNA levels unambiguously. This analysis confirmed that both MPA and guanine specifically modulate the levels of transcripts of the endogenous IMD2 gene (Fig. 2A). In addition to this specific feedback response, various investigators (8, 12, 13, 30) observed that IMD2 was also repressed under conditions of nutrient limitation (Fig. 2B). Two cis elements required for the correct transcriptional regulation of IMD2 have been mapped. Indeed, the GRE was required for appropriate expression and repression by guanine of an IMD2-lacZ fusion gene (10) (Fig. 2C), while repression induced by nutrient limitation was associated with a different cis element, located in the coding region of IMD2 and called the nutrient-sensing element (NSE) (12). Consequently, a fusion containing only the IMD2 promoter fused to the lacZ gene was not regulated when nutrients became limiting, as in postdiauxic phase (12) (Fig. 2D).
FIG. 2.
Repression of IMD2 by nucleotide feedback or by nutrient limitation occurs through separate cis elements. (A and B) Fifty micrograms of total cellular RNA from wild-type Y350 cells grown in the presence of MPA and guanine (Gua) (A) or wild-type Y350 cells grown to exponential phase (E) or beyond the diauxic shift phase (PD) (B) was analyzed by S1 nuclease digestion for the levels of transcripts of IMD2 and DED1 or ACT1 (as internal controls). (C to F) The β-galactosidase (βGal) activities of the lacZ fusions expressed from the indicated plasmids transformed into wild-type strain Y350 grown in the presence or absence of Gua or MPA (C and E) and grown to the indicated optical density at 600 nm (OD600) (D and F) were measured. wt, wild type; mut, mutant.
The importance of the NSE for the repression and activation of IMD2 by guanine and MPA was evaluated by analyzing lacZ fusion constructs that carried (P855) or lacked (P1200) the necessary coding region (Fig. 2E). It was clear that the coding region was not important for the regulation of IMD2 through the feedback mechanism induced by guanine. Conversely, mutation of the GRE had no impact on the repression of IMD2 when cells entered postdiauxic phase (Fig. 2F). Therefore, the two described forms of regulation of IMD2 clearly take place through distinct cis elements.
In the presence of MPA, ppr2 interferes with IMD2 expression through the NSE but not the GRE.
Even though the repression of IMD2 by metabolic downregulation and the induction of IMD2 by MPA occur through different cis elements, the former has been shown to preclude the latter (12). Indeed, once yeast cells engage in the metabolic downregulation of IMD2, they can no longer sense the guanylic nucleotide starvation induced by MPA and activate IMD2 (12). Consequently, any starvation condition is expected to slow down transcription through the NSE and make impossible induction through the GRE in response to guanylic nucleotide limitation. For example, we found that deoxynucleoside triphosphate starvation induced by hydroxyurea treatment resulted in decreased IMD2 expression (10) (Fig. 3A). As expected, the downregulation of IMD2 expression due to hydroxyurea required the IMD2 coding sequence and was abolished in the short IMD2 fusion devoid of the NSE (Fig. 3A). Similarly, mutations in the gene encoding TFIIS (PPR2/DST1) (16), a recognized transcription elongation factor, strongly decreased induction by MPA when the NSE was present in the fusion (P855) but had no effect in the absence of the NSE (P1200) (Fig. 3B). Since ppr2/dst1 mutants are hypersensitive to MPA, we interpret this result as indicating that in the absence of TFIIS and in the presence of MPA, cells sense some form of metabolic stress. This signal is transmitted to the growth response element within the coding sequence (NSE) of IMD2, leading to decreased expression and abolished induction of IMD2, most probably contributing to sensitivity to MPA or 6-AU. The twofold induction of P855 still occurring in the ppr2 mutant at an extremely low MPA concentration (Fig. 3B) was lost at higher MPA doses (data not shown), in agreement with the findings of Shaw and Reines (29). Importantly, even in a wild-type strain, IMD2 induction was almost precluded at MPA doses of higher than 30 μg/ml, which strongly affected growth (data not shown).
Taken together, these experiments demonstrate that under certain conditions, such as in strains hypersensitive to MPA, the regulation of IMD2 by MPA and guanine through the GRE needs to be studied in a context devoid of the NSE.
IMD2 expression is tightly correlated with MPA resistance.
The IMD2 GRE is highly unusual. Indeed, it looks like a TATA sequence but is positioned far away (202 bp) from the IMD2 5′ transcription start site (Fig. 4A). Moreover, its mutation does not abolish IMD2 expression, as would be expected for a normal TATA box, but rather leads to a loss of IMD2 regulation by either MPA or guanine and to constitutive expression somewhat higher than the basal level of IMD2 expression from the wild-type GRE (10). To elucidate the function of the GRE, we first attempted to identify the TATA box in the IMD2 promoter. We hence mutated other AT-rich sequences, in particular, the three most closely related to TATA boxes, within the IMD2 promoter region (TATA1, TATA2, and TATA3; Fig. 4A), either alone or in combination with the GRE, and analyzed the expression of lacZ fusion constructs (data not shown). None of these elements was important for the regulation of IMD2 expression by MPA or guanine. However, mutation of TATA2 increased both basal and induced IMD2 expression, while mutation of TATA3 slightly decreased both basal and activated IMD2 expression. Thus, the expression of IMD2 was affected by mutations in TATA2 and, to a lesser extent, in TATA3, but only the GRE was absolutely necessary for the regulated expression of IMD2.
The importance of the GRE and the TATA2 element for the appropriate expression and regulation of IMD2 needed to be confirmed for the endogenous IMD2 gene. We thus created strains in which the endogenous IMD2 gene carried mutations in the GRE and the TATA2 element, either alone or in combination (see Materials and Methods). S1 nuclease analysis of the IMD2 mRNA levels confirmed our previous findings with fusion constructs. That is, the GRE box was necessary for the regulation of IMD2 transcript levels by both MPA and guanine (compare Y1648 with Y1649 in Fig. 4B), while the TATA2 box mutation resulted in increased levels of the IMD2 transcript (compare Y1648 with Y1651 in Fig. 4B). Importantly, the major transcription start site was the same for all strains (Fig. 4C). Some transcription initiated at several downstream sites, particularly under conditions of higher expression (Y350 or Y1651 in the presence of MPA) or when both the GRE and the TATA2 sequences were mutated (Y1652).
These strains allowed us to determine the importance of IMD2 expression levels for the resistance to both MPA and 6-AU. Figure 4D shows that strain Y1649, in which the IMD2 GRE had been mutated, making IMD2 no longer inducible by MPA, was MPA sensitive and, to a lesser extent, 6-AU sensitive. In contrast, strain Y1651, in which IMD2 TATA2 had been mutated, resulting in higher expression of IMD2, was more resistant than the parental wild-type strain to MPA and, to a lesser extent, to 6-AU. Strain Y1652, in which both the GRE and the TATA2 sequences of IMD2 were mutated, was sensitive to MPA. Taken together, these results show that the expression of IMD2 and its capacity to be highly induced in the presence of MPA correlate well with resistance to MPA and, to a lesser extent, to resistance to 6-AU. It is interesting that IMD2 was maximally induced by 0.1 μg of MPA/ml (data not shown) but that 100 μg of 6-AU/ml was needed to obtain the same induction (Fig. 4D). However, the induction of IMD2 by both drugs was dependent upon the same GRE (data not shown).
The GRE is a bona fide TATA element of the IMD2 gene.
The results presented so far define the GRE as an element important for the regulation of IMD2 by guanylic nucleotide levels but do not establish whether it indeed functions as a TATA element. To address this issue, the GRE was mutated to a canonical TATA element; it was clear that this alteration had no effect on GRE function (Fig. 5A).
In a second approach, ChIP experiments carried out with strains either wild type or mutant for the GRE and with TBP antibodies revealed that the modulation of the association of TBP with the wild-type IMD2 promoter correlated with transcription levels, increasing with MPA and decreasing with guanine (Y1648 in Fig. 5B). In contrast, when the GRE was mutated, no modulation of the low level of cross-linking of TBP to the mutated IMD2 promoter was detectable in the presence of MPA or guanine (Y1649 in Fig. 5B). The fold regulation of TBP binding to the promoter did not reflect directly the fold regulation of IMD2 transcript levels, but this finding might reflect a limitation inherent to our ChIP experiments, since we have observed this finding for several promoters under many conditions (data not shown). It was surprising that while the basal expression of IMD2 was higher in mutant strain Y1649 (Fig. 4B), less TBP was found associated with the sequences surrounding the GRE (Fig. 5B). However, it is very possible that in the mutant strain, the alternative core promoter allowing expression was not located close enough to the GRE to be amplified to any appreciable extent in the ChIP experiments.
Finally, we created a strain carrying a mutated GRE corresponding to a mutated TATA element that can be recognized by an altered-specificity mutant of TBP (32). Using this IMD2 mutant strain (Y1653) and a wild-type IMD2 strain (Y1655), we transformed a plasmid overexpressing either wild-type TBP or the altered-specificity mutant. We found that the wild-type IMD2 strain (Y1655) was normally regulated, regardless of the presence in the plasmid of a gene encoding wild-type or mutant TBP. The presence of endogenous wild-type TBP was most likely responsible for the correct regulation of the wild-type construct, despite the concomitant expression of the mutant TBP. Furthermore, while IMD2 transcript levels in cells with the mutated GRE and wild-type TBP were not regulated by guanine and MPA, such regulation was restored with the altered-specificity mutant of TBP (Fig. 5C). The fact that the regulation was not totally restored might reflect some form of competition between the endogenous TBP present in the cell (required for viability) and the mutant TBP encoded by the plasmid (possibly for assembly into TFIID).
Taken together, these results clearly establish that the GRE functions as a TATA element in the IMD2 promoter. Furthermore, this TATA element is necessary for the regulation of IMD2 gene expression by MPA or guanine.
The distant position of the GRE ensures low levels of IMD2 expression.
Why is the IMD2 TATA box located so far away from the transcription initiation site? To understand the role of the distance in the regulation of IMD2, we removed increasing amounts of sequence between the GRE and the transcription initiation site. We were able to show that shortening of the distance between the GRE and the initiation site gradually increased the expression of IMD2-lacZ fusions but did not affect the regulation by guanine (Fig. 6). Thus, the combination of a 100-bp fragment containing the GRE TATA box and the first 52 nucleotides of the IMD2 promoter was sufficient to create the regulated expression of IMD2. However, further deletion of these 52 nucleotides (P1524) abolished guanine regulation (Fig. 6). The importance of this 52-nucleotide region was confirmed with a reporter construct lacking only these 52 nucleotides (P1637), since mRNA levels expressed from this construct were similar whether cells were grown in the presence or in the absence of guanine (data not shown). We characterized the 52-nucleotide region important for the regulated expression of IMD2. However, when this region was divided into three largely overlapping segments and used to replace the complete sequence in P1623, none of the segments was sufficient to restore the regulation of IMD2 (data not shown). Thus, no short element within the first 52 nucleotides is sufficient in combination with the GRE to install regulated expression of the IMD2 gene. These observations were confirmed by S1 nuclease analysis of transcript levels produced by the reporter constructs (data not shown).
Therefore, the long distance between the IMD2 TATA box and the transcription start site ensured a low level of IMD2 expression. In contrast, no single upstream activating sequence (UAS) or upstream repressing sequence (URS) element could be found within the IMD2 promoter.
TFIID is important for the recruitment of TBP to the IMD2 promoter in response to guanylic nucleotide starvation.
The results presented so far suggest that the regulation of IMD2 gene transcription involves an interaction among distantly positioned elements: a TATA element (the GRE) located 202 bp upstream of the transcription start site, a 52-nucleotide stretch just upstream of the transcription initiation site, and the NSE located within the IMD2 coding sequence. These findings led us to investigate the roles of different complexes that affect chromatin structure in the regulation of IMD2 gene expression. We used strains carrying mutations in SNF2 (for a review, see reference 28), ISW1, ISW2, and both ISW1 and ISW2 (33). In these strains, both the extent and the kinetics of regulation by MPA or guanine were similar to those in the wild-type strains, except for the basal expression of IMD2, which was slightly increased in the snf2 mutant (data not shown). We next examined the importance of the SAGA complex (15). We found that IMD2 was appropriately expressed in cells lacking Gcn5p (data not shown). Moreover, we analyzed a strain lacking Spt7p, where the SAGA complex is disrupted (data not shown), or a strain lacking Spt3p, which is thought to be important for the SAGA complex interaction with TBP. In these spt3Δ and spt7Δ mutants (both MPA hypersensitive [9]), IMD2 induction by MPA was partially impaired but, as for the ppr2 mutant, this effect was mostly indirect, since it was lost in the IMD2 reporters devoid of the NSE (data not shown).
Genome-wide studies with several mutant yeast TAFIIs have suggested that basal transcript levels for the IMD genes are decreased when TFIID is mutated (18). TAF1 is a component of TFIID (but not SAGA), another complex that contributes to the recruitment of TBP to promoters. We thus analyzed specifically with S1 nuclease assays the regulation of IMD2 expression in a temperature-sensitive taf1 mutant (taf1-4) (7). IMD2 transcript levels were decreased in the taf1-4 mutant at a permissive temperature but were still inducible by MPA, albeit to a lesser extent than in wild-type cells (Fig. 7A, right panel; basal levels were visible only on a longer exposure). However, TFIID function was abolished in this mutant only at a restrictive temperature. Thus, we shifted the taf1-4 cells to the restrictive temperature for 10 min and then treated the cells with MPA for 1 h. To analyze the inducibility of the IMD2 promoter in response to guanylic nucleotide levels independently of the dominant metabolic downregulation of IMD2 by the NSE, we used a reporter construct in which the IMD2 promoter with its ATG was fused to lacZ (P1200; see above). We found that the IMD2 reporter construct was not at all inducible by MPA at a restrictive temperature in the conditional taf1-4 mutant strain, while under the same conditions, it was inducible in the wild-type strain (Fig. 7B).
These results suggest that the upregulation of IMD2 by MPA requires TAF1 and is most likely mediated by TFIID binding to the GRE. To confirm this suggestion, we performed ChIP experiments. Indeed, the association of TAF1 with the IMD2 promoter, like that of TBP, was induced by MPA (Fig. 7C).
DISCUSSION
Specific characterization of IMD2 expression.
In this study, we have undertaken a careful characterization of the regulation of the IMD2 gene, one of the four IMD genes of S. cerevisiae. We used an analysis that allowed us to distinguish among the transcripts originating from the four IMD genes (22). In addition, to define elements important in IMD2 regulation, we introduced mutations in the chromosomal IMD2 locus. Indeed, in previous work by various investigators (10, 29, 30), conclusions about IMD2 gene regulation have been obtained by use of fusion constructs with lacZ or luc reporter genes carried by episomal plasmids. We now know that the enzyme levels produced from some IMD2-lacZ reporter constructs (containing internal deletions in the IMD2 promoter) do not always coincide with transcript levels (data not shown). This fact is apparently due to an “ATG” located at −70 in the promoter region that only in some promoter deletion configurations seems to affect translation of the IMD2 reporter fusions (data not shown). Furthermore, when endogenous transcript levels were previously analyzed by Northern blot analysis, the probes used could not distinguish among the four highly homologous IMD genes (10, 29, 30). This result could have been misleading, since it is clear that IMD2 and IMD3 are expressed at similar levels (10, 12) and furthermore that their relative responses to MPA and 6-AU are different. Indeed, the overexpression of IMD2 leads mostly to MPA resistance, while the overexpression of IMD3 and IMD4 leads mostly to resistance to 6-AU (9). Finally, conclusions have also been drawn by use of strains carrying deletions of one or the other IMD gene (3). The problem in the interpretation of those experiments is that the imd2 deletion strain used carried a large chromosomal disruption including many other genes.
The GRE is the TATA box of the IMD2 gene.
We found here that the previously defined GRE (10) is indeed important for regulation of the endogenous IMD2 gene. This element is essential for the induction of IMD2 by MPA and the repression of IMD2 by guanine. Our very new and exciting finding is that the GRE is in fact the TATA element of the IMD2 gene. First, we show that the sequence can be mutated into a totally consensus TATA sequence without a loss of function while, in contrast, single point mutations destroying the TATA sequence lead to a loss of IMD2 gene regulation. Second, regulated binding of TBP to the IMD2 promoter region, in response to MPA and guanine, requires the GRE sequence. Finally, and most importantly, the regulation of IMD2 can be restored by a mutated GRE sequence when cells are transformed with a plasmid expressing an altered-specificity TBP that can recognize the mutated GRE sequence.
Several promoter elements determine low levels of IMD2 expression.
The definition of GRE as the TATA element of IMD2 was unexpected, since it is located 202 bp upstream of the transcription start site. This long distance between the GRE and the transcription start site is not important for the response of IMD2 to guanylic nucleotide levels but instead ensures a low level of expression of IMD2. We have also identified another element that contributes to the low basal expression of IMD2, namely, a TATA-like sequence (TATA2) which is localized at nucleotide −20 relative to the transcription start site and which functions as a repressive element for basal, repressed, and induced levels of IMD2 transcription. This mapped element is certainly responsible for previous observations made by others where a 36-nucleotide deletion of promoter elements around that transcription start site led to increased expression of a reporter construct (30). Moreover, the IMD2 coding sequence also contains the NSE cis element responsible for IMD2 postdiauxic shift repression. Taken together, our results indicate that IMD2 is generally in a “repressed” state and can be repressed further under certain conditions. Because we have observed a slight increase in IMD2 basal expression in an snf2 mutant (data not shown), we can imagine that the general repression of IMD2 may be related to a particular structure of the chromatin. More studies will be needed to test such a model.
Considering the distance of the GRE from the transcription initiation site, it is unclear how it functions to recruit the transcription machinery to the initiation site, and it also seems aberrant that no sequence closer to the start site functions instead of this element. In any event, it is clear that when the GRE is mutated, other elements within the promoter can function to mediate transcription initiation, leading to a basal and unregulated level of expression that is higher than that initiated from the GRE. However, this alternate core promoter is obviously not fully functional or accessible when the GRE is intact, perhaps because of the structure imposed upon the IMD2 promoter by the functional GRE. This masking effect is probably related to the way in which a functional preinitiation complex is recruited to the correct distant initiation site. Whatever the nature or position of this alternate core promoter element, it cannot functionally replace the GRE, since it cannot be regulated by either guanine or MPA. One can expect it to be a TATA-less sequence, similar to the TATA-less core promoters of the HIS3 and HIS4 genes which, in contrast to their TATA box promoters, cannot be activated by Gcn4p (21, 27).
The combination of multiple elements within the IMD2 promoter and the GRE is necessary for appropriate regulation by guanylic nucleotide levels.
In the IMD2 promoter, besides the TATA box, no cis-acting UAS or URS could be found. Many unsuccessful attempts were undertaken to look for cis elements and, by defining genetic screens, to identify trans-acting factors. Further analysis of specific possible regulators, according to their known functions, such as Rtf1p (31), Mot1p (2, 6, 19), the Ccr4-Not complex (5, 7, 19, 20), or the NC2 regulator (4, 14, 17, 19, 23), was also negative (data not shown). One other unfruitful hypothesis was that the other Imd proteins would have a regulatory role for IMD2, which was not the case (data not shown).
Even though our results seem to establish that no specific single cis-acting sequence other than the GRE exists within the IMD2 promoter, the introduction of the GRE into a heterologous CYC1-lacZ reporter fusion gene does not lead to guanine and MPA regulation (data not shown). Therefore, the GRE is necessary but not sufficient to confer MPA and guanine regulation.
We have found that the first 52 nucleotides upstream of the transcription start site are necessary and sufficient to mediate IMD2 regulation in combination with a 100-bp fragment including the GRE. There is clearly something specific about this sequence, since these 52 nucleotides cannot be replaced by other elements of the IMD2 promoter (data not shown). In addition, we could not define any single element within this sequence that can functionally replace the 52 nucleotides. It seems that multiple elements within this sequence act together to confer specificity. Perhaps a particular structure is conferred by these multiple elements and the establishment of this structure is sensitive to guanylic nucleotide levels. What such a regulatory structure could be is at present totally unclear to us, since neither Snf2p, Isw1p, Isw2p, nor Gcn5p is important for IMD2 regulation.
The binding of TBP to the GRE and the function of TFIID are essential elements of IMD2 regulation.
Our analyses of mutants have confirmed the crucial importance of TBP binding to the GRE and of TFIID for the regulated expression of IMD2. Regulation is unlikely to be mediated by the levels of TBP, since the overexpression of TBP or mutation of the Kap114p importin (rendering TBP mostly cytoplasmic) (24) had no effect on IMD2 expression or regulation (data not shown). It must be noted that the position of the binding site for TBP at the 5′ end of the promoter and the long distance to the site of transcription initiation are very exceptional and suggest that the assembly of the transcription initiation machinery may also be quite exceptional. It seems likely that TBP is recruited in the form of TFIID, since TFIID is necessary for IMD2 expression and activation, and TAF1 association at the IMD2 promoter follows that of TBP. The absence of any other identifiable single UAS or URS cis-acting element suggests that the recruitment of TFIID to the core promoter essentially depends upon its inherent capacity to bind or the accessibility of its DNA sequence, without the help of trans-acting factors bound to other cis elements. It is therefore very interesting that the synthesis of guanylic nucleotides with such broad and essential roles in cells seems to have kept regulation features simple, without UAS or URS elements. Whether the levels of guanylic nucleotides affect a chromatin structure, allowing TFIID to bind to the GRE or to form a functional preinitiation complex over the distant site of transcription initiation, or whether they directly affect TFIID itself is still an open question.
Does regulation by guanylic nucleotide levels require a particular promoter organization?
It has been observed by analyzing the yeast proteome after MPA treatment that only the three Imd proteins are induced (11), indicating that MPA treatment does not have broad effects on gene expression. Thus, the mechanism leading to the induction of the IMD genes in response to guanylic nucleotide levels is likely to be very specific. TFIID obviously is not a component specific to the transcription of the IMD genes. This apparent paradox could be tentatively explained by considering the unusual IMD gene promoters. IMD3 is regulated by MPA and guanine, like IMD2 (data not shown), and the GRE is conserved in the same position in the IMD3 promoter and is absolutely essential for the regulated expression of IMD3 (data not shown). Furthermore, the promoters of these two genes are generally very similar (61%), a fact that is rather unusual for noncoding regions, unless it is indeed related to the very specific regulation of these genes. Indeed, if regulation requires multiple elements that act together, the promoters will necessarily be quite conserved overall, rather than just sharing a given short regulatory sequence.
The regulated recruitment of TBP to the GRE is abolished by metabolic shutdown of IMD2 by the element in the coding sequence.
The notion of a particular structure at the IMD2 promoter that more or less allows TFIID to associate productively with the core promoter is reinforced by our finding that IMD2 initiation is shut off by the NSE in the coding sequence (12). This NSE, located 260 nucleotides downstream of the transcription initiation site, controls TBP binding to its binding site 462 nucleotides upstream and thereby transcription initiation (12). It seems reasonable to imagine that this entire DNA region is presented in a particular structure that can be either permissive or not permissive for initiation. Exactly how this regulation is achieved is still not known since, as indicated above, proteins implicated in chromatin remodeling, such as Isw1, Isw2, Swi2, or Gcn5, are clearly not critical for IMD2 -regulation (data not shown). The relationship between the unusually distant TATA box and the regulatory element within the coding sequence is certainly an exciting new item to investigate.
Acknowledgments
We thank Kevin Struhl for plasmids encoding wild-type or altered-specificity mutant TBP. We thank Steve Buratowski and Lisete Fernandes for helpful comments and Cécile Deluen and Eve Lenssen for critical reading of the manuscript.
This work was supported by grants from the CNRS (UMR5095) and ARC (4749) to B.D.-F., by a fellowship to M.E.-H from the Portuguese Government (FCT SFRH/BPD/5725/2001), and by a grant (3100-059199.99) from the Swiss National Science Foundation to M.A.C.
REFERENCES
- 1.Allison, A. C., and E. M. Eugui. 2000. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 47:85-118. [DOI] [PubMed] [Google Scholar]
- 2.Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. MOT1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. [DOI] [PubMed] [Google Scholar]
- 3.Barton, A. B., H. Bussey, R. K. Storms, and D. B. Kaback. 1997. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: characterisation of the 54 kb right terminal CDC15-Floi-PHO11 region. Yeast 13:1251-1253. [DOI] [PubMed] [Google Scholar]
- 4.Cang, Y., D. T. Auble, and G. Prelich. 1999. A new regulatory domain on the TATA-binding protein. EMBO J. 18:6662-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collart, M. A., and K. Struhl. 1994. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 6.Davis, J. L., R. Kunisawa, and J. Thorner. 1992. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1879-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler, M. Lemaire, N. Paquet, and M. A. Collart. 2002. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 9.Desmoucelles, C., B. Pinson, C. Saint-Marc, and B. Daignan-Fornier. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug mycophenolic acid. J. Biol. Chem. 277:27036-27044. [DOI] [PubMed] [Google Scholar]
- 10.Escobar-Henriques, M., and B. Daignan-Fornier. 2001. Transcriptional regulation of the yeast GMP synthesis pathway by its end products. J. Biol. Chem. 276:1523-1530. [DOI] [PubMed] [Google Scholar]
- 11.Escobar-Henriques, M., A. Balguerie, C. Monribot, H. Boucherie, and B. Daignan-Fornier. 2001. Proteome analysis and morphological studies reveal multiple effects of the immunosuppressive drug mycophenolic acid specifically resulting from guanylic nucleotide depletion. J. Biol. Chem. 276:46237-46242. [DOI] [PubMed] [Google Scholar]
- 12.Escobar-Henriques, M., M. A. Collart, and B. Daignan-Fornier. 2003. Transcription initiation of the yeast IMD2 gene is abolished in response to nutrient limitation through a sequence in its coding region. Mol. Cell. Biol. 23:6279-6290. [DOI] [PMC free article] [PubMed]
- 13.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goppelt, A., and M. Meisterernst. 1996. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 24:4450-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 16.Huber, J. C., A. Guyonvarch, B. Kammerer, F. Exinger, P. Liljelund, and F. Lacroute. 1983. Complete sequence of a eukaryotic regulatory gene. EMBO J. 2:2071-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inostroza, J. A., F. H. Mermelstein, I. Ha, W. S. Lane, and D. Reinberg. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70:477-489. [DOI] [PubMed] [Google Scholar]
- 18.Lee, T. I., H. C. Causton, F. C. P. Holstege, W.-C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 19.Lemaire, M., J. Xie, M. Meisterernst, and M. A. Collart. 2000. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 36:163-173. [DOI] [PubMed] [Google Scholar]
- 20.Liu, H.-Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahadevan, S., and K. Struhl. 1990. TC, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol. Cell. Biol. 10:4447-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster, and M. A. Collart. 2000. The essential function of NOT1 lies within the CCR4-NOT complex. J. Mol. Biol. 303:131-143. [DOI] [PubMed] [Google Scholar]
- 23.Meisterernst, M., and R. G. Roeder. 1991. Family of proteins that interact with TFIID and regulate promoter activity. Cell 67:557-567. [DOI] [PubMed] [Google Scholar]
- 24.Morehouse, H., R. M. Buratowski, P. A. Silver, and S. Buratowski. 1999. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA 96:12542-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 26.Otero, G., J. Fellows, Y. Li, T. de Bizemont, N. M. G. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 27.Pellman, D., M. E. McLaughlin, and G. R. Fink. 1990. TATA-dependent and TATA-independent transcription at the HIS4 gene of yeast. Nature 348:82-85. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, C. L., and J. W. Tamkun. 1995. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem. Sci. 20:143-146. [DOI] [PubMed] [Google Scholar]
- 29.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw, R. J., J. L. Wilson, K. T. Smith, and D. Reines. 2001. Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast. J. Biol. Chem. 276:32905-32916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strubin, M., and K. Struhl. 1992. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68:721-730. [DOI] [PubMed] [Google Scholar]
- 33.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann, A. G., J. J. Gu, J. Laliberte, and B. S. Mitchell. 1998. Inosine-5′-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog. Nucleic Acid Res. Mol. Biol. 61:181-209. [DOI] [PubMed] [Google Scholar]