Abstract
Mediator is a general cofactor implicated in the functions of many transcriptional activators. Although Mediator with different protein compositions has been isolated, it remains unclear how Mediator facilitates activator-dependent transcription, independent of its general stimulation of basal transcription. To define the mechanisms of Mediator function, we isolated two forms of human Mediator complexes (Mediator-P.5 and Mediator-P.85) and demonstrated that Mediator-P.5 clearly functions by enhancing activator-mediated recruitment of RNA polymerase II (pol II), whereas Mediator-P.85 works mainly by stimulating overall basal transcription. The coactivator function of Mediator-P.5 was not impaired when TATA-binding protein (TBP) was used in place of TFIID, but it was abolished when another general cofactor, PC4, was omitted from the reaction or when Mediator-P.5 was added after pol II entry into the preinitiation complex. Moreover, Mediator- P.5 is able to enhance TBP binding to the TATA box in an activator-dependent manner. Our data provides biochemical evidence that Mediator functions by facilitating activator-mediated recruitment of pol II and also promoter recognition by TBP, both of which can occur in the absence of TBP-associated factors in TFIID.
Initiation of transcription in eukaryotes requires many proteins, including gene-specific transcription factors, protein cofactors, and the general transcription machinery (52, 91). Gene-specific transcription factors (or activators) are necessary to turn on specific gene expression by recruiting components of the general transcription machinery, which includes TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and RNA polymerase II (pol II), as well as chromatin cofactors, such as ATP-dependent chromatin remodeling complexes and histone acetyltransferases. This communication between activators and the general transcription machinery also requires at least one of the three general cofactors: Mediator, TATA-binding protein (TBP)-associated factors (TAFs) in TFIID, and upstream stimulatory activity (USA)-derived protein components, such as positive cofactor 4 (PC4).
TAFs in TFIID act collectively as transcriptional coactivator to enhance activator-dependent transcription from chromatin (68, 97), to contact the activation domain (26, 33) or the DNA-binding domain (14) of activator, to facilitate the recruitment of the initiation form of pol II to the promoter region (94), or to induce DNA wrapping on the TFIID-bound promoter region (72). TAFs also enhance promoter recognition through multiple protein-DNA interactions with Initiator (87) and downstream core promoter elements (9). In addition, the enzymatic activities of TFIID, due to the presence of TAFII250, also lead to acetylation of histones H3 and H4 (63), phosphorylation of PC4 (45) and the RAP74 component of TFIIF (18), as well as ubiquitin activation and conjugation of histone H1 (76). However, TAFs are not universally required for activator function (11, 22, 47, 60, 64, 73, 88, 93, 96), nor are they needed for transcriptional repression by some gene-specific transcriptional repressors (21, 35).
USA contains both negative cofactor NC2 and positive cofactors, such as PC1, PC2, PC3, and PC4, modulating the level of transcription (42). PC4, an unusual single-stranded and double-stranded DNA-binding protein with a relatively small molecular size of approximately 15 kDa, can substitute for USA to mediate activator-dependent transcription (24, 49, 96). Interestingly, PC4 also acts as a repressor to suppress basal transcription in the absence of an activator (58, 90, 93). Repression of basal transcription by PC4 occurs prior to the formation of a complete preinitiation complex and can be alleviated by an increasing amount of TFIID, TFIIH, and pol II holoenzyme (45, 58). This dual activity of PC4 is dose dependent and can work synergistically with other USA-derived cofactors, such as PC2 and PC3 (57).
Mediator is also a critical general cofactor implicated in activator-dependent transcription. It has been genetically defined and also biochemically isolated based on its associations with pol II or activators or as a free entity of protein complex (7, 10, 52, 59, 67, 70, 77). The protein compositions of Mediator complexes, although varying slightly depending on the methods of purification, appear to be similar from different isolations variously named TRAP/SMCC (22, 29, 38), DRIP (78, 79), ARC (69, 85), CRSP (71, 81), PC2 (57), NAT (84), and Mediator (8, 28, 40, 41, 46, 51, 56, 62, 74, 89). Thus, a set of Mediator components apparently forms a core to associate dynamically with other peripheral proteins. The structures of Mediator complexes from different species are also highly conserved, with three visible modules forming distinct conformations when in association with pol II or activator (4, 7, 16, 20, 71, 85).
Mediator can enhance basal and activator-dependent transcription as well as stimulate the kinase activity of TFIIH (46). However, the presence of cyclin-dependent kinase 8 (cdk8) and cyclin C in some forms of mammalian Mediator complexes appears to play an inhibitory role in activated transcription (84) and also negatively regulates TFIIH kinase activity (3). Yeast homologues of the cdk8/cyclin C pair (i.e., Srb10/Srb11) has been shown to inhibit transcription by phosphorylating the carboxy-terminal domain (CTD) of the largest subunit (RPB1) of pol II, occurring prior to preinitiation complex formation (32). Thus, the cdk8 and cyclin C pair may act as a negative regulator in transcription either as a heterodimer or as part of Mediator components, consistent with the recent biochemical isolation of a dissociable inhibitory module found in some forms of yeast Mediator complexes (6). Nevertheless, several cdk8/cyclin C-containing human Mediator complexes are also active in supporting activator-dependent transcription with either naked DNA or chromatin templates (8, 29, 38, 41, 69). It remains unclear whether the differences in the reported activity of the cdk8/cyclin C pair in Mediator function is due to the heterogeneity of the complexes used in the transcription reactions or the presence of cdk8/cyclin C-inhibitory activity in some of the crude fractions used in the assays. Clearly, modulation of basal transcription by Mediator further complicates its effect on activator-dependent transcription.
To define the mechanisms of Mediator function in the initiation of transcription and to address the functional redundancy or synergy among the three general cofactors (Mediator, TAFs, and PC4), we first purified two forms of human Mediator (Mediator-P.5 and Mediator-P.85). When tested in a transcription system reconstituted with only recombinant proteins (TFIIB, TFIIE, TFIIF, and PC4) and highly purified epitope-tagged multiprotein complexes (TFIID, TFIIH, and pol II), we found that Mediator-P.5 clearly functioned by enhancing activator-mediated recruitment of pol II, whereas Mediator-P.85 worked mainly by stimulating overall basal transcription. The coactivator function of Mediator-P.5 depends on the presence of PC4 but is independent of TAFs. Furthermore, the stimulating effect would be abolished if Mediator-P.5 was added after pol II entry, indicating a critical role of Mediator in enhancing preinitiation complex assembly at the pol II step. Moreover, in the absence of TAFs, Mediator-P.5 is able to enhance promoter recognition by TBP, as illustrated by DNase I footprinting, in an activator-dependent manner. Interestingly, depletion of cdk8-containing polypeptides did not affect the function of Mediator-P.5. Our data thus provide convincing biochemical evidence that Mediator indeed functions by facilitating activator-mediated recruitment of pol II and also promoter recognition by TBP, both of which can occur in the absence of TAFs in TFIID.
MATERIALS AND METHODS
Plasmid constructions.
The retroviral vector carrying FLAG-tagged human Med7 cDNA, pBn-Fo:hMed7, was constructed by cloning an NdeI-KpnI-digested PCR fragment spanning the coding region of the hMed7 cDNA (66) (gift from R. D. Kornberg) into pFLAG°(AS)-7 (92) to create pFo:hMed7-7, in which the resulting FLAG-tagged human Med7 cDNA was excised and cloned into the BamHI and EcoRI sites of pBabe Neo (65) to create pBn-Fo:hMed7. Plasmid pF:hMed7-11d, used for expression of human Med7 in bacteria, was generated by cloning the NdeI-EcoRI fragment of pFo:hMed7-7 into the NdeI and EcoRI sites of pF:E2-11d (35).
The DNA template, pG5MLT-mutTATA, which contains nucleotide substitutions on the adenovirus major late TATA box by changing TATAAAA to TGTGGGA, was created in pG5MLT (83) by PCR amplification with primer pairs spanning the mutated nucleotides according to the QuikChange site-directed mutagenesis protocol (Stratagene). The other transcription templates, pMLΔ53, p2E2(IR)Δ53, and pΔMLP, have already been described (reference 94 and references therein).
Establishment of the hMED7-7 cell line.
The HeLa-derived hMED7-7 cell line, which constitutively expresses FLAG-tagged human Med7, was established by retrovirus-mediated gene transfer with pBn-Fo:hMed7 following described procedures (95).
Protein purification.
To purify human Mediator complexes (Mediator-P.5 and Mediator-P.85), we prepared nuclear extracts from 40 liters of hMED7-7 cells according to published protocols (17). P11 ion-exchange chromatography was then carried out as described previously (45) with 50 ml of hMED7-7 nuclear extracts and a 30-ml phosphocellulose P11 column. The 0.5 and 0.85 M KCl fractions (termed P.5 and P.85, respectively) were dialyzed against BC300 (12) for 5 h, and centrifuged at 14,000 rpm with a Beckman JA-25.50 rotor to remove insoluble material. Immunoaffinity purification of Mediator complexes was conducted by incubating 250 μl of anti-FLAG M2 monoclonal antibody-conjugated agarose beads (Sigma) with 10 ml of the P.5 or P.85 fraction prepared from hMED7-7 or HeLa nuclear extracts at 4°C for 6 to 12 h. The protein-bound beads were washed four times with BC300 plus 0.2% NP-40, with a 5-min rotation at 4°C included between each wash, and eluted with 200 μl of elution buffer (BC300 plus 0.2% NP-40 and 0.2 mg of FLAG peptide/ml) after incubation at 4°C for 1 h. Elutions were repeated for a total of four times.
Bacterially expressed FLAG-tagged human Med7 was purified from BL21(DE3)pLysS harboring pF:hMed7-11d as described previously (13). Purification of recombinant FLAG-tagged TFIIB, FLAG-tagged TBP, FLAG-tagged TFIIEα, FLAG-tagged TFIIEβ, six-histidine-tagged RAP30, six-histidine-tagged RAP74, PC4, and Gal4-VP16 was performed as described previously (93, 96). FLAG-tagged E2 protein was purified from Sf9 insect cells infected with recombinant baculoviruses harboring FLAG-tagged human papillomavirus type 11 E2 cDNA (35). FLAG-tagged multiprotein complexes TFIID, TFIIH, and pol II were purified from HeLa-derived cell lines expressing FLAG-tagged TBP, FLAG-tagged p62 of human TFIIH, and FLAG-tagged RPB9 of human pol II, respectively (12, 93, 96).
Immunodepletion of pol II and cdk8 from Mediator-P.5 and Mediator-P.85.
To remove pol II from Mediator-P.5 and Mediator-P.85, we first generated anti-pol II antibody-conjugated beads by covalently linking 8WG16 monoclonal antibody (86), which recognizes the CTD of the RPB1 subunit of pol II, to protein A-Sepharose CL-4B (Amersham Pharmacia Biotech) as described previously (94). Immunodepletion was then conducted by incubating 1.2 ml of immunoaffinity-purified Mediator complexes (∼60 nM), which contains approximately 2.4 μg of FLAG-tagged Med7, as estimated by quantitative Western blotting with known amounts of bacterially expressed FLAG-tagged human Med7 protein, with 25 μl of immobilized 8WG16 beads (1 mg/ml) at 4°C for 8 h. The supernatant was collected after passing through a microcentrifuge tube filter (Spin-X, Costar). Immunodepletion was conducted twice to ensure a complete removal of pol II from Mediator-P.5 and Mediator-P.85.
Immunodepletion of cdk8 and cyclin C from Mediator-P.5 was similarly performed by incubating 1 ml of pol II-depleted Mediator-P.5 with 25 μl of immobilized anti-cdk8 antibodies (2 mg/ml; Santa Cruz) at 4°C for 8 h and processed as described above for a total of two times.
In vitro transcription.
A standard two-step in vitro transcription reaction was performed with individually purified transcription factors and cofactors as described previously (93, 94). Briefly, 1 nM each supercoiled pG5MLT and p2E2(IR)Δ53 (or pMLΔ53) templates was preincubated with 1 nM TBP or TFIID in the absence or presence of 10 nM TFIIB, 0.24 nM pol II, 1.6 nM TFIIF, 0.8 nM TFIIE, 0.7 nM TFIIH, 75 nM Gal4-VP16, 50 nM E2, 400 nM PC4, or 0.7 nM Mediator-P.5 (or Mediator-P.85) at 30°C for 20 min. The remaining transcription components and NTPs were then added, with or without 10 nM pΔMLP challenge template, to initiate transcription. The reaction was continued at 30°C for 60 min and processed as described previously. Transcription signals were quantified by Typhoon 9200 PhosphorImager (Amersham Biosciences). Unless otherwise specified, fold activation in each set of reactions is defined as the signal intensity from each activator-binding site-containing template relative to that from the same DNA template performed in the presence of PC4 but without activator and Mediator (i.e., the first lane of each reaction set).
Measurement of transcription factor binding on immobilized DNA template.
Transcription with immobilized pG5MLT or pG5MLT-mutTATA template was conducted by first linking a 700-bp pG5MLT (or pG5MLT-mutTATA) promoter fragment, generated by PCR amplification with a sense primer (5′ AACTCGACTGCAGCATATGTATCATACACATACG 3′) annealing to a sequence upstream of the five Gal4-binding sites and a biotinylated antisense primer (5′ biotin-CGATTCATTAATGCAGCTGG 3′) which hybridizes to a region downstream of the 380-bp G-less cassette, to streptavidin Dynabeads (Dynal, Inc.) according to the manufacturer's instructions. Three microliters of the immobilized pG5MLT or pG5MLT-mutTATA template (∼25 ng of DNA/μl of beads) was incubated with 16.5 μl of transcription cocktail (0.11 M HEPES [pH 7.8], 2.3 mg of bovine serum albumin/ml, 20 mM MgCl2, 23 mM dithiothreitol, and 22.5 U of RNasin) at 4°C for 1 h to block nonspecific protein-binding sites on the beads. Sixty-three microliters of protein mix in BC100, which contains approximately threefold more protein than is used in a standard transcription assay, was then added and incubated at 27°C for 30 min. One-seventh of the mixture was used for transcription analysis, which was carried out at 30°C for 30 min after the addition of NTPs. The rest of the immobilized templates with bound proteins were washed with 200 μl of BC80 twice. The protein-bound beads were finally mixed with 10 μl of 1× protein sample buffer and analyzed by Western blotting with different anti-protein antibodies.
DNase I footprinting.
The DNA fragment containing five Gal4-binding sites linked to the adenovirus major late promoter spanning −53 to +10 preceding a G-less cassette of 380 nucleotides, used for DNase I footprinting, was generated by the end-labeling reaction with [γ-32P]ATP using T4 polynucleotide kinase on the immobilized pG5MLT template (368 ng). The 32P-labeled DNA fragment was washed three times with Tris-EDTA and cleaved off the beads with EcoRI (100 U). The supernatant, separated from the beads by magnet, was then removed and used for DNase I footprinting as described previously (35) with some modifications. Briefly, 0.75 nM 32P-labeled DNA fragment was incubated with 56 nM TBP and 20 nM Gal4-VP16, in the presence or absence of 1.2 nM Mediator-P.5, at 30°C for 30 min in the transcription buffer. The reaction mixture was digested with 0.03 U of DNase I (Gibco-BRL) at room temperature for 2 min, followed by addition of an equal volume of stop solution (0.2 M NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, 100 μg of tRNA/ml), and then incubated with 100 μg of proteinase K/ml at 50°C for 30 min. After ethanol precipitation, the final DNA was analyzed on a 6% polyacrylamide-urea sequencing gel. DNA bands were imaged by a Typhoon 9200 scanner (Amersham Pharmacia Biotech).
RESULTS
Isolation of two forms of human Mediator complexes.
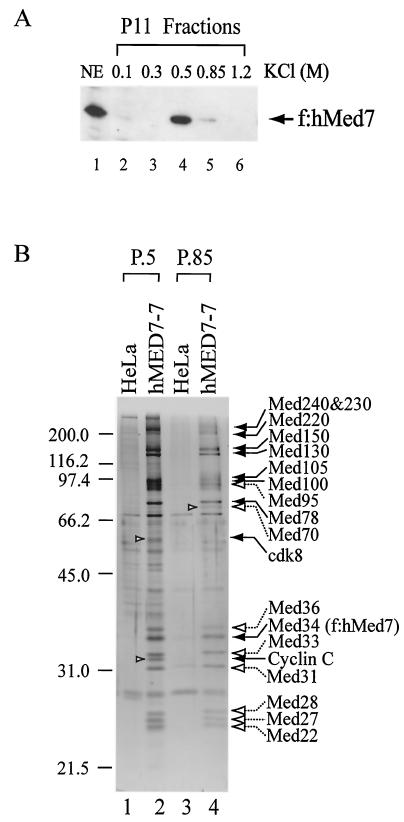
To isolate human Mediator, we established a clonal HeLa-derived cell line (hMED7-7) expressing the FLAG-tagged Med7 (f:hMed7) component of human Mediator. Med7, also named Med34 based on a unifying nomenclature (77), is a universal subunit found in all Mediator complexes identified in different organisms, including yeast, Drosophila, C. elegans, plants, mice, and humans (7). Fractionation of hMED7-7 nuclear extracts by P11 ion-exchange chromatography (Fig. 1A) revealed that the majority of f:hMed7 was present in the 0.5 M KCl fraction (P.5), with some also being found in the 0.85 M KCl fraction (P.85). Mediator was then isolated via immunoaffinity purification and peptide elution, respectively, from the P.5 and P.85 fractions derived from hMED7-7 nuclear extracts. Control purification was conducted in parallel with similar fractions from HeLa nuclear extracts. Many polypeptides corresponding to protein components identified in previously described Mediator complexes, now indicated with the unifying names (77), were found in both Mediator-P.5 and Mediator-P.85 complexes, except the specific presence of cyclin C, cdk8, Med240, and Med230 in Mediator-P.5 and Med70 in Mediator-P.85 (Fig. 1B). The protein composition of Mediator-P.5 appears to be similar to that of ARC-L (85) and TRAP/SMCC (22, 29, 38), whereas Mediator-P.85 seems to be comparable to CRSP (71, 81) and PC2 (57).
FIG. 1.
Isolation of two human Mediator complexes. (A) Detection of epitope-tagged human Med7 in P11 fractions. Nuclear extracts (NE) from HeLa-derived hMED7-7 cells, which express FLAG-tagged human Med7 (f:hMed7), were fractionated over a phosphocellulose P11 ion-exchange column and step eluted with BC buffer (12) containing different concentrations of KCl as indicated. Western blotting was performed with anti-FLAG M2 monoclonal antibody (Sigma). (B) Silver staining of purified human Mediator complexes. Mediator-P.5 and Mediator-P.85 were purified, respectively, from P11 0.5 M (P.5) and 0.85 M (P.85) KCl fractions of hMED7-7 nuclear extracts. Control purification was conducted in parallel with similar fractions derived from HeLa cells. Protein size markers (in kilodaltons) are indicated on the left. The assignment of Mediator components was based on comparison of various Mediator complexes purified from different laboratories with a newly proposed unifying nomenclature (77) and further confirmed by Western blotting with available antibodies against TRAP240/Med240, TRAP230/Med230, TRAP220/Med220, TRAP170/CRSP150/Med150, CRSP130/Med130, ARC105/Med105, TRAP100/Med100, TRAP80/Med78, cdk8, and cyclin C (data not shown). Polypeptides confirmed by Western blotting are indicated by solid arrows, whereas proteins corresponding to previously characterized Mediator components based on the migration patterns are pointed by open arrows with dashed lines. Positions of cyclin C, cdk8, and Med70 are depicted on the gel by open arrowheads.
A nonredundant role of PC4 and Mediator in transcriptional activation.
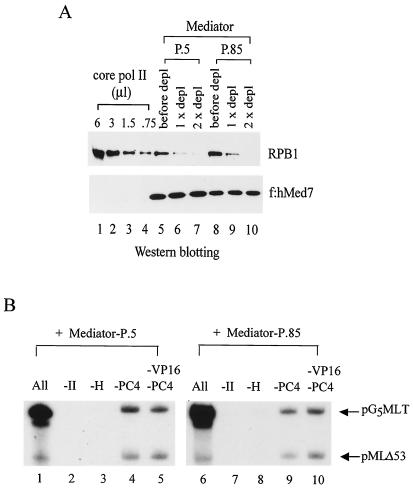
Since pol II was frequently copurified with Mediator and might accidentally contribute to the general stimulation of basal transcription by Mediator, we conducted immunodepletion on the purified Mediator complexes with immobilized 8WG16 monoclonal antibody, which recognizes the CTD present in the RPB1 subunit of pol II, to remove any potential pol II contamination. Indeed, the majority of copurified pol II, even though it was not visually detected by silver staining (see Fig. 1B), was removed after two rounds of immunodepletion (Fig. 2A). To examine whether these pol II-depleted Mediator complexes still retained any pol II activity as found in the original Mediator-P.5 and Mediator-P.85 preparations (data not shown), we conducted a transcription assay using a highly purified in vitro transcription system reconstituted with recombinant TFIIB, TFIIE, TFIIF, and epitope-tagged TFIID, TFIIH, and pol II (96). To monitor basal transcription, we used pMLΔ53 template, which contains the adenovirus major late core promoter linked to a G-less cassette of approximately 280 nucleotides. For activator-dependent transcription, we also included recombinant PC4 and Gal4-VP16 in the reaction and scored the level of transcription from pG5MLT template, which has five copies of the Gal4-binding site preceding the adenovirus major late core promoter in front of a 380-nucleotide G-less cassette. As shown in Fig. 2B, these pol II-depleted Mediator complexes could not substitute for pol II in transcribing pG5MLT and pMLΔ53 templates (compare lanes 1 and 2 and lanes 6 and 7), indicating that pol II activity has been completely removed from Mediator-P.5 and Mediator-P.85. Unlike a previous report (29), TFIIH is essential in our transcription system in the presence of Mediator (Fig. 2B, lanes 1 to 3 and 6 to 8). Interestingly, when PC4 was left out from the reaction with or without a simultaneous removal of Gal4-VP16, only basal transcription was detected from both templates (Fig. 2B, compare lanes 1, 4, and 5 and lanes 6, 9, and 10). This experiment not only demonstrated that PC4 is essential for activator-dependent transcription, but also illustrated a nonredundant role of PC4 and Mediator in transcriptional activation. Clearly, the coactivator function of PC4 could not be substituted by the activity provided by either form of Mediator in our highly purified human transcription system devoid of any cross-contaminating activity among individually purified general transcription factors and pol II.
FIG. 2.
The coactivator function of Mediator depends on the presence of pol II, TFIIH, and PC4. (A) Immunodepletion of pol II from Mediator-P.5 and Mediator-P.85. Anti-pol II CTD antibodies (8WG16) were used to remove pol II from purified Mediator complexes. The amounts of pol II and Mediator remaining after two rounds (1x and 2x) of immunodepletion (depl) were revealed by Western blotting with anti-CTD 8WG16 (for RPB1 detection) and anti-FLAG M2 (for f:hMed7 detection) antibodies, respectively. Purified FLAG-tagged human pol II (9.6 nM, in which 0.75 μl is normally used for one transcription reaction) was loaded in parallel for evaluating the efficiency of immunodepletion. (B) Mediator activity is dependent on exogenous pol II, TFIIH, and PC4. In vitro transcription was reconstituted with recombinant TFIIB, TFIIE, TFIIF, PC4, Gal4-VP16 (VP16), and epitope-tagged multiprotein complexes (TFIID, TFIIH, and pol II) in the presence (+) of Mediator-P.5 or Mediator-P.85. Individual components (pol II, TFIIH, or PC4) were then left out (−) from the reactions as indicated. The pG5MLT template contains five Gal4-binding sites linked to the adenovirus major late core promoter preceding a G-less cassette of approximately 380 nucleotides. The pMLΔ53 template is devoid of activator-binding sites but contains the same adenovirus major late core promoter linked to a shorter G-less cassette (∼280 nucleotides).
Mediator-P.5 enhances activator-dependent transcription, whereas Mediator-P.85 increases overall basal transcription.
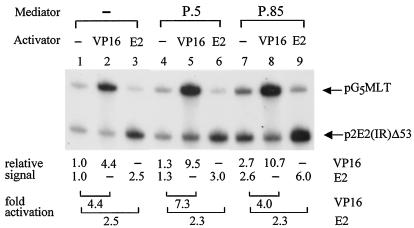
The removal of pol II activity from our purified Mediator complexes allowed us to unambiguously define the role of Mediator-P.5 and Mediator-P.85 on basal and activated transcription without further complications caused by minor pol II activity often found in Mediator preparations. For activator specificity, we also tested the effect of Mediator in supporting activation by the full-length human papillomavirus type 11 (HPV-11) E2 protein, which activates transcription from p2E2(IR)Δ53 template containing two copies of the E2-binding site located upstream of the adenovirus major late core promoter linked to a 280-nucleotide G-less cassette (36, 94). As shown in Fig. 3, addition of Mediator-P.5 to the reconstituted transcription system further enhances activation mediated by Gal4-VP16 but not by HPV-11 E2 (compare lanes 1 to 3 with lanes 4 to 6), indicating a differential requirement of Mediator for activator-dependent transcription. When Mediator-P.85 was examined in parallel, we also detected an increase of transcription by both Gal4-VP16 and E2 (Fig. 3, lanes 7 to 9). The enhanced signal, however, resulted from an increase of basal transcription (Fig. 3, compare lanes 1 and 7). A Mediator-like activity similar to that of Mediator-P.85 in stimulating basal transcription has also been recently detected in HeLa nuclear extracts as well as the derived P.85 fraction by immunodepletion studies with antibodies against specific components of human Mediator (5, 62).
FIG. 3.
Mediator-P.5 enhances activation mediated by Gal4-VP16, whereas Mediator-P.85 stimulates overall basal transcription. In vitro transcription was performed with individually purified transcription components (TFIIB, TFIID, TFIIE, TFIIF, TFIIH, pol II, and PC4) in the absence (−) or presence of Gal4-VP16 (VP16), E2, Mediator-P.5, or Mediator-P.85, as indicated. Fold activation is defined as the signal intensity, quantified by PhosphorImager (Amersham Biosciences), from each activator-binding site-containing template relative to that from the same DNA template performed in the absence of activator and Mediator (i.e., lane 1). The p2E2(IR)Δ53 template contains two human papillomavirus E2-binding sites linked to the same core promoter with a shorter G-less cassette (∼280 nucleotides).
Mediator-P.5 facilitates activator-mediated recruitment of pol II.
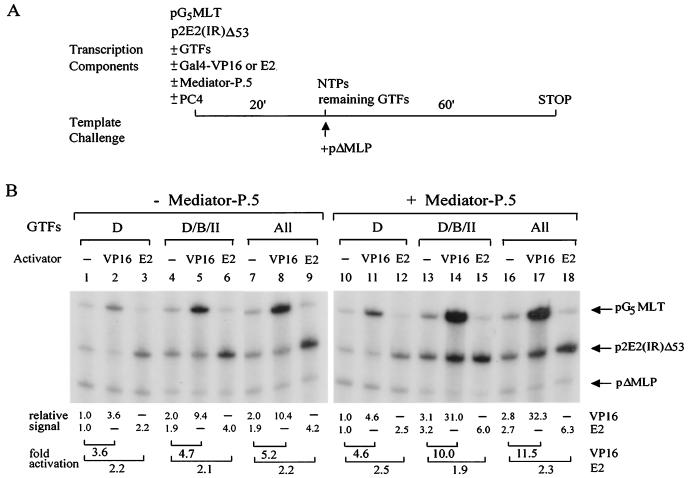
Since Mediator-P.5 is the predominant form of human Mediator complexes (Fig. 1A) and seems to be involved in activator-dependent transcription (Fig. 3), we decided to dissect its role in preinitiation complex assembly. Order-of-addition and template challenge experiments, used previously to define the mechanisms of TFIID- and TBP-mediated activation (94), were conducted by dividing the transcription process into two steps. G-less cassette templates responding to either Gal4-VP16 (pG5MLT) or HPV-11 E2 [p2E2(IR)Δ53] were preincubated with TFIID or other protein factors for 20 min. The remaining transcription components and nucleoside triphosphates (NTPs) were then added, together with a 10-fold excess of an adenovirus major late promoter-driven G-less template (pΔMLP), to initiate transcription (see the outline in Fig. 4A). The pΔMLP template was used to titrate away any unbound proteins and to challenge the stability of protein complexes preformed on pG5MLT and p2E2(IR)Δ53 templates. As shown in Fig. 4B, inclusion of Mediator-P.5 with TFIID during the preincubation step had little effect on basal and activator-stimulated transcription (compare lanes 1 to 3 with lanes 10 to 12), suggesting that Mediator-P.5 works mainly after TFIID binding to the promoter region. Additional supplementation with TFIIB during the preincubation step also did not reveal the coactivator function of Mediator-P.5 (data not shown). Intriguingly, when pol II was included with TFIID and TFIIB during the preincubation step, a strong enhancement of transcription with Mediator-P.5 was observed by Gal4-VP16, but not by E2 (Fig. 4B, lanes 4 to 6 versus lanes 13 to 15). Addition of TFIIF, TFIIE, and TFIIH during preincubation did not further enhance transcription (Fig. 4B, compare lanes 13 to 15 with lanes 16 to 18). We concluded that Mediator-P.5 works mainly by facilitating pol II recruitment by specific transcriptional activators during preinitiation complex assembly.
FIG. 4.
Mediator-P.5 acts by enhancing activator-facilitated recruitment of pol II to the preinitiation complex. (A) Diagram of order-of-addition and template challenge experiments. (B) Mediator-P.5 enhances pol II recruitment by Gal4-VP16. In vitro transcription was performed as outlined in panel A by preincubating pG5MLT and p2E2(IR)Δ53 DNA templates with TFIID (D), together with TFIIB (B) and pol II (II), or the rest (All) of general transcription factors (GTFs), in the absence (−) or presence (+) of PC4, activator, or Mediator-P.5, at 30°C for 20 min. The remaining transcription components and ribonucleoside triphosphates (NTPs) were then added, together with a 10-fold excess of pΔMLP challenge template, which contains only the adenovirus major late core promoter linked to a G-less cassette of approximately 200 nucleotides, to initiate transcription. The reaction was continued at 30°C for 60 min and processed as described in Materials and Methods. Fold activation is defined as the signal intensity relative to that detected in lane 1 for reactions performed in the absence of Mediator-P.5 and in lane 10 for reactions performed in the presence of Mediator-P.5.
Removal of cdk8-associated polypeptides does not impair the coactivator function of Mediator-P.5.
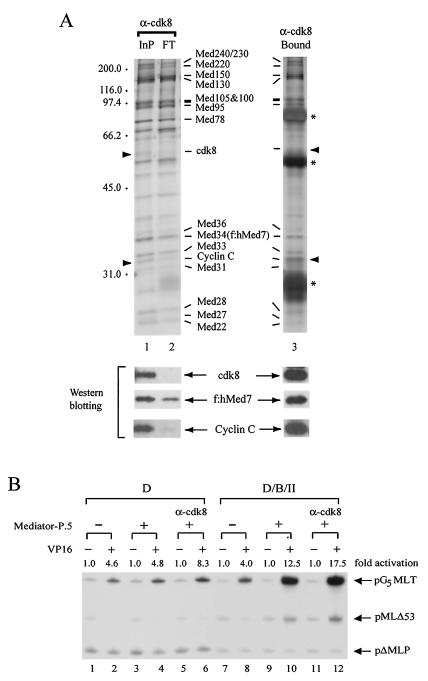
Since cdk8 and cyclin C are considered to be negative regulators of eukaryotic transcription (see the introduction), we asked whether removal of cdk8 and cyclin C would significantly change the coactivator function of Mediator-P.5. Immunodepletion was thus conducted on purified Mediator-P.5 with anti-cdk8 antibodies. The immunodepleted protein profile of Mediator-P.5 was nearly identical to that of undepleted Mediator-P.5 (Fig. 5A, top panel, lanes 1 and 2), except that cdk8 and cyclin C were significantly reduced, as detected by Western blotting (Fig. 5A, bottom panel). Interestingly, the anti-cdk8 antibody-bound proteins contained not only the cdk8 and cyclin C pair, but also the full complement of Mediator-P.5 (Fig. 5A, compare lane 3 with lanes 1 and 2), suggesting that there are two Mediator-P.5 complexes: one with cdk8 and cyclin C and the other without the protein pair. This cdk8/cyclin C-depleted Mediator-P.5 exhibited a slight increase of coactivator activity relative to the undepleted Mediator-P.5 preparation, again mainly at the step of the activator-facilitated recruitment of pol II (Fig. 5B, compare lanes 9 and 10 with lanes 11 and 12). Therefore, removal of the cdk8/cyclin C-containing Mediator complex, which likely corresponds to ARC-L, did not impair the coactivator function of Mediator-P.5, consistent with the observation that ARC-L does not support transcription (85). Interestingly, the cdk8/cyclin C-depleted Mediator-P.5 also exhibited a slight enhancement of transcription at the TFIID step (Fig. 5B, compare lanes 3 and 4 with lanes 5 and 6), suggesting that Mediator-P.5 may also facilitate promoter recognition by TATA-binding factors (see below).
FIG. 5.
The coactivator function of Mediator-P.5 was not impaired following the removal of cdk8-associated polypeptides. (A) Immunodepletion of cdk8-associated polypeptides from Mediator-P.5. Anti-cdk8 (α-cdk8) antibodies were used to remove cdk8-associated polypeptides from Mediator-P.5. The input (InP), flowthrough (FT), and bound Mediator complexes were analyzed by silver staining (top panels) and Western blotting (bottom panels) with the indicated antibodies. Positions of cyclin C and cdk8 are indicated by solid arrowheads. Cross-linked heavy- and light-chain immunoglobulin bands are depicted by asterisks. (B) Coactivator function of Mediator-P.5 before and after cdk8 depletion. In vitro transcription was performed as outlined in the legend for Fig. 4A, except that pMLΔ53 template was used in the absence (−) or presence (+) of Mediator-P.5 without or with (α-cdk8) immunodepletion with anti-cdk8 antibodies. Fold activation in each pair of reactions is defined as the signal increase from pG5MLT performed in the presence of Gal4-VP16 relative to that in its absence.
The coactivator function of Mediator-P.5 is independent of TAFs.
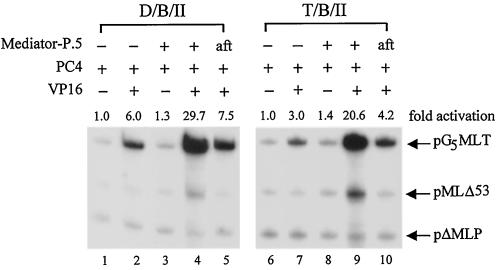
In our previous study (94), we demonstrated that TAFs in TFIID could also enhance activator-facilitated recruitment of pol II. It is likely that TAFs are not required in Mediator-P.5 function if both play a similar role in facilitating the recruitment of pol II by activators. To explore this possibility, we performed order-of-addition and template challenge experiments using either TFIID or TBP as the TATA-binding factor. Indeed, Mediator-P.5 showed a nearly comparable level of coactivator function when TBP was used in place of TFIID (Fig. 6, lanes 1 to 4 versus lanes 6 to 9). This finding suggested that TAFs in TFIID were not essential for Mediator function. Nevertheless, the presence of TAFs appeared to enhance the overall level of activation (Fig. 6, compare lanes 1 and 2 with lanes 6 and 7 and lanes 3 and 4 with lanes 8 and 9). It is important to note that the enhanced signals at the same position as pMLΔ53 transcript (Fig. 6, lanes 4 and 9) are likely due to paused complexes on the pG5MLT template observed at a high level of transcription, as the same signals were similarly detected in the absence of the pMLΔ53 template (data not shown). Consistent with a stimulation of activator-facilitated recruitment of pol II, the coactivator function of Mediator-P.5 was abolished when Mediator-P.5 was added after pol II preincubation (Fig. 6, lanes 1 and 2 versus lanes 3 and 5 and lanes 6 and 7 versus lanes 8 and 10). This experiment further demonstrated that Mediator-P.5 works by enhancing activator-facilitated recruitment of pol II, which can occur in the absence of TAFs.
FIG. 6.
The coactivator function of Mediator-P.5 can occur in the presence or absence of TAFs. In vitro transcription was performed as outlined in the legend for Fig. 4A, with TFIID (D) or TBP (T) as the TATA-binding factor, without (−) or with Mediator-P.5 added either during (+) or after (aft) preincubation. Fold activation in each set of reactions is defined as the signal intensity from pG5MLT relative to that performed in the absence of Gal4-VP16 and Mediator-P.5 (i.e., the first lane of each reaction set).
Recruitment of Mediator is an activator-dependent event and correlates with an enhanced recruitment of pol II to the promoter region.
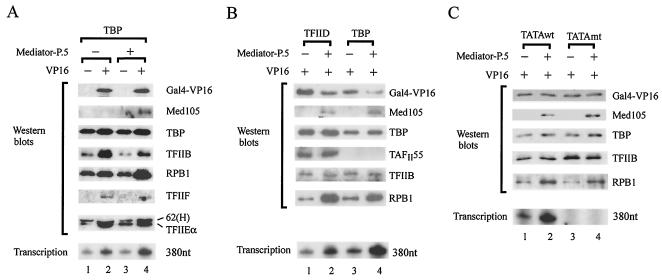
To examine whether the enhancement of transcription by Mediator-P.5 indeed correlates with the recruitment of pol II, we performed reconstituted transcription assays with immobilized pG5MLT linear template using TBP as the TATA-binding factor. Following in vitro transcription, one-seventh of each sample was analyzed for transcription signal, whereas the other six-sevenths was used for Western blotting with antibodies against various components of the general transcription machinery. As shown in Fig. 7A, recruitment of TFIIB, but not TBP or pol II, was dramatically enhanced in the presence of Gal4-VP16 (lanes 1 and 2), consistent with the view that acidic transcriptional activators like Gal4-VP16 appear to enhance transcription by facilitating the recruitment of TFIIB (55). Clearly, recruitment of downstream factors, such as TFIIF, TFIIE, and TFIIH, after pol II entry was also increased (Fig. 7A, lanes 1 and 2). Inclusion of Mediator-P.5 without activator did not lead to its recruitment to the promoter region, nor did its presence alter the amounts of general transcription factors already recruited (Fig. 7A, compare lanes 1 and 3). Interestingly, in the presence of Gal4-VP16, Mediator could then be recruited to the transcription template, with a concomitant increase of pol II recruitment and an enhancement of the transcription signal (Fig. 7A, compare lanes 3 and 4). Our data thus indicate that functional recruitment of Mediator is an activator-dependent event and correlates well with an enhanced recruitment of pol II to the promoter region.
FIG. 7.
Activator-facilitated recruitment of pol II to the promoter region is enhanced in the presence of Mediator-P.5. (A) Activator-dependent recruitment of Mediator to the promoter region. In vitro transcription and Western blotting were conducted with immobilized pG5MLT template in the absence (−) or presence (+) of Mediator-P.5 and Gal4-VP16, as described in Materials and Methods. Antibodies used for Western blotting analysis are indicated on the right of individual strips. (B) Enhancement of activator-facilitated recruitment of pol II by Mediator can occur in the presence or absence of TAFs. In vitro transcription and Western blotting were conducted with immobilized pG5MLT template in the absence (−) or presence (+) of Mediator-P.5 and Gal4-VP16, as described in the legend for panel A. (C) Assembly of transcription complexes can occur in the absence of a functional TATA box. Immobilized template assays were performed with pG5MLT (TATAwt) or pG5MLT-mutTATA (TATAmt) as described in the legend for panel A.
A comparative study was then similarly conducted using either TFIID or TBP as the TATA-binding factor. As expected, addition of Mediator-P.5 enhanced formation of a 380-nt transcript (Fig. 7B, bottom panel). In analyzing the proteins eluted from the immobilized template, we found that the amount of pol II, but not the other general transcription components, was significantly enhanced in the presence of Mediator-P.5, irrespective of the use of TFIID or TBP in the reactions (Fig. 7B). A slight reduction of Gal4-VP16 in the presence of Mediator-P.5 (Fig. 7B, lanes 2 and 4) is likely due to experimental variations, as the amount of Gal4-VP16 in general remains constant (e.g., see Fig. 7C, lanes 1 to 4, Gal4-VP16 panel). Although the presence of TAFs in TFIID could enhance activator-facilitated recruitment of pol II (94), which might slightly increase the amounts of pol II and Gal4-VP16 retained on the immobilized template (Fig. 7B, compare lanes 2 and 4), TAFs could also suppress transcription mediated by TBP when an equivalent amount of TFIID and TBP was used in the transcription assays (22, 93, 96). This might explain why less transcription was detected by TFIID-mediated activation in comparison with TBP-mediated transcription (Fig. 7B, bottom panel).
The assembly of transcription complexes on the immobilized templates may be recruited via core promoter-binding factors (41) or by upstream activators (80, 82), depending on the amounts of proteins used in the assays. To examine whether recruitment of Mediator-P.5 is indeed an activator-dependent event, we used an excess amount of Gal4-VP16 to saturate all activator-binding sites on the immobilized templates and performed transcriptional recruitment assays using immobilized DNA templates containing either a wild-type or mutated TATA box. As shown in Fig. 7C, when nucleotide substitutions were introduced into the TATA box of the pG5MLT template (i.e., changing TATAAAA to TGTGGGA), we still detected the assembly of the transcription components on the TATA-mutated template, even though transcript was not observed (Fig. 7C). This finding suggests that transcription complexes can nevertheless be assembled through non-TATA sequences, likely via upstream activator-binding sites (80, 82), independently of the transcription process. Our data are consistent with recruitment assays performed with other TATA-mutated templates (80, 82). When immobilized template assays were performed with nonsaturating amounts of Gal4-VP16, transcription complex assembly occurs in a TATA box-dependent manner (41).
Mediator and PC4 act through distinct steps during preinitiation complex formation in TAF-dependent and TAF-independent activation.
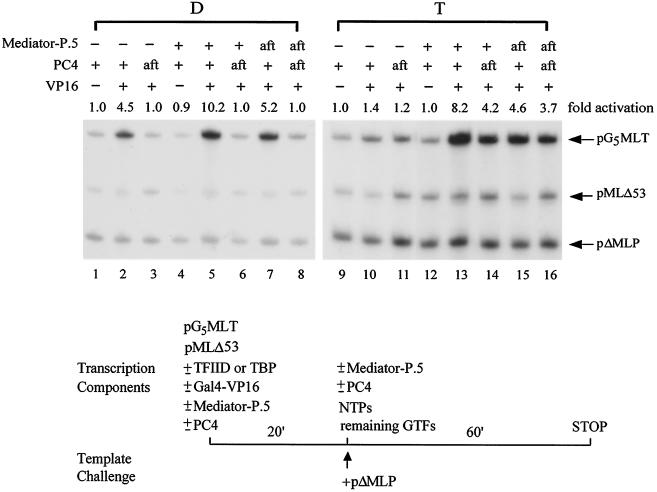
Since promoter recognition by TFIID in TAF-dependent activation and pol II entry into the preinitiation complex in TAF-independent (i.e., TBP-mediated) activation represent two major regulated steps by transcriptional activators (19, 50, 54, 94) and Mediator-P.5 appears to work in a manner similar to TAFs in facilitating activator-mediated recruitment of pol II, we asked whether the activator-regulated step in TAF-independent activation would shift from pol II entry to promoter recognition in the presence of Mediator-P.5. To explore this intriguing possibility, we performed an order-of-addition and template challenge experiment by preincubating pG5MLT and pMLΔ53 DNA templates with either TFIID or TBP, in the absence or presence of Gal4-VP16. Mediator- P.5 and PC4 were added either during or after preincubation. The remaining transcription components and NTPs, together with the challenge template (pΔMLP), were then added to initiate transcription. As shown in Fig. 8, addition of activator, in the absence of Mediator-P.5, at the promoter recognition step enhances TAF-dependent (lanes 1 versus 2), but very little, if any, TAF-independent (lanes 9 versus 10), activation. This stimulation was abolished when PC4 was added after preincubation (lanes 2 versus 3), indicating that the coactivator function of PC4 acts at the promoter recognition step by TFIID. Inclusion of Mediator-P.5 during preincubation significantly enhanced TAF-independent activation (compare lanes 12 and 13 with lanes 9 and 10) but also stimulated approximately twofold TAF-dependent activation (lanes 4 and 5 versus lanes 1 and 2; see also Fig. 5, lanes 2 and 6). Surprisingly, addition of PC4 and Mediator-P.5, individually or together, after the preincubation step completely eliminated the coactivator function of PC4 and Mediator-P.5 in TAF-dependent activation (lanes 5 to 8 versus lanes 2 and 3) but not in TAF-independent activation (lanes 13 to 16 versus lanes 10 and 11). These data indicate that (i) the coactivator function of Mediator-P.5 and PC4 was eliminated after preincubation of DNA templates with TFIID, but not with TBP; (ii) Mediator-P.5 and PC4 could still enhance activator function after promoter recognition by TBP, indicating that Mediator and PC4 were able to enhance activator-facilitated recruitment of downstream factors in the absence of TAFs; (iii) Mediator could not functionally replace TAFs in switching the activator-regulated step from pol II entry to promoter recognition in TAF-independent activation (Fig. 8), indicating that Mediator and TAFs, although they share some common coactivator activity in activator-facilitated recruitment of pol II, do not function equivalently during preinitiation complex assembly; and (iv) in the absence of TAFs, Mediator is able to enhance promoter recognition by TBP, in addition to its role in facilitating pol II entry to the preinitiation complex.
FIG. 8.
The coactivator function of Mediator-P.5 and PC4 was eliminated after preincubation of DNA templates with TFIID, but not with TBP. In vitro transcription was performed as outlined, without (−) or with Mediator-P.5 and PC4 added either during (+) or after (aft) preincubation.
Mediator-P.5 is able to enhance promoter recognition by TBP in an activator-dependent manner.
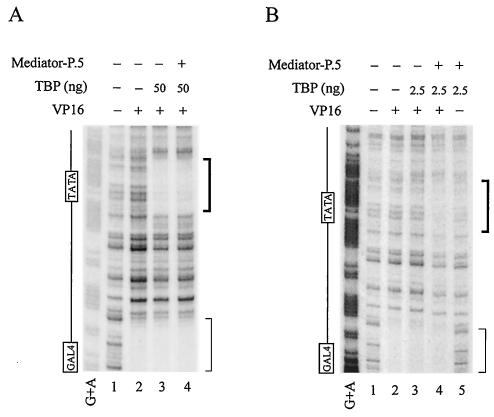
To directly demonstrate whether Mediator-P.5 could indeed enhance promoter recognition by TBP, we carried out DNase I footprinting with a promoter fragment containing five Gal4-binding sites linked to the adenovirus major late promoter preceding a 380-nucleotide G-less cassette. As shown in Fig. 9A, distinct footprints over the TATA box and the activator-binding sites were not further extended by the presence of Mediator-P.5, as all the TBP- and Gal4-VP16-binding sites had been saturated. When a limiting amount of TBP, which was unable to generate a clear protection over the TATA box, was used in the footprinting assay, Mediator-P.5 could indeed promote TATA box recognition by TBP (Fig. 9B, compare lanes 3 and 4). This enhancement of promoter recognition by TBP in the presence of Mediator-P.5 was significantly reduced when Gal4-VP16 was omitted (Fig. 9B, lanes 4 and 5), suggesting that Mediator-P.5 is able to enhance promoter recognition by TBP in an activator-dependent manner.
FIG. 9.
Mediator-P.5 enhances promoter recognition by TBP in an activator-dependent manner. (A) DNase I footprinting performed with a high concentration of TBP. DNase I footprinting was performed with a 32P-labeled DNA fragment containing five Gal4-binding sites linked to the adenovirus major late promoter preceding a 380-nucleotide G-less cassette in the absence (−) or presence (+) of Mediator- P.5, TBP, and Gal4-VP16 (VP16), as described in Materials and Methods. The protected regions surrounding the TATA box and the Gal4-binding site are bracketed with thick and thin lines, respectively. (B) DNase I footprinting performed with a low concentration of TBP. The footprinting assay was performed as described in the legend for panel A, except using 0.18 nM DNA, 2.8 nM TBP, 10 nM Gal4-VP16, and 0.005 U of DNase I.
DISCUSSION
The isolation of human Mediator complexes and the availability of a human transcription system reconstituted with only recombinant proteins and epitope-tagged multiprotein complexes without cross-contaminating activities allowed us to unambiguously define the mechanisms of Mediator function and to further address functional cooperativity and redundancy among three general cofactors, namely, Mediator, PC4, and TAFs. The significance of this work lies in the following facts. (i) This study provides biochemical evidence to convincingly demonstrate that Mediator functions as a bridging factor between activators and pol II, without complications caused by the existence of other protein cofactors whose presence in vivo or in transcription systems containing some partially purified fractions may accidentally mask the intrinsic activity of Mediator. (ii) This work documents that when assayed under a nonrepressed condition, the coactivator function of Mediator, which is separate from its general stimulation on basal transcription, strictly depends on PC4 but is independent of TAFs. (iii) This study shows that Mediator recruitment to the promoter region is an activator-dependent process in both TAF-dependent and TAF-independent activation. (iv) This is the first functional illustration that in the absence of TAFs, Mediator is able to enhance promoter recognition by TBP, in addition to its role in facilitating pol II entry to the preinitiation complex.
Multiple forms of Mediator complexes.
In yeast, the core of Mediator can interact with a dissociable repressor module containing Srb8, Srb9, Srb10, and Srb11 (6) to generate diversity of Mediator complexes (7). A similar module consisting of humanMed230/TRAP230/ARC240, Med240/TRAP240/ARC250,cdk8, and Cyclin C was also found in Mediator-P.5 but was significantly reduced or undetectable in Mediator-P.85 (Fig. 1B). Clearly, multiple forms of Mediator complexes are present in eukaryotes, including yeast (44, 56), Drosophila (28), C. elegans (51), and humans (57, 62, 85, 89; this study). They may share some common properties due to the presence of core polypeptides, but they also likely show distinct functions as conferred by the association with other protein modules or polypeptides. In this work, we found that stimulation of basal transcription and enhancement of activator-dependent transcription are two separate activities intrinsic to distinct forms of human Mediator complexes. This conclusion was also supported by experiments using immunodepletion of transcription activity from nuclear extracts with antibodies against specific components of Mediator (5, 62). Interestingly, two similar but distinct forms of human Mediator (CRSP and ARC-L) exhibit a dramatic difference in coactivator function and pol II association (71, 85). Only the smaller form of Mediator complex, CRSP, is highly active in transcription and is able to interact with the CTD of pol II. Our Mediator-P.85, whose protein composition appears similar to that of CRSP, did not exhibit coactivator activity as reported by CRSP in our highly purified reconstituted transcription system. Nevertheless, the enhancement of basal transcription exhibited by our Mediator-P.85 is consistent with the finding of a basal-stimulating activity by a Mediator complex detected in the P.85 fraction of HeLa nuclear extracts (62). The functional difference between Mediator-P.5 and CRSP may be due to the use of different transcription templates and activators in distinct transcription conditions, which remains to be further explored. In contrast, the larger form of Mediator complex, ARC-L, is transcriptionally inactive, which is likely due to a specific presence of the repressor module containing ARC240, ARC250, cdk8, and cyclin C in ARC-L (84), in agreement with the biochemical characterization of a repressor form of cdk8-containing Mediator complex (83). However, in other studies, removal of cdk8-associated polypeptides from purified Mediator did not significantly alter the functional property of the undepleted complex (69, 89; this study), indicating either a nonrepressing or an inactive role of the cdk8 module in the complex. This discrepancy remains to be further investigated.
Activator-facilitated recruitment of the transcription complex.
Recruitment of general transcription factors and cofactors by gene-specific activators is an important event leading to the initiation of transcription. In a previous study, we found that promoter recognition by TFIID in TAF-dependent activation and recruitment of pol II for preinitiation complex formation in TAF-independent activation constitute two major steps regulated by transcriptional activators (94). When Mediator was additionally included in this transcription system, it worked mainly by enhancing activator-facilitated recruitment of pol II in both TAF-dependent and TAF-independent activation (Fig. 4 to 6 and data not shown). Consistent with a recent study performed with HeLa nuclear extracts (82), recruitment of Mediator to the promoter region is an activator-dependent process in vivo (15, 75) and in vitro (Fig. 7A and 9B). This finding further documents the interesting observation that an activator is necessary to target the nonspecific DNA-binding activity of general cofactors (PC4 and Mediator) and chromatin-modifying complexes (SWI/SNF and GCN5) to specific promoter regions and thereby enhance transcription of activated genes (1, 2, 31). The assembly of the transcription complex, however, was not dependent on the presence of a functional TATA box and, instead, is likely to be recruited through upstream activator-binding sites (80) (Fig. 7C). Yet, in TATA-less promoters such as the human TAFII55 gene (99, 100), activator-facilitated recruitment of the general cofactors and pol II complexes may also be mediated through the initiator and downstream core promoter elements. Thus, it will be of great interest to examine whether transcription complexes are assembled on TATA-less promoters through a distinct assembly pathway.
Functional cooperativity among three general cofactors.
The coactivator function of the three general cofactors (Mediator, TAFs, and PC4) appears nonredundant, as mutations on individual subunits of the complexes, even within the same module, show a differential effect on genome-wide gene expression patterns (34). This is in part due to recruitment of discrete general cofactors by different transcription activators. In this study, we found that Mediator-P.5 could enhance activation by Gal4-VP16, but not by HPV-11 E2 protein, indicating that E2 may recruit general cofactors distinct from Mediator for gene activation. Indeed, Mediator was not found in the E2-cellular complexes isolated from a clonal human cell line conditionally expressing HPV-11 E2. Instead, E2 assembles into a large multiprotein complex with TFIID and other bromodomain-containing chromatin-modifying cofactors (S.-Y. Wu, S. Y. Hou, and C.-M. Chiang, unpublished data). This selective recruitment of Mediator and TFIID for activation by Gal4-VP16 and HPV-11 E2, respectively, further reinforces the view that general cofactors are differentially required for gene activation in vivo (11, 23, 25, 27, 37, 39, 51, 61, 64, 82, 88, 98).
In our completely defined human cell-free transcription system, TAFs are not required for Mediator-facilitated activation by Gal4-VP16 (Fig. 6 to 8), a result consistent with immunodepletion experiments performed with HeLa nuclear extracts (73) and with in vitro transcription assays reconstituted with some partially purified fractions and DNA templates responding to activation by thyroid hormone receptor (22). The involvement of TAFs nonetheless becomes obvious when transcription assays were conducted in a suppressive environment imposed by the use of chromatin templates or by the presence of nonchromosomal inhibitory activities (5, 30, 41, 48, 50, 53, 54, 68, 81, 97). Undoubtedly, TAFs can work cooperatively with Mediator to enhance the overall level of activation, although the mechanism of synergy remains to be further elucidated.
The previous finding that TAFs also work by enhancing activator-facilitated recruitment of the initiation form of pol II into the preinitiation complex (94) reveals a functional similarity between TAFs and Mediator in activator-dependent transcription. Surprisingly, the coactivator function of Mediator was apparently blocked when Mediator was added after preincubation of TFIID with DNA templates (Fig. 8). Presumably, TAFs induce a topological change in promoter structure (72), which somehow restricts the access of preformed Mediator-pol II complexes to the promoter region. Alternatively, TAFs and Mediator may compete for the same binding surfaces on pol II. In our studies, Mediator could not functionally replace TAFs in switching the activator-regulated step from pol II entry to promoter recognition in TAF-independent activation (Fig. 8), indicating that Mediator and TAFs, although they share some common coactivator activity in activator-facilitated recruitment of pol II, do not function equivalently during preinitiation complex assembly. The ability of TAFs to function collectively as a sequence-specific core promoter-binding factor in binding to initiator and downstream promoter elements also distinguishes the properties of TAFs from those exhibited by Mediator and PC4.
In this work, we found that PC4 is essential for the coactivator function of Mediator in our cell-free transcription system (Fig. 2 and 8) and can work cooperatively with Mediator as described previously for the synergy between PC2 and other USA-derived coactivators (57). PC4 apparently acts at the promoter recognition step in TAF-dependent activation, as reported previously (43), but can also function at a later step(s) in TAF-independent activation (Fig. 8). It will be of great interest to see how these general cofactors work synergistically with other types of transcriptional coactivators and corepressors in facilitating transcription from both naked DNA and chromatin templates, which will further shed light on the functional properties of Mediator, PC4, and TAFs in eukaryotic transcription.
Acknowledgments
We thank R. D. Kornberg for human Med7 cDNA; D. Reinberg for pG5MLT and pΔMLP DNA templates; R. G. Roeder for antibodies against TRAP240, TRAP230, TRAP220, TRAP170, TRAP150, TRAP100, and TRAP80; and R. Tjian for anti-ARC105, anti-CRSP150, and anti-CRSP130 antibodies. We are also grateful to D. Luse for advice on immobilized DNA template and P. de Haseth, D. Luse, E. Stavnezer, and D. Samols for comments on the manuscript.
C.-M.C. is a Mt. Sinai Health Care Foundation Scholar. This work was supported by grants GM59643 and CA81017 from the National Institutes of Health.
REFERENCES
- 1.Acevedo, M. L., and W. L. Kraus. 2003. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor α-dependent transcription with chromatin templates. Mol. Cell. Biol. 23:335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 4.Asturias, F. J., Y. W. Jiang, L. C. Myers, C. M. Gustafsson, and R. D. Kornberg. 1999. Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283:985-987. [DOI] [PubMed] [Google Scholar]
- 5.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 7.Boube, M., L. Joulia, D. L. Cribbs, and H.-M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 9.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, M., and J. A. Jaehning. 1997. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 25:4861-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Z., and J. L. Manley. 2000. Robust mRNA transcription in chicken DT40 cells depleted of TAFII31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol. 20:5064-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang, C.-M., H. Ge, Z. Wang, A. Hoffmann, and R. G. Roeder. 1993. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12:2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, C.-M., and R. G. Roeder. 1993. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Res. 6:62-64. [PubMed] [Google Scholar]
- 14.Chiang, C.-M., and R. G. Roeder. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531-536. [DOI] [PubMed] [Google Scholar]
- 15.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the Mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 16.Davis, J. A., Y. Takagi, R. D. Kornberg, and F. J. Asturias. 2002. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell 10:409-415. [DOI] [PubMed] [Google Scholar]
- 17.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 19.Dorris, D. R., and K. Struhl. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 20:4350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson, Y. W. Jiang, Y. Li, R. D. Kornberg, and F. J. Asturias. 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97:14307-14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fondell, J. D., F. Brunel, K. Hisatake, and R. G. Roeder. 1996. Unliganded thyroid hormone receptor α can target TATA-binding protein for transcriptional repression. Mol. Cell. Biol. 16:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fondell, J. D., M. Guermah, S. Malik, and R. G. Roeder. 1999. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl. Acad. Sci. USA 96:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freiman, R. N., S. R. Albright, S. Zheng, W. C. Sha, R. E. Hammer, and R. Tjian. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293:2084-2087. [DOI] [PubMed] [Google Scholar]
- 24.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 25.Ge, K., M. Guermah, C.-X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 26.Gill, G., E. Pascal, Z. H. Tseng, and R. Tjian. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gim, B. S., J. M. Park, J. H. Yoon, C. Kang, and Y.-J. Kim. 2001. Drosophila Med6 is required for elevated expression of a large but distinct set of developmentally regulated genes. Mol. Cell. Biol. 21:5242-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu, J.-Y., J. M. Park, E. J. Song, G. Mizuguchi, J. H. Yoon, J. Kim-Ha, K.-J. Lee, and Y.-J. Kim. 2002. Novel mediator proteins of the small mediator complex in Drosophila SL2 cells. J. Biol. Chem. 277:27154-27161. [DOI] [PubMed] [Google Scholar]
- 29.Gu, W., S. Malik, M. Ito, C.-X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3:97-108. [DOI] [PubMed] [Google Scholar]
- 30.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 32.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 33.Hoey, T., R. O. Weinzierl, G. Gill, J.-L. Chen, B. D. Dynlacht, and R. Tjian. 1993. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72:247-260. [DOI] [PubMed] [Google Scholar]
- 34.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 35.Hou, S. Y., S.-Y. Wu, T. Zhou, M. C. Thomas, and C.-M. Chiang. 2000. Alleviation of human papillomavirus E2-mediated transcriptional repression via formation of a TATA binding protein (or TFIID)-TFIIB-RNA polymerase II-TFIIF preinitiation complex. Mol. Cell. Biol. 20:113-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou, S. Y., S.-Y. Wu, and C.-M. Chiang. 2002. Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J. Biol. Chem. 277:45619-45629. [DOI] [PubMed] [Google Scholar]
- 37.Ito, M., H. J. Okano, R. B. Darnell, and R. G. Roeder. 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 21:3464-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito, M., C.-X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z.-Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 39.Ito, M., C.-X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 40.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, K. M., J. Wang, A. Smallwood, C. Arayata, and M. Carey. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16:1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser, K., and M. Meisterernst. 1996. The human general co-factors. Trends Biochem. Sci. 21:342-345. [PubMed] [Google Scholar]
- 43.Kaiser, K., G. Stelzer, and M. Meisterernst. 1995. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 14:3520-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee, and Y.-J. Kim. 2001. The structural and functional organization of the yeast Mediator complex. J. Biol. Chem. 276:42003-42010. [DOI] [PubMed] [Google Scholar]
- 45.Kershnar, E., S.-Y. Wu, and C.-M. Chiang. 1998. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J. Biol. Chem. 273:34444-34453. [DOI] [PubMed] [Google Scholar]
- 46.Kim, Y.-J., S. Björklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 47.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466-469. [DOI] [PubMed] [Google Scholar]
- 48.Komarnitsky, P. B., B. Michel, and S. Buratowski. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13:2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kretzschmar, M., K. Kaiser, F. Lottspeich, and M. Meisterernst. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78:525-534. [DOI] [PubMed] [Google Scholar]
- 50.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 51.Kwon, J. Y., J. M. Park, B. S. Gim, S. J. Han, J. Lee, and Y.-J. Kim. 1999. Caenorhabditis elegans mediator complexes are required for developmental-specific transcriptional activation. Proc. Natl. Acad. Sci. USA 96:14990-14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 53.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 54.Li, X.-Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 55.Lin, Y.-S., and M. R. Green. 1991. Mechanism of action of an acidic transcriptional activator in vitro. Cell 64:971-981. [DOI] [PubMed] [Google Scholar]
- 56.Liu, Y., J. A. Ranish, R. Aebersold, and S. Hahn. 2001. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276:7169-7175. [PubMed] [Google Scholar]
- 57.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 58.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. USA 95:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 60.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 61.Metzger, D., E. Scheer, A. Soldatov, and L. Tora. 1999. Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 18:4823-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mittler, G., E. Kremmer, H. T. M. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizzen, C. A., X.-J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 64.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 65.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Myers, L. C., C. M. Gustafsson, D. A. Bushnell, M. Lui, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 1998. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 68.Näär, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Näär, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 70.Näär, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 71.Näär, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oelgeschläger, T., C.-M. Chiang, and R. G. Roeder. 1996. Topology and reorganization of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 73.Oelgeschläger, T., T. Yong, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 74.Park, J. M., B. S. Gim, J. M. Kim, J. H. Yoon, H.-S. Kim, J.-G. Kang, and Y.-J. Kim. 2001. Drosophila mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol. 21:2312-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 76.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 77.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 78.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Näär, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 79.Rachez, C., Z. Suldan, J. Ward, C.-P. B. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 82.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 83.Sun, X., D. Ma, M. Sheldon, K. Yeung, and D. Reinberg. 1994. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 8:2336-2348. [DOI] [PubMed] [Google Scholar]
- 84.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 85.Taatjes, D. J., A. M. Näär, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 86.Thompson, N. E., D. B. Aronson, and R. R. Burgess. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265:7069-7077. [PubMed] [Google Scholar]
- 87.Verrijzer, C. P., K. Yokomori, J.-L. Chen, and R. Tjian. 1994. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science 264:933-941. [DOI] [PubMed] [Google Scholar]
- 88.Walker, S. S., J. C. Reese, L. M. Apone, and M. R. Green. 1996. Transcription activation in cells lacking TAFIIs. Nature 383:185-188. [DOI] [PubMed] [Google Scholar]
- 89.Wang, G., G. T. Cantin, J. L. Stevens, and A. J. Berk. 2001. Characterization of Mediator complexes from HeLa cell nuclear extract. Mol. Cell. Biol. 21:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Werten, S., G. Stelzer, A. Goppelt, F. M. Langen, P. Gros, H. T. M. Timmers, P. C. V. der Vliet, and M. Meisterernst. 1998. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 17:5103-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]
- 92.Wu, S.-Y., and C.-M. Chiang. 1996. Establishment of stable cell lines expressing potentially toxic proteins by tetracycline-regulated and epitope-tagging methods. BioTechniques 21:718-725. [DOI] [PubMed] [Google Scholar]
- 93.Wu, S.-Y., and C.-M. Chiang. 1998. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J. Biol. Chem. 273:12492-12498. [DOI] [PubMed] [Google Scholar]
- 94.Wu, S.-Y., and C.-M. Chiang. 2001. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276:34235-34243. [DOI] [PubMed] [Google Scholar]
- 95.Wu, S.-Y., and C.-M. Chiang. 2001. Expression and purification of epitope-tagged multisubunit protein complexes from mammalian cells, p. 16.22.1-16.22.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Hoboken, N.J. [DOI] [PubMed]
- 96.Wu, S.-Y., E. Kershnar, and C.-M. Chiang. 1998. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 17:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu, S.-Y., M. C. Thomas, S. Y. Hou, V. Likhite, and C.-M. Chiang. 1999. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J. Biol. Chem. 274:23480-23490. [DOI] [PubMed] [Google Scholar]
- 98.Zhou, J., J. Zwicker, P. Szymanski, M. Levine, and R. Tjian. 1998. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 95:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou, T., and C.-M. Chiang. 2001. The intronless and TATA-less human TAFII55 gene contains a functional initiator and a downstream promoter element. J. Biol. Chem. 276:25503-25511. [DOI] [PubMed] [Google Scholar]
- 100.Zhou, T., and C.-M. Chiang. 2002. Sp1 and AP2 regulate but do not constitute TATA-less human TAFII55 core promoter activity. Nucleic Acids Res. 30:4145-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]