Abstract
The fission yeast checkpoint protein Crb2, related to budding yeast Rad9 and human 53BP1 and BRCA1, has been suggested to act as an adapter protein facilitating the phosphorylation of specific substrates by Rad3-Rad26 kinase. To further understand its role in checkpoint signaling, we examined its localization in live cells by using fluorescence microscopy. In response to DNA damage, Crb2 localizes to distinct nuclear foci, which represent sites of DNA double-strand breaks (DSBs). Crb2 colocalizes with Rad22 at persistent foci, suggesting that Crb2 is retained at sites of DNA damage during repair. Damage-induced Crb2 foci still form in cells defective in Rad1, Rad3, and Rad17 complexes, but these foci do not persist as long as in wild-type cells. Our results suggest that Crb2 functions at the sites of DNA damage, and its regulated persistent localization at damage sites may be involved in facilitating DNA repair and/or maintaining the checkpoint arrest while DNA repair is under way.
Eukaryotic cells respond to DNA damage by activating checkpoint signaling pathways conserved from yeast to humans (26, 51). Current models of DNA damage checkpoint signaling envision distinct groups of sensor, adapter, and effector proteins acting in a sequential manner to effect checkpoint responses (20, 35). Sensor proteins act at the top of the pathways by recognizing the DNA lesions. Adapter proteins then transduce the signal from the sensors to downstream effectors (Table 1).
TABLE 1.
Checkpoint signaling proteins in S. pombe and S. cerevisiae
| Type of protein | Protein(s) found in:
|
|
|---|---|---|
| S. pombe | S. cerevisiae | |
| Sensor | Rad3 and Rad26 | Mec1 and Lcd1 |
| Rad1, Rad9, and Hus1 | Rad17, Ddc1, and Mec3 | |
| Rad17 | Rad24 | |
| Adapter | Crb2 | Rad9 |
| Mrc1 | Mrc1 | |
| Effector | Chk1 | Chk1 |
| Cds1 | Rad53 | |
Three checkpoint protein complexes have been suggested to play roles in sensing DNA damage. The components of these complexes in fission yeast are the “checkpoint Rad proteins,” including Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1. The Rad3-Rad26 complex is a protein kinase orthologous to budding yeast Mec1-Lcd1 and mammalian ATR-ATRIP (8). Rad17 associates with four replication factor C (RFC) subunits to form a pentameric complex that is thought to load the ring-shaped Rad1-Rad9-Hus1 “sliding clamp” onto DNA (11, 39, 46). Recent studies showed that the ATR-ATRIP complex and the checkpoint sliding clamp are recruited to the chromatin at the sites of DNA damage through distinct mechanisms (16, 20, 34, 36, 52). The independent recruitment of these two complexes at sites of DNA damage suggests that proper recognition of DNA damage may require multiple checkpoint sensors with distinct functions.
In fission yeast, the checkpoint Rad proteins are required for both the replication checkpoint that senses stalled replication forks and activates the effector kinase Cds1 (Chk2) and the DNA damage checkpoint that activates the effector kinase Chk1 (6, 32). The different outputs of the two checkpoint pathways are likely to be determined by adapter checkpoint proteins, which provide specificity to the checkpoint responses initiated by a common set of sensor proteins (41). The fission yeast adapter protein involved in the DNA damage checkpoint is Crb2, which shares sequence similarity in its C-terminal tandem BRCT repeats with budding yeast Rad9 and mammalian 53BP1 and BRCA1 (37, 49). Crb2 is specifically involved in the DNA damage checkpoint and is required for the activation of the effector kinase Chk1. In response to DNA damage, Crb2 becomes hyperphosphorylated (37). This DNA damage-induced phosphorylation of Crb2 requires the checkpoint Rad proteins but not Chk1, suggesting that Crb2 acts downstream of the checkpoint Rad proteins but upstream of Chk1. The nature of the damage-induced phosphorylation of Crb2 and its significance in Crb2 function are not known. Studies of the Crb2-related protein in budding yeast, ScRad9, have provided more mechanistic details on the function of checkpoint adapters. The DNA damage-induced phosphorylation of ScRad9 by the sensor kinases is essential for its function (38). Only the phosphorylated ScRad9 can interact with the effector kinase Rad53 and promote the activation of Rad53 through in trans phosphorylation (10).
The fact that checkpoint sensor proteins translocate to sites of DNA damage implies that they carry out the signaling function at these sites (16, 21, 36). Therefore, it is reasonable to hypothesize that the immediate downstream components of the signaling cascade, the checkpoint adapter proteins, are also recruited to the sites of DNA damage. To explain the requirement of the checkpoint sliding clamp complex for the phosphorylation of adapter proteins, it has also been postulated that the checkpoint sliding clamp may recruit adapter proteins to sites of DNA damage where it can be phosphorylated by the sensor kinases (20, 35). We wished to examine these hypotheses by monitoring the localization of fission yeast adapter protein Crb2 in response to DNA damage.
Fluorescence microscopy analysis of budding yeast checkpoint proteins tagged with green fluorescent protein (GFP) has yielded valuable information about the checkpoint sensor proteins (21). However, no conclusive result on the localization of checkpoint adapter proteins was obtained. In this study, we created strains of fission yeast in which Crb2 was tagged with color variants of GFP. We found that Crb2 rapidly concentrates at discrete nuclear foci in response to DNA damage, and these foci represent the sites of DNA double-strand breaks (DSBs). Surprisingly, damage-induced Crb2 focus formation is independent of the checkpoint Rad proteins. However, the checkpoint Rad proteins are required for the persistence of Crb2 foci. We suggest that Crb2 is recruited to sites of DNA damage independent of checkpoint Rad proteins and the maintenance of Crb2 at sites of DNA damage is under the control of the checkpoint Rad proteins.
MATERIALS AND METHODS
Antibody.
A polyclonal antibody against Crb2 was generated with a glutathione S-transferase (GST) fusion of the N-terminal fragment of Crb2 (amino acids 1 to 275) purified from Escherichia coli as an antigen to immunize rabbits.
Yeast strains and plasmids.
The strains used in this study are listed in Table 2. The cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) coding sequences were from plasmids pDH3 and pDH5, provided by the Yeast Resource Center (YRC) at the University of Washington (http://depts.washington.edu/∼yeastrc/) (13). To tag Crb2 at its N terminus, two tandem copies of YFP or CFP coding sequence were inserted behind the start codon of the Crb2 open reading frame in a genomic fragment containing both the upstream and downstream intergenic regions. The constructs containing tagged Crb2 were integrated at the leu1 locus of a crb2Δ mutant. Rad22 was tagged by integrating at the rad22 locus a plasmid containing a C-terminal Rad22 fragment fused with two tandem copies of CFP. The HO site was constructed by annealing two 51-base-long oligonucleotides and inserting the resultant fragment into a plasmid containing a kanMX4 marker. The sequences of the two oligonucleotides used for constructing the HO site are GAATTCGGCCAGGTACCTTTCAGCTTTCCGCAACAGTATAAAGTACTGTAC and AGTACTTTATACTGTTGCGGAAAGCTGAAAGGTACCTGGCCGAATTCGTAC. The HO site in conjunction with the kanMX4 marker was inserted at the arg3 or lys1 locus by PCR methods (2, 17). The HO-expressing plasmid was made by inserting a 1.8-kb fragment containing the HO coding sequence (27) into the NruI site in pJR1-41XH (22), and the resulting plasmid was designated pLD102.
TABLE 2.
Genotypes of the S. pombe strains used in this study
| Strain | Genotype |
|---|---|
| KS1599 | h+leu1-32 |
| LLD3259 | h− leu1-32 ura4-D18 crb2-D2::ura4+ |
| LLD3260 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+ |
| LLD3261 | h+leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+ |
| LLD3262 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 |
| LLD3263 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 arg3::HO site-kanMX4 |
| LLD3264 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 lys1::HO site-kanMX4 |
| LLD3265 | leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 arg3::HO site-kanMX4 lys1::HO site-kanMX4 |
| LLD3266 | h+leu1-32::2xCFP-crb2+-leu1+ura4−crb2-D2::ura4+his3-D1 his7::dis1 promoter-GFP-LacI-NLS-his7+arg3::HOsite-kanMX4 lys1::lacO repeat-lys1+ |
| LLD3267 | h+leu1-32::2xCFP-crb2+-leu1+ura4 crb2-D2::ura4+his3-D1 his7::dis1 promoter-GFP-LacI-NLS-his7+lys1::HO site-kanMX4-lacO repeat |
| LLD3268 | h+leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad22-2xCFP-kanMX6 |
| LLD3269 | leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad22-2xCFP-kanMX6 his3-D1 arg3::HO site-kanMX4 |
| LLD3270 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad3::ura4+ |
| LLD3271 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad1::ura4+ |
| LLD3272 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad9::ura4+ |
| LLD3273 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad17::ura4+ |
| LLD3274 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+rad26::ura4+ |
| LLD3275 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+chk1::ura4+ade6-704 |
| LLD3276 | leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 arg3::HO site-kanMX4 chk1::ura4+ |
| LLD3277 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4+his3-D1 arg3::HO site-kanMX4 rad3::ura4+ |
| LLD3278 | h− leu1-32::2xYFP-crb2+-leu1+ura4-D18 crb2-D2::ura4 cdc10-129 |
GFP fluorescence microscopy.
Unless otherwise noted, cells were grown at room temperature in Edinburgh minimal medium with necessary supplements (1). Cells grown at room temperature have stronger YFP-Crb2 signal and weaker background fluorescence. Cells were concentrated by centrifugation and kept on ice before microscopy. Microscopy was performed with a DeltaVision optical sectioning microscope model 283 equipped with a YFP/CFP filter set and a Photometrics CH350L cooled charge-coupled device camera. Images were acquired with a ×60, 1.4 NA objective and a ×1.5 optivar. Eight Z-sections at 0.5-μm intervals were photographed and projected into one image using softWoRx software. Between 80 and 200 cells were counted for each sample. To estimate the number of Crb2 foci induced by 1 Gy of gamma radiation, we subtracted the average number of Crb2 foci per cell in untreated cells from the average number of Crb2 foci per cell in cells treated with 1.5, 3, and 6 Gy of gamma radiation and extrapolated the values to the dose of 1 Gy based on the assumption that the number of damage-induced foci is directly proportional to the dose of gamma radiation.
RESULTS
Crb2 is recruited to sites of DSBs generated by HO endonuclease.
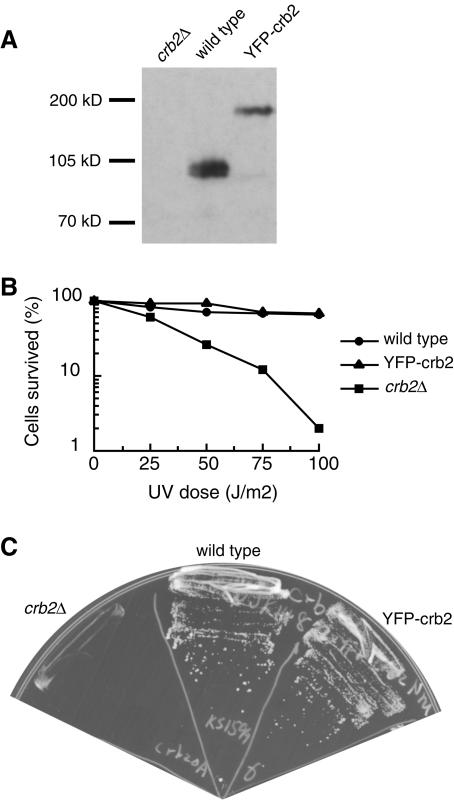
To monitor the localization of Crb2 in live cells we created a strain expressing Crb2 tagged with YFP at approximately the endogenous level (Fig. 1A). YFP-Crb2 is functional as the only copy of Crb2 in the cell because it confers the wild-type level of resistance to UV radiation and hydroxyurea (HU) (Fig. 1B and C). Consistent with the previous report (37), we found that Crb2 showed a diffuse nuclear distribution in most of the cells (Fig. 2A). A small fraction of the cells had nuclear foci—mostly one focus per cell. The percentage of these cells varied from 10 to 20%, depending on growth conditions. These foci are likely to result from spontaneous DNA damage. Spontaneous nuclear foci formed by other checkpoint proteins have been observed before (21).
FIG. 1.
YFP-Crb2 is expressed at approximately endogenous level and is functional as the only copy of Crb2 in the cell. (A) Western blot analysis of lysates made from crb2Δ, wild-type, and YFP-Crb2 strains using an anti-Crb2 polyclonal antibody. Equal amounts of lysate were loaded in each lane. (B) The YFP-Crb2 strain is not more sensitive to UV radiation than the wild type. (C) The YFP-Crb2 strain is not more sensitive to 10 mM HU than the wild type.
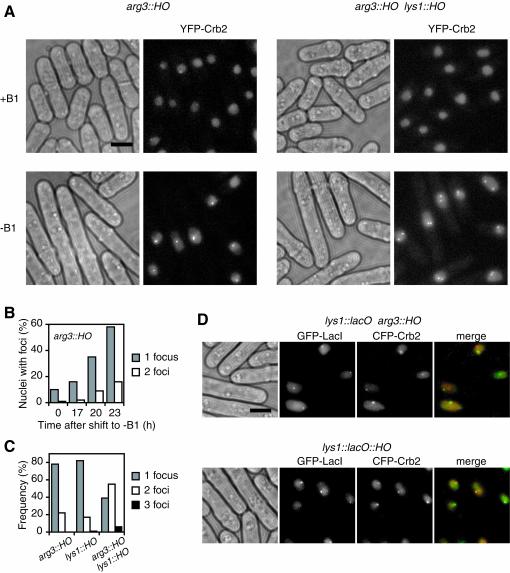
FIG. 2.
Crb2 localizes to sites of DSBs generated by HO endonuclease. (A) Expression of HO endonuclease leads to Crb2 nuclear focus formation in cells with HO sites. YFP-Crb2 was expressed under the crb2 promoter as the only copy of Crb2 in the cell, and HO was expressed under the inducible promoter. Cells with one HO site inserted at the arg3 locus or two HO sites at the arg3 and lys1 loci were grown under repression conditions (+B1) or induction conditions for 23 h (−B1). (B) Induction time course of YFP-Crb2 focus formation in cells with one HO site at the arg3 locus. (C) The number of foci per nucleus after 23 h of induction of HO correlated with the number of HO sites in the cell. More than 60% of nuclei contained foci for each strain. The percentages shown were derived from dividing the count of nuclei with a specific number of foci by the count of the nuclei with foci. (D) HO-induced CFP-Crb2 foci (green) colocalized with GFP-LacI (red) labeling the lys1 locus when an HO site was inserted at lys1 but not when an HO site was inserted at arg3. Scale bar, 5 μm.
In budding yeast, the HO endonuclease generates a single DSB in the genome at the MAT locus, and it has been employed to study cellular responses to the induction of DSBs (12). Heterologous expression of HO endonuclease in fission yeast can generate specific DSBs (27). To examine the localization of Crb2 in response to DSBs at different locations in the fission yeast genome, we inserted the HO cleavage site into arg3 and lys1 loci separately in different strains or together in the same strain. These two loci are located 1.5 Mb apart on chromosome I. HO endonuclease was expressed from an episomal plasmid under the control of the thiamine (B1)-repressible nmt1 promoter from the pREP41 plasmid (3). Under repression conditions, only the background level of Crb2 foci was detected in cells containing one or two HO sites (Fig. 2A). Removal of thiamine led to elongation of the cells and, concurrently, induction of Crb2 nuclear focus formation (Fig. 2A and B). Cell elongation was due to the checkpoint-dependent cell cycle arrest, because chk1 and rad3 null mutations abolished this phenotype (see Fig. 5C). Cells that did not form induced Crb2 foci were often not elongated. These cells may have lost the HO-expressing plasmid. Full induction of the nmt promoter takes at least 20 h at room temperature (43). The maximum percentage of cells containing Crb2 foci varied from 60 to 90% between transformants and was reached around 23 h postinduction. Cells containing foci stopped dividing but continued to elongate after 23 h and eventually died. Cell death is likely due to prolonged cell cycle arrest induced by persistent DSBs generated by continually expressed HO. HO expression resulted in the formation of a single Crb2 nuclear focus in most of the cells containing a single HO site at arg3 or lys1 loci (Fig. 2A and C). In contrast, cells containing two HO sites more frequently formed two Crb2 foci in response to HO expression. The correlation between the number of HO-induced foci and the number of HO sites suggests that Crb2 foci represent the sites of DSBs generated by HO endonuclease.
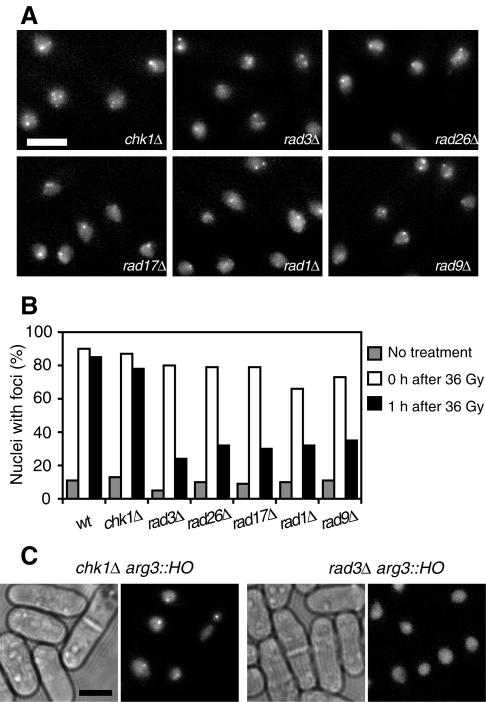
FIG. 5.
Checkpoint Rad proteins are required for the maintenance but not the establishment of the localization of Crb2 to sites of DSBs. (A) YFP-Crb2 focus formation immediately after gamma radiation is independent of Chk1, Rad3, Rad26, Rad17, Rad1, and Rad9. A dose of 36 Gy of gamma radiation was applied to the cells. (B) Gamma radiation-induced foci disappear faster in rad3Δ, rad26Δ, rad17Δ, rad1Δ, and rad9Δ mutants. (C) The rad3Δ mutant but not the chk1Δ mutant fails to form HO-induced YFP-Crb2 foci. Scale bar, 5 μm.
To directly demonstrate that Crb2 foci were at DNA lesion sites, we introduced the HO site into a fission yeast strain carrying the lac operator-repressor system, which allows visualization of the lys1 locus in live cells (23). The HO site is 2.8 kb away from the boundary of the LacO repeats. To simultaneously visualize Crb2 and the GFP-tagged lac repressor (GFP-LacI) that labels the lys1 locus, we tagged Crb2 with CFP. CFP-tagged Crb2 is also functional as the only copy of Crb2 in the cell and forms HO-induced nuclear foci like YFP-tagged Crb2 (unpublished data). When we expressed HO endonuclease in cells containing the HO site at the arg3 locus, no significant overlap was observed between the Crb2 foci and the LacI foci (Fig. 2D). Only 6% of the CFP-Crb2 foci colocalized with the GFP-LacI foci in cells with both CFP and GFP foci. In contrast, when HO was expressed in cells containing the HO site at the lys1 locus, 59% of the CFP-Crb2 foci colocalized with the GFP-LacI foci in cells with both CFP and GFP foci. The CFP-Crb2 foci that did not colocalize with GFP-LacI in these cells likely represented sites of spontaneous DNA damage. The localization of Crb2 foci to the sites of HO cleavage confirms that Crb2 is recruited to the sites of DNA damage.
Crb2 is recruited to sites of DSBs generated by gamma irradiation.
In the experiment system that we described above, it takes more than 20 h to fully induce the HO endonuclease with the nmt promoter, and the induction is not entirely synchronous among the cells. Once induced, the continuous expression of HO endonuclease will generate persistent DSBs. To analyze the relocalization of Crb2 in response to repairable DNA damage, we examined Crb2 focus formation in response to gamma irradiation. Fission yeast cells can withstand a much higher dose of gamma radiation than mammalian cells. A dose of 100 Gy of gamma radiation has little effect on the viability of the wild-type cells and results in about 10% survival of the most sensitive checkpoint mutants such as rad3Δ (4). We found that doses of gamma radiation far lower than the lethal level are sufficient to induce Crb2 focus formation within minutes after the treatment (Fig. 3A). The rapidity and sensitivity of Crb2 focus formation in response to gamma radiation suggest that Crb2 relocalization is an early event in the cellular response to DNA damage induced by gamma radiation.
FIG. 3.
Crb2 localizes to sites of DSBs generated by gamma radiation. (A) Gamma radiation induces YFP-Crb2 nuclear focus formation. (B) The number of Crb2 foci correlates with the dose of gamma radiation. Scale bar, 5 μm.
Ionizing radiation is known to produce a plethora of different types of molecular damage to DNA (47). Among them, DNA DSBs are the lesion most closely associated with the biological consequences of the radiation (14). Therefore, checkpoint proteins may be recruited only to sites of DSBs, and Crb2 foci may represent such sites. To address this possibility, we examined whether the number of foci match the number of DSBs. Due to the resolution limit, it is difficult to accurately count the number of foci when a nucleus contains more than three foci. Hence, we classified nuclei with foci into three categories (one focus, two foci, and three or more foci per nucleus) and plotted the percentage of each category against the radiation dose (Fig. 3B). Both the total percentage of the nuclei with foci and the number of foci per nucleus increased with the dose of gamma radiation. Based on these data, we roughly estimated that 1 Gy of gamma radiation generated 0.12 foci per cell (see Materials and methods). According to data obtained by gel electrophoresis methods, the mean value of DSB yield produced by low-linear-energy-transfer radiation is 5.5 × 10−9 DSBs per Gy per bp (31). The genome size of fission yeast is 13.8 Mb (50). Therefore, the calculated yields are about 0.076 DSB per Gy per cell in G1-phase cells and 0.15 DSB per Gy per cell in G2-M-phase cells. Most of the cells in an asynchronized fission yeast culture are in G2-M phases. Hence the calculated number of DSBs is similar to the number of Crb2 foci that we observed by fluorescence microscopy. Considering that gamma radiation produces other types of DNA damage, such as single-strand breaks and base damage at frequencies 1 order of magnitude higher than DSBs (47), the close match between the number of Crb2 foci and the number of DSBs suggests that Crb2 is specifically recruited to the biologically significant lesions, the DSBs.
Colocalization of Crb2 foci and Rad22 foci.
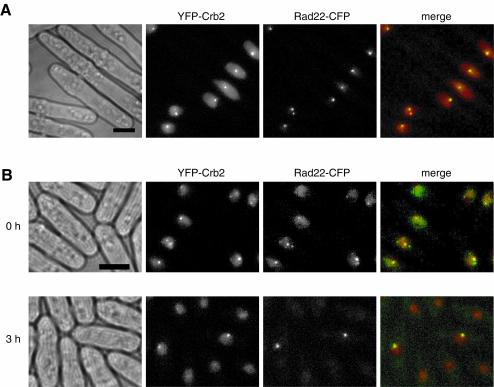
Sites of DNA damage recruit not only checkpoint signaling proteins, but also DNA repair proteins. Fission yeast Rad22, a Rad52 homolog, is a DNA repair protein required for homologous recombination (45). Chromatin immunoprecipitation analysis has shown that Rad22 binds DSBs at the mating locus in vivo (15). We tagged Rad22 with CFP and found that Rad22-CFP is functional and shows a diffuse nucleoplasmic distribution in most of the cells, with occasional observation of spontaneous nuclear foci. The spontaneous Rad22 foci may represent sites of DSBs formed when the leading-strand replication is stalled at the mating locus imprint (9) or sites of DNA breaks arising from erroneous replication. Treatments that damage DNA led to the formation of Rad22 nuclear foci. When we generated DNA damage with HO in cells expressing both YFP-Crb2 and Rad22-CFP, 82% of the Crb2 foci colocalized with Rad22 foci, and 97% of the Rad22 foci colocalized with Crb2 foci (Fig. 4A). This result suggests that both proteins are associated with the sites of persistent DNA damage created by HO endonuclease.
FIG. 4.
Crb2 colocalizes with Rad22 to sites of DSBs. (A) YFP-Crb2 foci (red) and Rad22-CFP (green) foci induced by HO overlapped with each other. (B) YFP-Crb2 and Rad22-CFP partially colocalize to gamma radiation-induced foci. Rad22-CFP foci became brighter after 3 h of recovery; therefore, the diffuse nucleoplasmic signal is no longer visible after normalization of the image to the intensity of foci. Scale bar, 5 μm.
Rad22 also forms dose-dependent foci in response to gamma radiation. When we applied 36 Gy of gamma radiation to cells expressing both YFP-Crb2 and Rad22-CFP, within minutes after the treatment, 47% of the Crb2 foci colocalized with Rad22 foci, and 63% of the Rad22 foci colocalized with Crb2 foci (Fig. 4B). This result suggests that both proteins are almost simultaneously recruited to many gamma radiation-induced DNA lesions. When cells are allowed to recover from the gamma radiation, both Crb2 foci and Rad22 foci gradually disappear. The fluorescence intensity of the Crb2 foci does not change significantly during the recovery. However, the Rad22 foci in recovering cells were brighter than the foci formed initially after radiation. Three hours after a treatment with 36 Gy of gamma radiation, 90% of the remaining Crb2 foci colocalized with Rad22 foci, and 92% of the Rad22 foci colocalized with Crb2 foci (Fig. 4B). These foci are likely to be DNA lesions that are difficult to repair, and as a result, a higher local concentration of DNA repair proteins accumulated at these sites. The persistent localization of Crb2 at DNA lesions undergoing repair may be needed to regulate DNA repair and/or to maintain the checkpoint signal.
Different genetic requirement for the initial and persistent localization of Crb2 to sites of DSBs.
Previous work has shown that DNA damage-induced phosphorylation of Crb2 depends on the checkpoint Rad proteins (37). The checkpoint Rad proteins are thought to recruit Crb2 to sites of DNA damage, where it in turn recruits checkpoint effectors such as Chk1 (20, 35). To test this hypothesis, we investigated whether the localization of Crb2 to sites of DNA damage requires the other checkpoint proteins.
The rapid formation of Crb2 foci in response to gamma radiation occurred in chk1, rad3, rad26, rad17, rad1, and rad9 null mutants (Fig. 5A and B). Therefore, none of the checkpoint proteins encoded by these genes is needed for the initial targeting of Crb2 to DNA lesions. However, during recovery, the rate of disappearance of foci varies among the mutants. One hour after a treatment with 36 Gy of gamma radiation, in wild-type and chk1Δ cells, the percentage of cells containing Crb2 foci remained at nearly the level seen immediately after the treatment (Fig. 5B). In contrast, more than half of the rad3Δ, rad26Δ, rad17Δ, rad1Δ, and rad9Δ cells had completely lost Crb2 foci at this point. DNA repair is not likely to occur faster in the checkpoint mutants than in the wild-type cells, so Crb2 foci may have disappeared in the rad mutants despite unrepaired DNA damage. This result suggests that the checkpoint Rad proteins but not Chk1 are required for the maintenance of Crb2 at the sites of DNA damage. Because the rad mutants are more sensitive to gamma radiation than wild-type and chk1Δ cells (4), to exclude the possibility that the rad mutants lose Crb2 foci due to the loss of viability, we examined the induction of Crb2 foci by a second dose of 36 Gy of gamma radiation 1 h after the cells received a first 36-Gy treatment. The rapid induction of Crb2 foci still occurred after the second treatment of the rad mutants (unpublished data); therefore, the faster disappearance of Crb2 foci in these cells is not due to changes that permanently prevent Crb2 from association with the sites of DSBs such as the degradation of Crb2.
Consistent with the notion that checkpoint Rad proteins control the persistent localization of Crb2 at sites of DSBs, we did not observe the formation of HO-induced foci at higher-than-background levels in rad3Δ and rad1Δ cells containing the HO site, even though most of these cells eventually died when HO was expressed (Fig. 5C) (unpublished data). The lengthy and asynchronous nature of the HO induction probably makes it impossible to observe any transient formation of Crb2 foci. The lack of HO-induced foci in rad3Δ and rad1Δ cells did not result from the absence of cell cycle arrest, because chk1Δ cells did not arrest, but nonetheless formed HO-induced foci (Fig. 5C).
Crb2 localization to sites of DSBs is regulated by cell cycle.
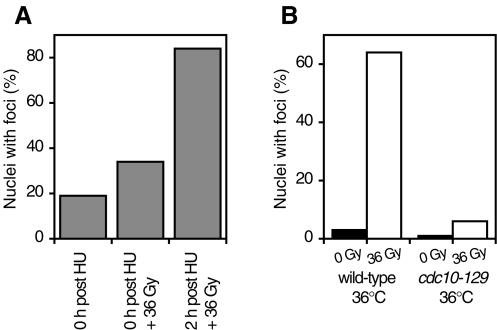
Fission yeast apparently limit the activation of the DNA damage checkpoint to certain cell cycle stages since HU- or cdc10-arrested cells cannot activate Chk1 in response to DNA damage (5, 19). Cdc10 is a transcription factor required for passage through Start (18, 25). Crb2 is essential for the activation of Chk1 (37). Therefore, we investigated whether the localization of Crb2 to sites of DNA damage is regulated by the cell cycle. Cells immediately released from HU arrest were not proficient in Crb2 focus formation in response to gamma radiation (Fig. 6A), whereas 2 h after release from HU arrest, gamma radiation-induced foci formed to a similar level as those in asynchronized cells, suggesting that the localization of Crb2 to DNA lesions is blocked at the early S phase of the cell cycle. Similarly, we found that arresting cells at G1 stage by shifting a cdc10 temperature-sensitive mutant to restrictive temperature also abolished the gamma radiation-induced Crb2 focus formation (Fig. 6B). The correlation between the cell cycle stages lacking Chk1 activation and the cell cycle stages when Crb2 foci cannot form suggests that the cell cycle regulation of DNA damage checkpoint signaling may be achieved by the control of Crb2 localization.
FIG. 6.
Localization of Crb2 to sites of DSBs is regulated by cell cycle. (A) Early-S-phase-arrested cells fail to form YFP-Crb2 foci in response to gamma radiation. Cells grown in medium containing 12 mM HU for 4 h were washed with medium and then immediately treated with 36 Gy of gamma radiation or allowed to grow for an additional 2 h and then irradiated. (B) G1-arrested cells fail to form YFP-Crb2 foci in response to gamma radiation. Wild-type or cdc10-129 cells were shifted from 25°C to 36°C for 4 h and then treated with 36 Gy of gamma radiation or mock treated.
DISCUSSION
One central question in the study of DNA damage checkpoint signaling is how the signaling molecules detect DNA damage and initiate the checkpoint responses (35). The findings that two checkpoint complexes are recruited to the sites of DNA damage independently (16, 21) indicate that the damage-sensing step of the signaling pathway may require an intricate system of multiple sensors. The data that we present here reveal the additional complexity of this system. Crb2 relocalization to the sites of DSBs is an early event in checkpoint signaling and is independent of other checkpoint proteins. Furthermore, we discovered that the recruitment and retention of Crb2 at DNA lesion sites have different genetic requirements.
Crb2 functions at sites of DSBs.
Microscopy analysis of the localization of proteins involved in DNA damage response has shown that many of them form nuclear foci in response to DNA damage (35). These foci are commonly thought to represent sites of DNA damage. However, evidence directly linking such foci to the sites of DNA damage is scarce. When human cell nuclei were treated with ultrasoft X rays in conjunction with an irradiation mask, the Mre11 protein was found to concentrate to regions of the nuclei that have been exposed to radiation (24). UVA microirradiation of human cells sensitized by the incorporation of halogenated thymidine analogues demonstrated that γ-H2AX, Rad50, Nbs1, BRCA1, and Rad51 clustered in irradiated regions of nuclei (29, 33, 42). These and other partial-volume irradiation studies demonstrated that the proteins involved in DNA damage response are recruited to regions of DNA damage but did not address the question of whether damage-induced foci represent individual DNA lesions. A study using GFP-tagged protein in budding yeast provided probably the best evidence that each damage-induced focus represents a DNA damage site (21). They found that a single HO-induced DSB resulted in a single Ddc1 or Lcd1 focus, whereas multiple foci formed when multiple DSBs were induced in the telomeric region. In the same study, the Crb2-related protein ScRad9 was found to form very faint damage-induced nuclear foci in a fraction of the cells, suggesting ScRad9 is also recruited to DNA lesion sites. The difficulty in detecting ScRad9 foci may be due to lower local concentration of ScRad9 compared to Ddc1 and Lcd1.
Several of our observations suggest that Crb2 nuclear foci represent sites of DSBs. First, Crb2 foci were induced by the expression of HO endonuclease, which generates specific DSBs in the genome (12). Second, the number of HO-induced Crb2 foci correlated with the number of DSBs produced by HO cleavage. Third, HO-induced Crb2 foci colocalized with the GFP-LacI foci that marked the sites of DSBs. Fourth, the number of Crb2 foci induced by gamma radiation was proportional to the dose of gamma radiation and consistent with the estimated number of DSBs induced by gamma radiation. Additionally, we observed significant colocalization between Crb2 foci and Rad22 foci, especially among the persistent foci. Rad22 plays an important role in DSB repair and is known to associate with sites of DSBs (15).
Ionizing radiation generates multiple types of DNA damage. Among them, base damage and single-strand breaks far outnumber DSBs (47). The match between the number of gamma radiation-induced Crb2 foci and the estimated number of DSBs implied that Crb2 is specifically recruited to the sites of DSBs. We also observed that UV radiation induced rapid formation of Crb2 nuclear foci in a dose-dependent manner, and lower numbers of foci were induced by UV compared to gamma radiation, which produces the same killing effect (unpublished data). These UV-induced foci may represent the small number of DSBs generated by UV radiation (30).
The biological significance of the relocalization of checkpoint proteins to the sites of DNA damage is not easy to assess. In budding yeast, point mutations in Lcd1, which abolish the ability of the Mec1-Lcd1 complex to localize to sites of DNA damage, result in a phenotype similar to that of lcd1Δ cells (36), suggesting that the recruitment of Mec1-Lcd1 kinase to DNA lesions is essential for its function. In this study, we found that G1-phase and early-S-phase cells fail to form Crb2 foci in response to gamma radiation. Cells in these cell cycle stages are also unable to activate Chk1 in response to DNA damage (5, 19). This correlation suggests that relocalization of Crb2 to sites of DNA damage may be essential for the checkpoint response.
In budding yeast, two checkpoint complexes, the ATR-like kinase Mec1-Lcd1 and the checkpoint sliding clamp, can be recruited to the sites of DNA damage independent of each other and also independent of the Crb2-related protein ScRad9 (16, 21). It is likely that the counterparts of these two complexes in fission yeast, Rad3-Rad26 kinase and the Rad1-Rad9-Hus1 complex, are also recruited to the sites of DNA damage. In fission yeast, Crb2 is specific for the DNA damage checkpoint pathway, whereas Rad3-Rad26 kinase and the Rad1-Rad9-Hus1 complex are also required for the replication checkpoint (6, 32). It is possible that Crb2 provides specificity to the DNA damage checkpoint pathway due to its ability to recognize DSBs.
It is not clear whether the checkpoint proteins associate with chromatin at sites of DNA damage through direct binding to aberrant DNA structures or through interaction with other proteins, such as the repair factors. There is evidence that some checkpoint proteins possess in vitro DNA binding activity. Purified ATM preferentially binds DNA ends (40), and purified ATR preferentially binds UV-damaged DNA (44). Recombinant Lcd1 can bind DNA in vitro and Lcd1 was proposed to be the DNA binding subunit of the Mec1-Lcd1 complex (36). BRCA1, which shares structural and functional homology with Crb2, also has been shown to bind DNA directly (28). We have found that Crb2 purified from either fission yeast or E. coli has in vitro DNA binding ability (unpublished data). However, we cannot exclude the possibility that, in vivo, the recruitment of Crb2 to DSBs requires other proteins.
The checkpoint Rad proteins regulate the persistent localization of Crb2 at DNA lesions during repair.
It has been shown that, in budding yeast, checkpoint proteins Ddc1 and Lcd1 remain associated with the persistent DNA damage sites generated by HO nuclease for a prolonged period (21). We have shown here that Crb2 colocalizes with Rad22 in persistent foci, which are likely to be DNA lesions undergoing repair. These results suggest that checkpoint proteins remain attached at the sites of DNA damage during the DNA repair process. This persistent association may be essential for the maintenance of a checkpoint signal while repair is under way. Since Crb2 has been implicated in the regulation of DNA repair (7), it is also possible that Crb2 may interact with repair proteins at persistent loci. These possibilities are not mutually exclusive.
We found that checkpoint Rad proteins are required for the persistence of damage-induced Crb2 foci, suggesting a mechanism for these proteins to regulate DNA repair and/or the maintenance of checkpoint arrest. Although an earlier study using a temperature-sensitive allele of rad3 suggested that Rad3 is required to initiate but not maintain the DNA damage checkpoint arrest (19), we have found that inactivation of Rad3 by using the same allele leads to premature release from cell cycle arrest (unpublished data). The requirement for the checkpoint Rad proteins to maintain the persistent localization of Crb2 at sites of DNA damage may reflect the change in DNA structures during DNA repair. The repair of DSBs involves the resection of DNA ends that leads to the formation of the 3′-ended single-stranded DNA (48). We hypothesize that Crb2 by itself can associate with DSBs but not with extensively resected DNA ends. Rad3-Rad26 kinase and the checkpoint sliding clamp are needed to maintain the association of Crb2 with DNA lesion sites once the DNA ends have been processed to a certain extent. Based on this model, in wild-type cells, Crb2 is released from the chromatin only after the completion of DNA repair, whereas in the rad mutants, Crb2 prematurely dissociates from the DNA lesion sites after the extensive processing of DSBs.
Crb2 is hyperphosphorylated in response to DNA damage, and this phosphorylation is dependent on the checkpoint Rad proteins (37). It is tempting to speculate that this phosphorylation event is needed to maintain the localization of Crb2 at sites of persistent damage. The damage-induced phosphorylation may stabilize the association of Crb2 with chromatin by increasing its affinity for DNA or proteins localized at the damage sites. In order to assess the significance of the damage-induced phosphorylation, it will be necessary to map the sites of phosphorylation in Crb2.
Conclusions.
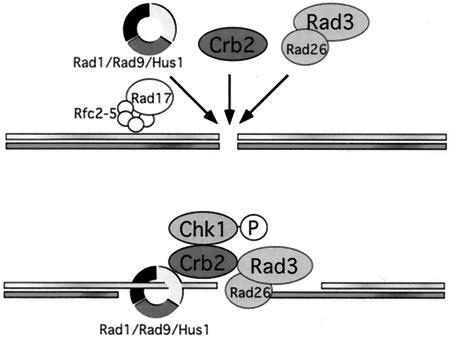
Based on our results, we propose a new model for the initiation and maintenance of the DNA damage checkpoint in fission yeast (Fig. 7). The general checkpoint protein Rad1 and Rad3 complexes and the DNA damage specific checkpoint protein Crb2 are recruited to DSBs independent of each other. The independent targeting of Crb2 to DSBs ensures the eliciting of specific checkpoint responses, such as the phosphorylation of Chk1. After the processing of DSBs, Rad1 and Rad3 complexes are required for the persistent localization of Crb2 to the DNA lesions, which may be involved in the maintenance of checkpoint signals during DNA repair.
FIG. 7.
A model for the initiation and maintenance of DNA damage checkpoint in fission yeast. The Rad1-Rad9-Hus1 complex, Rad3-Rad26 complex, and Crb2 are recruited to DSBs independent of each other. After the processing of DSBs, Rad1 and Rad3 complexes facilitate the persistent localization of Crb2 to the DNA lesions.
Acknowledgments
We thank S. Subramani for the DNA construct encoding HO endonuclease, M. Yanagida for the lys1::lacO strain, J. E. Haber for advice on the HO system, C. H. McGowan for critical reading of the manuscript, and the Yeast Resource Center at the University of Washington for plasmids encoding YFP and CFP. L.-L. Du is a fellow of the Leukemia and Lymphoma Society.
This work was supported by National Institutes of Health grants to P. Russell. T. M. Nakamura was supported by the Damon Runyon Cancer Research Foundation (DRG-1565).
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. Mcleod, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 3.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, N. J., D. A. Holtzman, G. Flaggs, K. S. Keegan, A. DeMaggio, J. C. Ford, M. Hoekstra, and A. M. Carr. 1996. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 15:6641-6651. [PMC free article] [PubMed] [Google Scholar]
- 5.Brondello, J.-M., M. N. Boddy, B. Furnari, and P. Russell. 1999. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 19:4262-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 7.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 9.Dalgaard, J. Z., and A. J. Klar. 2001. Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17:153-157. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 11.Griffith, J. D., L. A. Lindsey-Boltz, and A. Sancar. 2002. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 277:15233-15236. [DOI] [PubMed] [Google Scholar]
- 12.Haber, J. E. 2002. Uses and abuses of HO endonuclease. Methods Enzymol. 350:141-164. [DOI] [PubMed] [Google Scholar]
- 13.Hailey, D. W., T. N. Davis, and E. G. Muller. 2002. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 351:34-49. [DOI] [PubMed] [Google Scholar]
- 14.Iliakis, G. 1991. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. BioEssays 13:641-648. [DOI] [PubMed] [Google Scholar]
- 15.Kim, W. J., S. Lee, M. S. Park, Y. K. Jang, J. B. Kim, and S. D. Park. 2000. Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J. Biol. Chem. 275:35607-35611. [DOI] [PubMed] [Google Scholar]
- 16.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 17.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowndes, N. F., C. J. McInerny, A. L. Johnson, P. A. Fantes, and L. H. Johnston. 1992. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature 355:449-453. [DOI] [PubMed] [Google Scholar]
- 19.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 21.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno, M. B., A. Duran, and J. C. Ribas. 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16:861-872. [DOI] [PubMed] [Google Scholar]
- 23.Nabeshima, K., T. Nakagawa, A. F. Straight, A. Murray, Y. Chikashige, Y. M. Yamashita, Y. Hiraoka, and M. Yanagida. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9:3211-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 25.Nurse, P., and Y. Bissett. 1981. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292:558-560. [DOI] [PubMed] [Google Scholar]
- 26.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 27.Osman, F., E. A. Fortunato, and S. Subramani. 1996. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142:341-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paull, T. T., D. Cortez, B. Bowers, S. J. Elledge, and M. Gellert. 2001. Direct DNA binding by Brca1. Proc. Natl. Acad. Sci. USA 98:6086-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 30.Peak, J. G., and M. J. Peak. 1990. Ultraviolet light induces double-strand breaks in DNA of cultured human P3 cells as measured by neutral filter elution. Photochem. Photobiol. 52:387-393. [DOI] [PubMed] [Google Scholar]
- 31.Prise, K. M., G. Ahnstrom, M. Belli, J. Carlsson, D. Frankenberg, J. Kiefer, M. Lobrich, B. D. Michael, J. Nygren, G. Simone, and B. Stenerlow. 1998. A review of dsb induction data for varying quality radiations. Int. J. Radiat. Biol. 74:173-184. [DOI] [PubMed] [Google Scholar]
- 32.Rhind, N., and P. Russell. 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10:749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos-Mattjus, P., B. T. Vroman, M. A. Burtelow, M. Rauen, A. K. Eapen, and L. M. Karnitz. 2002. Genotoxin-induced Rad9-Hus1-Rad1 (9-1-1) chromatin association is an early checkpoint signaling event. J. Biol. Chem. 277:43809-43812. [DOI] [PubMed] [Google Scholar]
- 35.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 36.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 37.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 39.Shiomi, Y., A. Shinozaki, D. Nakada, K. Sugimoto, J. Usukura, C. Obuse, and T. Tsurimoto. 2002. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-1-1 and Rad17-RFC. Genes Cells 7:861-868. [DOI] [PubMed] [Google Scholar]
- 40.Smith, G. C., R. B. Cary, N. D. Lakin, B. C. Hann, S. H. Teo, D. J. Chen, and S. P. Jackson. 1999. Purification and DNA binding properties of the ataxia-telangiectasia gene product ATM. Proc. Natl. Acad. Sci. USA 96:11134-11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 42.Tashiro, S., J. Walter, A. Shinohara, N. Kamada, and T. Cremer. 2000. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 150:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tommasino, M., and K. Maundrell. 1991. Uptake of thiamine by Schizosaccharomyces pombe and its effect as a transcriptional regulator of thiamine-sensitive genes. Curr. Genet. 20:63-66. [DOI] [PubMed] [Google Scholar]
- 44.Unsal-Kacmaz, K., A. M. Makhov, J. D. Griffith, and A. Sancar. 2002. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. USA 99:6673-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Bosch, M., K. Vreeken, J. B. Zonneveld, J. A. Brandsma, M. Lombaerts, J. M. Murray, P. H. Lohman, and A. Pastink. 2001. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461:311-323. [DOI] [PubMed] [Google Scholar]
- 46.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, J. F. 1988. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 35:95-125. [DOI] [PubMed] [Google Scholar]
- 48.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willson, J., S. Wilson, N. Warr, and F. Z. Watts. 1997. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 25:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 52.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]