Abstract
Recent work has shown that histone methylation is an important regulator of transcription. While much is known about the roles of histone methyltransferases (HMTs) in the establishment of heterochromatin, little is known of their roles in the regulation of actively transcribed genes. We describe an in vivo role of the Saccharomyces cerevisiae HMT, Set2. We identified SET2 as a gene necessary for repression of GAL4 basal expression and show that the evolutionarily conserved SACI, SACII, and SET domains of Set2 are necessary for this repression. We confirm that Set2 catalyzes methylation of lysine 36 on the N-terminal tail of histone H3. Conversion of lysine 36 to an unmethylatable arginine causes a decrease in the repression of GAL4 transcription, as does a Δset2 mutation. We further show that lysine 36 of histone H3 at GAL4 is methylated and that this methylation is dependent upon the presence of SET2.
The DNA of eukaryotic cells is organized into two different states: a highly condensed form called heterochromatin that is generally transcriptionally inactive and a less condensed form called euchromatin that is generally transcriptionally active. In both cases the fundamental unit of chromatin is the nucleosome, composed of 146 bp of DNA wrapped around a histone octamer consisting of two copies of each histone protein, H2A, H2B, H3 and H4 (39). Ordered nucleosomes are generally repressive but can be modified in a number of ways to alter rates of transcription (36). Posttranslational modifications of histone N-terminal tails are important mechanisms by which transcription of open chromatin is modulated (9, 32).
Methylation of specific lysines of histone proteins can target chromatin for activation or repression of transcription (11, 43). The effects of methylation are believed to be a result of either direct changes in protein structure or, in some cases, creation of a binding site for proteins with a chromodomain (2, 15, 24). For example, methylation of lysine 9 of histone H3 recruits HP1 via its chromodomain and establishes heterochromatin in flies and mammals (2, 15). Similarly, in Schizosaccharomyces pombe, the histone methyltransferase (HMT) Clr4 methylates H3 lysine 9 and this serves as a binding site for the HP1-like protein, Swi6 (24).
The highly conserved SET domain is found in many proteins implicated in modulating chromatin structure, including members of the Polycomb and Trithorax groups of proteins, and proteins involved in position effect variegation and gene silencing in budding and fission yeasts (8). The SET domain is associated with lysine HMT activity (23, 25, 33, 34, 37, 41), and it interacts with the N-terminal tails of histones, signaling proteins, components of nucleosome remodeling factors, and proteins involved in DNA damage repair (4, 10, 29).
Saccharomyces cerevisiae has six proteins with a SET domain, two of which have been characterized. Set1 catalyzes methylation of lysine 4 of H3 and has been correlated with transcriptional activity (3, 12, 27, 30). Set2 methylates lysine 36 of H3 and represses transcription when tethered to a reporter gene (33). Several recent papers have shown that Set2 binds to RNA polymerase II and specifically to the elongating form of the C-terminal tail of the largest subunit (17, 18, 40). The functional significance of this binding and of H3 lysine 36 methylation is not known. Here we describe experiments showing that methylation by Set2 is required for repression of basal transcription of a specific gene, GAL4.
MATERIALS AND METHODS
Plasmid construction.
Unless specified, all PCRs were performed with Pfu Turbo DNA polymerase (Stratagene) according to the manufacturer's instructions. The DNA sequence of each cloned PCR product was determined to ensure the fidelity of the polymerase.
Using primers 5′-GCGCGGATCCATGTCGAAGAACCAAAGTG-3′ and 5′-TTAAGCGGTGAGCCAATATGAGTCGACGCGC-3′, the recombinant glutathione S-transferase (GST)-Set2 expression vector was constructed by amplifying the first 900 bases of the SET2 open reading frame (ORF) from yeast genomic DNA. Using the BamHI and SalI sites, the amplicon was then subcloned into pGEX-KG, resulting in an in-frame N-terminal fusion with GST. Primer pair JL154 (5′-GATGGCGTCAACCATGCCTATGACGAGGATTCTGACTGT-3′) and JL155 (5′-ACAGTCAGAATCCTCGACATAGGCATGGTTGACGCCATC-3′) and primer pair JL113 (5′-GCCAGATTTTGCAATCACTCTGCCAGCCCCAATGCATATGTT-3′) and JL114 (5′-AACATATGCATTGGGGCTGGCAGAGTGATTGCAAAATCTGGC-3′) were subsequently used (together with GST-Set2 as a template for site-directed mutagenesis) to generate pJWL129 (GST-Set2 C82Y) and pJWL108 (GST-Set2 C201A), respectively.
Recombinant Drosophila histone expression vectors pJWL124 (H3 K9R), pJWL110 (H3 K27R), and pJWL112 (H3 K36R) were constructed by site-directed mutagenesis using a wild-type H3 expression plasmid, pdH3, as a template (16), and amplification was performed using primer pair JL144 (5′-ACCAAGCAAACTGCTCGCAGATCGACTGGTGGAAAGGCG-3′) and JL145(5′-CGCCTTTCCACCAGTCGATCTGCGAGCAGTTTGCTTGGT-3′),primer pair JL111 (5′-GCTACTAAGGCCGCTCGCAGGAGTGCTCCAGCCACCGGA-3′) and JL112 (5′-TCCGGTGGCTGGAGCACTCCTGCGAGCGGCCCTAGTAGC-3′), and primer pair JL135 (5′-GCGATAGCGGTGGGGCTTCCTCACACCTCCGGTGGCTGG-3′) and JL136 (5′-CCATCTACCGGTGGTGTTAGGAAGCCTCACAGATATAAG-3′), respectively. pJWL113 (H3 K27,36R) was constructed using pJWL110 as a template and primer set JL135 and JL136.
The yeast histone expression plasmid pJWL116 (pRS314 hht2 K36R-HHF2) is derived from pRM200 (pRS314 HHT2-HHF2). Using the EcoRI and SalI sites, the EcoRI-SalI fragment containing HHT2 and HHF2 from pRM200 was subcloned into pUC18 to generate pASB121. pASB121 was then used as a template together with primers JL137 (5′-CAATCTACCGGTGGTGTTAGGAAGCCTCACAGATATAAG-3′) and JL138 (5′-CTTATATCTGTGAGGCTTCCTAACACCACCGGTAGATGG-3′) for site-directed mutagenesis to make the H3 K36R mutation. Using the EcoRI and SalI sites, the insert containing hht2 K36R-HHF2 was reintroduced into the pRM200 backbone to generate pJWL116.
The yeast SET2 plasmid pBM3352 contains the yeast SET2 ORF, 600 bp upstream of the initiating methionine, and 200 bp downstream of the stop codon. pBM3354 is identical to pBM3352 with the exception of C82Y and D83Q amino acid conversions. pBM3352 was used as a template together with primers JL154 and JL155 for site-directed mutagenesis to generate pJWL130. Using the SacI and XhoI sites, the SET2-containing fragments from pBM3352, pBM3354, and pJWL130 were then subcloned into pRS412 to generate pJWL133, pJWL132, and pJWL134, respectively.
The yeast plasmid pBM3670 used for cloning mutagenized SET domain fragments of SET2 had the DNA encoding amino acids (aa) 42 to 263 of Set2 deleted and replaced with a HIS3 gene flanked by MluI sites. The HIS3 gene was amplified using primers OM1358 (5′-CCTCAAAGGCTTTTTGATCAAGAGCCTGATCTCACAGAGGAAGCATTGACGCGTGGCCTCCTCTAGTACACTC-3′) and OM1360 (5′-GGCATCAGCGATGTTCTGGGGCAATAAAGATGCCGCATCTGTTTGACGCGTGCGCGCCTCGTTCAGAATG-3′). ThePCR product was then introduced with pBM3352 into YM6119 (MATα ura3-52 his3Δ200 ade2 lys2 leu2 trp1-901 met-gal80-538 Δset2::GFP∼HIS3 ΔUAS-GAL4∼URA3), and transformants were selected for growth on synthetic dextrose (SD)-His-Trp. A few clones were recovered from yeast and transformed into Escherichia coli DH5α. Clones were sequenced in their entirety to confirm the correct integration of the HIS3 gene, and a single transformant was chosen and designated pBM3670.
Genetic screens.
Selection for repressors of basal GAL4 transcription was performed by plating YM3108 (MATa ura3-52 his3Δ200 ade2 lys2 trp1-901 met-gal80-538 CANR LEU2::GAL1-lacZ ΔUAS-GAL4∼URA3) on minimal medium with complete amino acids and 2% galactose. Gal+ revertants were isolated at a frequency of about 10−6. Three suppressor mutants were isolated and found to be recessive, unlinked to GAL4, and in the same complementation group. The strongest suppressor, designated YM3242, was plated to 5-FOA, and resistant colonies were isolated. This was necessary to remove the URA3 marker used in the ΔUAS gal4 integration. The resulting strain, YM4068, was then transformed with a YCp50 genomic library, and transformants were screened for the Gal− phenotype by replica plating to SD-Ura and synthetic complete (SC)-Ura with 2% galactose (28). Plasmids from Gal− isolates were then isolated from yeast and recovered in E. coli DH5α. Many plasmids with similar restriction patterns were isolated, and the plasmid with the smallest insert (pBM2302) was digested to generate the smallest complementing fragment (designated 1-1). The DNA sequence of this fragment was determined and was found to contain ORF YLJ168C. Using gap repair, this ORF was then isolated from the set2-1 mutant YM3242 and sequenced to identify the C82Y set2-1 mutation.
Using standard methods, mutagenesis of SET2 was performed on pBM3352 by transformation into the E. coli XL1Red mutator strain. Transformants were washed from the plates and pooled into one large culture. This pool of transformants was grown overnight at 37°C in Luria broth with 50 μg of ampicillin/ml, and plasmid DNA was produced from a large-scale preparation. This library of mutants served as a template for PCR using OM1100 (5′-GGGCAATTGATGTCGAAGAACCAAAGT-3′) and OM1233 (5′-GGGCAATTGGCTTAATTTCTTTAACTTTAGCCA-3′). The PCR product was used to gap repair an MluI digest of pBM3670 in strain YM6119. Transformants were selected for growth on SC-Trp with 2% galactose. Plasmids from Gal+ transformants were recovered from yeast and subsequently transformed into E. coli DH5α. The set2 ORFs were sequenced in their entirety.
Yeast strain construction.
GAL4 reporter strains for the experiments were derived from RMY200 (MATa ade2-101 his3Δ200 lys2-801 trp1Δ901 ura3-52 hht1-hhf1::LEU2 hht2-hhf2::HIS3 [pRM200]) (see Fig. 4). First, pRM200 was replaced with pMS329 (pRS316-HHF2-HHT2) to create strain yJL126. Using the kanMX6 knockout cassette (19), the SET2 ORF was then deleted from strain yJL126 to create yJL99. This strain was transformed with pRM200 and pJWL116 to Trp+ and FOAR, generating yJL103 and yJL116, respectively. yJL103 and yJL116 were transformed with the SacI-KpnI fragment from pBM2012 (selecting for Ura+) to create yJL109 and yJL121. pBM2012 is identical to pBM1974 except that the upstream activation sequence (UAS) region (bases −139 to −414) of the GAL4 promoter has been deleted. Correct recombination of the ΔUAS gal4::cat reporter at the endogenous GAL4 gene in yJL109 and yJL121 was confirmed by Southern blotting. yJL109 and yJL121 were each transformed with pRS412, pJWL133, pJWL132, and pJWL134. Multiple transformants were then assayed for chloramphenicol acetyltransferase (CAT) activity.
FIG. 4.
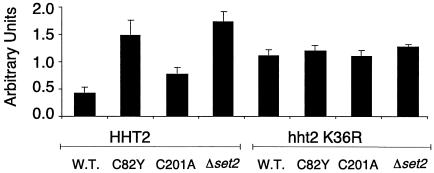
Methyltransferase activity of Set2 and the presence of lysine 36 on histone H3 are required for the repression of ΔUAS gal4::cat. CAT assays were performed on extracts from ΔUAS gal4::cat reporter strains containing SET2, set2 C82Y, set2 C201A, and Δset2 alleles in an HHT2 or hht2 K36R background. The data show derepression of ΔUAS gal4::cat with enzymatically defective Set2 mutants. In addition, significant derepression of the reporter was seen with the introduction of an unmethylatable arginine at residue 36 on H3. The bar graph represents the averages of four independent experiments. W.T., wild type.
Strains used in chromatin immunoprecipitations were derived from YM4192 [MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 met 15 gal80-538 CANR ΔUAS (−139 to −187) gal4::cat∼URA3 LEU2::GAL1-lacZ] (5). BCY1 was made by using the kanMX6 knockout cassette to delete SET2 from YM4192 (19).
Expression and purification of recombinant proteins.
Plasmids expressing recombinant GST-Set2, GST-Set2 (C82Y), GST-Set2 (C201A), and GST-Clr4 were transformed into BL21(DE3) Codon+ RIL (Stratagene catalog no. 230245). Expression was performed in 250-ml cultures of Luria broth with 50 μg of ampicillin/ml and 34 μg of chloramphenicol/ml. The cultures were grown at room temperature to an optical density at 600 nm of approximately 0.2 and then chilled on ice. Ethanol and IPTG (isopropyl-β-d-thiogalactopyranoside) were added to achieve final concentrations of 3% (vol/vol) and 0.45 mM, respectively. The cultures were induced at 18°C overnight with gentle mixing. The cells were pelleted at 5,000 × g for 10 min at 4°C, resuspended in 20 ml of ice-cold 1× phosphate-buffered saline, and then lysed by sonication. Insoluble material was removed by centrifugation at 12,000 × g for 20 min at 4°C. Fusion proteins were purified on a column using 1 ml of glutathione Sepharose (Amersham catalog no. 17-5132-01) according to the manufacturer's instructions. Eluted proteins were dialyzed in 4 liters of 1× phosphate-buffered saline at 4°C. Glycerol and dithiothreitol were added to the purified protein to achieve final concentrations of 10% (vol/vol) and 1 mM, respectively. Protein concentrations were estimated with polyacrylamide gel electrophoresis using bovine serum albumin standards. The purified proteins were frozen in liquid nitrogen and stored at −80°C.
Recombinant Drosophila histones were expressed from pdH3, pdH4, pJWL110, pJWL112, and pJWL124 as described previously (16). Approximately equal quantities of wild-type and mutant H3 and wild-type H4 were purified, solubilized, and renatured into H3/H4 tetramers according to established procedures (20). The concentration of the tetramers was estimated with polyacrylamide gel electrophoresis using bovine serum albumin standards.
Enzyme assays.
Methyl transfer reactions were performed at 30°C for 1 h in 50-μl volumes containing 50 mM Tris-Cl (pH 8.5), 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 250 mM sucrose, 1.1 μCi of S-adenosyl-l-[methyl-3H]methionine (Amersham catalog no. TRK865) (82.0 Ci/mmol), 1.5 μg of recombinant Drosophila H3/H4, and 5 μg of GST-Set2, GST-Set2(C82Y), GST-Set2(C201A), GST-Clr4, or GST. Proteins were precipitated by the addition of 1 ml of cold 20% trichloroacetic acid and incubation on ice for 30 min, filtered using glass filters on a vacuum manifold, and washed three times with 3 ml of 5% trichloroacetic acid followed by three washes with 3 ml of methanol. The filters were allowed to air dry and added to 4 ml of Ecoscint. Tritium was quantified using a liquid scintillation counter.
Extracts for the CAT assays of ΔUAS gal4::cat reporter strains were made as described previously (6). A total of 20 μg of protein from the extracts was assayed using a FAST CAT (deoxy) assay kit (Molecular Probes catalog no. F-6616) according to the manufacturer's instructions. Activity is expressed as the amount of acetylated BODIPY FL 1-deoxychloramphenicol fluorescence.
Chromatin immunoprecipitation.
Samples were processed for chromatin immunoprecipitation as described previously (13). In brief, YM4192 and BCY1 were grown in 200 ml of yeast extract-peptone-dextrose to an A600 of ∼0.8 and then fixed with formaldehyde. Whole-cell extracts were made from fixed cells and sonicated to shear the chromatin to an average size of 400 bp. Using 1 μl of anti-H3 methyl lysine 36 antibody (a generous gift from B. Strahl) and protein A-Sepharose, a total of 1/10 of the sheared chromatin was then immunoprecipitated in a 200-μl volume. Following immunoprecipitation, the cross-links were reversed and the DNA fragments were extracted for PCR.
PCR amplification of target loci was accomplished using Herculase polymerase (Stratagene) and the following primer sets: for ACT1, 5′-CCAATTGCTCGAGAGATTT-3′ and 5′-CATGATACCTTGGTGTCTTG-3′; for HMR-E, 5′-TGCAAAAACCCATCAACCTTG-3′ and 5′-ACCAGGAGTACCTGCGCTTA-3′; for cat, 5′-CTTGCCCGCCTGATGAATGCTC-3′ and 5′-GCCTTGCGTATAATATTTGCC-3′; and for the GAL4 promoter, 5′-AGTTTCAAAACGTCCGCGTC-3′ and 5′-CGATAGAAGACAGTAGCTTC-3′. Products were resolved on a 1.5% agarose gel and visualized with ethidium bromide.
RESULTS
Set2 is a repressor of GAL4 transcription.
The GAL4 promoter, which does not seem to contain a TATA box, is regulated by three cis-acting elements: a UAS necessary for maximal GAL4 expression, an upstream essential sequence that is essential for GAL4 expression, and Mig1 binding sites that mediate glucose repression of GAL4 expression (5). Mutants from which the GAL4 UAS has been deleted have a low basal level of GAL4 expression that is insufficient to support the utilization of galactose (5). These mutants revert to a Gal+ phenotype at a frequency of approximately 10−6 per plated cell. The mutations responsible for suppression of the Gal− phenotype caused by the UAS deletion are all recessive, implying that they affect genes encoding negative regulators of GAL4 transcription.
The gene corresponding to one of the mutant complementation groups was isolated and found to be SET2. The strongest mutant allele was designated set2-1. Assays were conducted to evaluate the effects of set2-1 on GAL4 expression levels by using strains with a gal4-cat reporter gene and measuring CAT activity. Consistent with our expectation that Set2 is a negative regulator of GAL4 expression, the set2-1 allele significantly increased expression of a UAS-less GAL4 gene (0.2 U of CAT activity [31] compared to 0.02 U for a comparable SET2 strain). This mutation does not increase activity from an intact GAL4 promoter (1.4 U compared to 1.6 U for a wild-type SET2 strain), suggesting that Set2 affects basal, but not activated, GAL4 expression. Set2 does not appear to be a global repressor of gene expression, because the set2-1 mutation did not affect expression of three other genes we tested (GCN4, CTS1, and HIS3) and Ty1 (data not shown).
The SET and SAC domains of Set2 are necessary for the repression of GAL4.
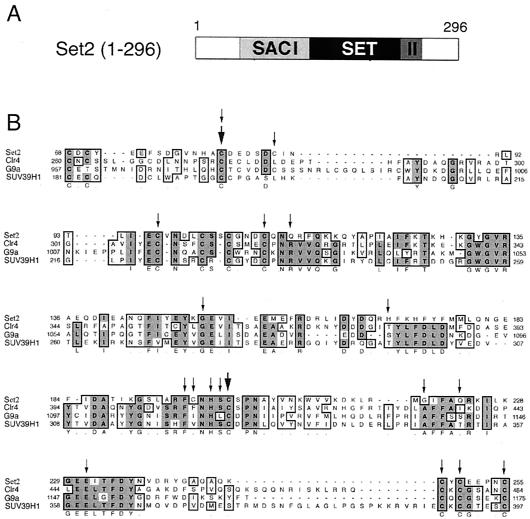
The Set2 protein contains a highly conserved SET domain, flanked by two SAC (SET domain-associated cysteine-rich) domains (Fig. 1). SAC domains are found in many, but not all, SET-containing methyltransferases (25, 37) and have been shown to be essential for in vitro activity (24). The set2-1 allele results in a C82Y conversion in the SACI domain of Set2 protein (Fig. 1). Because the set2-1 mutation resides in the SAC domain, we wanted to identify other residues in the SAC and SET domains that are necessary for repression of GAL4 expression. To do this, we mutagenized the SAC and SET domains and selected nonfunctional set2 mutants by the method used to isolate set2-1. The region of SET2 encoding the SAC and SET domains (aa 1 to 296) was amplified using mutagenic PCR, and the product was inserted into a SET2-containing plasmid (pBM3670) by recombination (gap repair; see Materials and Methods for details). Plasmids containing nonfunctional set2 genes were selected as Gal+ transformants and rescued in E. coli DH5α, and their set2 DNA sequences were determined.
FIG. 1.
Mutations that affect SET2 function cluster at conserved residues within the SAC and SET domains. (A) The first 296 aa of Set2 contain a SET domain flanked by two SAC domains, I and II. (B) Lineup of the SAC and SET domains from a selection of SET domain-containing proteins that have been demonstrated to have methyltransferase activity. The lineup was made with the ClustalW alignment program followed by hand editing. A consensus sequence is shown below the lineup. Mutations isolated in this study are designated by small arrows above the sequence. The large arrows identify amino acids that were also mutated in the E. coli Set2 expression plasmid and tested for HMT activity.
Most of the recovered mutations affected highly conserved amino acids in the SAC and SET domains (Fig. 1). Mutated residues in the SACI domain were predominantly cysteines (including cysteines 82, 88, 97, and 109). SACII domain mutations also predominantly affected cysteines (including cysteines 248, 250, and 255). Amino acid conversions in the SET domain, while more variable, were most often found in highly conserved amino acids (including residues G151, F196, H199, S200, G219, and E231) but also in less conserved residues (H170, C197, and Q223).
Biochemical characterization of the SAC and SET domains of Set2.
We found that purified recombinant GST-Set2 has robust methyltransferase activity on chicken erythrocyte histones. Analysis of the methylated histones by triton acid urea electrophoresis revealed that the predominant substrates are H3 and, to a much lesser extent, H4 (data not shown).
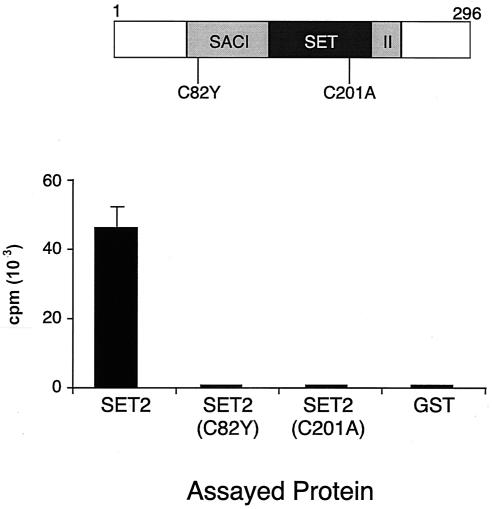
To confirm that the HMT activity detected was due to Set2, we introduced two single-amino-acid changes in highly conserved cysteines located in the SAC and SET domains. We chose to mutate C82 (because it is mutated in set2-1) as well as C201 (because previous studies determined that the corresponding mutation abolished the enzymatic activity of the Suv39 h1 and G9a HMTs [26, 34]). Each of these mutants was assayed for its ability to methylate recombinant H3/H4 tetramers. Both mutations resulted in a complete loss of HMT activity, showing that the in vitro activity we detected was due to Set2 (Fig. 2). The fact that the set2-1 mutation abolished in vitro activity suggests that Set2 enzymatic activity is essential for repression of GAL4.
FIG. 2.
Mutations in the SET and SAC domains result in loss of GST-Set2 methyltransferase activity. (Upper panel) The locations of the two amino acid changes made to highly conserved cysteine residues in the GST-Set2 expression plasmid are indicated. (Lower panel) GST-Set2, GST-Set2 (C82Y), GST-Set2 (C201A), and a GST control protein were purified and assayed for methyltransferase activity. The bar graphs represent averages of three independent assays.
The catalytic domain of Set2 predominantly methylates lysine 36 on H3.
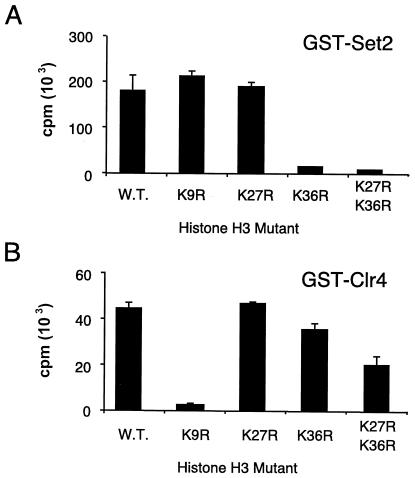
We used GST-Set2 to methylate recombinant H3 and determined that lysine 36 was methylated (data not shown), confirming a previous report (33). To complement this analysis, we undertook a mutational analysis of the H3 N-terminal tail. Single lysines 9, 27, and 36 and lysines 27 plus 36 on recombinant H3 were converted to arginine. The resulting mutated H3 proteins were expressed and mixed with wild-type recombinant H4 and then used as the substrates in methyltransferase reactions with GST-Set2. In agreement with the results from the sequence data, GST-Set2 does not transfer methyl groups to histones with lysine 36 on H3 converted to arginine (Fig. 3). As a control, the well-characterized Clr4 methyltransferase (24) was able to methylate H3 except when lysine 9 is converted to arginine (Fig. 3).
FIG. 3.
GST-Set2 preferentially methylates lysine 36 of H3. (A) GST-Set2 was used to methylate recombinant wild-type (W.T.) and mutant histone H3/H4 tetramers. Levels of GST-Set2 activity on the various histone H3 mutants are indicated. Activity was lost when lysine 36 was mutated to the unmethylatable arginine. (B) Clr4 was used as a control to monitor the quality of the histone tetramers. Methylation reactions were performed as described for Fig. 2 except that recombinant GST-Clr4 was used instead of GST-Set2. Consistent with published results, GST-Clr4 was unable to methylate the H3 K9R mutant. For each panel, the bar graph represents the averages of two experiments.
Histone H3 lysine 36 is essential for GAL4 repression by SET2.
To test the effect of the histone H3 lysine 36 mutation on GAL4 transcription, a ΔUAS gal4::cat reporter was introduced into strains which contained four different SET2 alleles: SET2, set2 C82Y, set2 C201A, and Δset2. Each strain also contained either wild-type HHT2 or an hht2 K36R mutation. Extracts from multiple transformants of each reporter strain were tested for CAT activity.
While the Δset2 mutation caused a significant increase in CAT activity in a HHT2 strain, the methyltransferase-defective set2 alleles differed somewhat in their abilities to repress cat expression. The set2 C82Y allele had the same levels of cat expression as the Δset2 allele, providing additional evidence that the methyltransferase activity of Set2 is necessary for its repressive effects on ΔUAS gal4::cat. The set2 C201A allele was found to have ∼50% repressive ability (Fig. 4), while this mutant protein was completely defective in vitro (Fig. 2). Possibly this allele is less defective in vivo than it is in vitro (see Discussion).
The hht2 K36R mutation caused a significant increase in CAT activity in the ΔUAS gal4::cat reporter strain. This increase in activity is independent of the SET2 allele, demonstrating that the ability to methylate lysine 36 is epistatic to SET2 and placing the hht2 K36R allele downstream of SET2 in the pathway of repression of ΔUAS gal4::cat. It is interesting that the Δset2 hht2 K36R strain has lower CAT activity than the Δset2 HHT2 strain (Fig. 4). This could be due to a global effect on gene expression because of the presence of the mutated H3 throughout the genome.
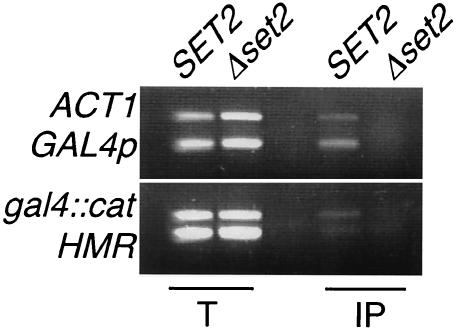
We next used chromatin immunoprecipitation to show that histones in the ΔUAS gal4::cat chromatin are a direct target of Set2 methylation. Using an antibody specific to H3 methyl lysine 36, we were able to selectively precipitate the ΔUAS gal4::cat promoter and coding region from extracts of a SET2 strain but not from those of a Δset2 strain (Fig. 5). The loss of methylation at lysine 36 in the Δset2 strain demonstrates a direct involvement of Set2 in the modification of the ΔUAS gal4::cat region. These data support previous results demonstrating that Set2 methylates H3 at the promoter and coding region of GAL4 (40). In our experiments HMR-E was not precipitated in a SET2 background and served as a negative control. It is interesting that we also precipitated sequences from the ACT1 ORF in a SET2 strain, suggesting that ACT1 is also a target of Set2 methylation.
FIG. 5.
Set2 methylates histone H3 lysine 36 at the GAL4 locus. Chromatin from whole-cell extracts from ΔUAS gal4::cat SET2 and ΔUAS gal4::cat Δset2 strains was immunoprecipitated using an antibody specific for H3 methyl lysine 36. Primers were used to amplify sequences of the ΔUAS gal4 promoter as well as those of the cat ORF. Primers for HMR-E and the ACT1 ORF were also used.
DISCUSSION
We first identified SET2 in a selection for genes involved in the basal repression of GAL4. Three set2 mutants were isolated, the strongest of which was designated set2-1, which changes a highly conserved cysteine residue (C82) in the catalytic domain of Set2 to tyrosine. This suggests that Set2 represses basal transcription of GAL4 through its methyltransferase activity.
We identified other residues in the catalytic domain that are necessary for GAL4 repression (Fig. 1). Conserved cysteine residues found in the SACI and SACII domains and highly conserved residues located in the SET domain are important for Set2 function. The structures of several SET domain proteins have been determined recently (7, 14, 21, 22, 35, 38, 42), and two of the solved structures, S. pombe Clr4 and Neurospora crassa Dim-5, contain cysteine-rich SAC domains. The structures show that cysteines in the N-terminal SAC domain have a structural role in coordinating the binding of a triangular zinc cluster, while the cysteine corresponding to C201 in Set2 has been suggested to contact the C-terminal cysteine-rich SAC domain to form a cofactor-substrate binding site (22, 42). Given this structural information, we believe that the C82Y mutant alters the zinc cluster structure and that the enzyme loses activity because of structural changes. On the other hand, the C201A mutation should cause minimal structural changes but the purified protein may be unable to form an intact cofactor or substrate binding site in vitro. It is possible that such a binding site can be restored to some degree in vivo in the presence of other proteins. These structure-based interpretations are consistent with our results shown in Fig. 4. In addition, three of the mutations recovered in the selection, C97, H199, and Q112 (similar to Clr4 R320), were found to ablate HMT activity in previously characterized HMT enzymes, leading us to believe that the defects in set2 affect catalysis rather than protein-protein interactions (24, 25, 41).
We found that the catalytic domain of Set2 has HMT activity in vitro (Fig. 2) and, in agreement with a recent report (33), that the prominent site of methylation is lysine 36 on histone H3. We showed that GST-Set2 cannot methylate a histone H3 substrate when lysine 36 is converted to an unmethylatable arginine, confirming its preference for lysine 36 (Fig. 3).
We believe that the HMT activity of Set2 on H3 lysine 36 is responsible for its basal repression of GAL4 for four reasons. First, the set2-1 mutant (C82Y) isolated in our original screening, as well as the C201A mutant, is catalytically inactive in vitro (Fig. 2). Second, these catalytically inactive mutants have a significantly reduced ability to repress the ΔUAS gal4::cat reporter gene (see Results and Fig. 4). Third, the ability of Set2 to repress GAL4 expression is dependent on the availability of lysine 36 on H3 for methylation, because the hht2 K36R change causes a loss of repression of the ΔUAS gal4::cat reporter gene that is the same whether the SET2 gene is present or has been deleted (Fig. 4). Finally, the chromatin immunoprecipitation experiments show that SET2 is directly responsible for methylation of lysine 36 at the GAL4 gene (Fig. 5). The combination of genetic and biochemical evidence strongly suggests that repression of GAL4 by Set2 is mediated by methylation of lysine 36 on histone H3.
The repressive effects of Set2 methylation on transcription in vivo are in agreement with a previous report (33). In that report, LexA-Set2 was found to repress transcription 20-fold when tethered to a CYC1-lexAop-lacZ reporter. The differences in the level of repression by Set2 at GAL4 and CYC1-lexAop-lacZ may be due to differences in the recruitment of Set2 to these promoters. In agreement with our results, a C201A mutation in LexA-Set2 resulted in a 50% loss in repression (33). This partial effect could be due to the different activity levels of the mutant protein in vitro (where it was completely defective; Fig. 2) versus that seen in vivo. Or perhaps Set2 has a repression function independent of its methylation activity.
It is not easy to reconcile our results regarding the repression of basal transcription of GAL4 with the numerous recent reports that Set2 binds to the elongating form of RNA polymerase II (17, 18, 40). Perhaps Set2 acts as a backup system for transcriptional repression. Under conditions of repression, transcriptional repressors bind to the promoter region of regulated genes, preventing the assembly and subsequent clearance of an RNA polymerase II transcription complex. But occasionally, “leaky” transcription can occur under repressive conditions. Perhaps Set2 methylates the promoter and the coding part of the gene when this leaky transcription occurs, thus marking the chromatin and preventing subsequent transcription. It is also possible that the methylation of lysine 36 by Set2 has different functions at promoters than at coding regions of genes.
The actual repression mechanism resulting from lysine 36 methylation is still not known. One model is that the methylation of lysine 36 causes an alteration of nucleosome structure that is repressive in nature. To test this hypothesis, we conducted MNase protection assays on hht2 K36R Δset2 and HHT2 SET2 strains at the GAL4 promoter. We found no difference in digestion patterns, suggesting that nucleosome positioning had not been altered in the absence of methylation (data not shown). It is still possible that K36 methylation changes chromatin structure in a way that cannot be detected with MNase assays. A second model is that methylation of lysine 36 might recruit a chromodomain-containing protein that acts as a repressor of transcription. This would be similar to the mechanism used for the establishment of heterochromatin by HP1 binding to methyl lysine 9 on H3 (2, 15, 24).
In summary, we have shown that Set2 methylation is involved in the repression of basal transcription of GAL4. The fate of the methylated histones under conditions of transcriptional activation is unknown. It is possible that methylation of K36 at GAL4 is permanent and that its repressive effects are overcome through the recruitment of transcriptional activators. It is also possible that a demethylating enzyme exists. Finally, there may be a mechanism whereby methylated histones are replaced by unmethylated ones upon transcriptional activation (1).
Acknowledgments
We thank Michael Grunstein for yeast strains and plasmids, Robert Haltiwanger for expert technical advice, Brian Strahl for the antibody to H3 methyl lysine 36, and Brian Colin for technical assistance.
J.L. was supported by a predoctoral fellowship from the Institute for Cell and Developmental Biology at Stony Brook University. The research was supported by NIH grants to R.S. (GM28220), M.J. (GM32540), and R.-M.X. (GM63716).
REFERENCES
- 1.Bannister, A., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui, X., I. De Vivo, R. Slany, A. Miyamoto, R. Firestein, and M. L. Cleary. 1998. Association of SET domain and myotubularin-related proteins modulates growth control. Nat. Genet. 18:331-337. [DOI] [PubMed] [Google Scholar]
- 5.Griggs, D. W., and M. Johnston. 1993. Promoter elements determining weak expression of the GAL4 regulatory gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:4999-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griggs, D. W., and M. Johnston. 1991. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. USA 88:8597-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs, S. A., J. M. Harp, S. Devarakonda, Y. Kim, F. Rastinejad, and S. Khorasanizadeh. 2002. The active site of the SET domain is constructed on a knot. Nat. Struct. Biol. 9:833-838. [DOI] [PubMed] [Google Scholar]
- 8.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 10.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 12.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 14.Kwon, T., J. H. Chang, E. Kwak, C. W. Lee, A. Joachimiak, Y. C. Kim, J. Lee, and Y. Cho. 2003. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 22:292-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 16.Levenstein, M. E., and J. T. Kadonaga. 2002. Biochemical analysis of chromatin containing recombinant Drosophila core histones. J. Biol. Chem. 277:8749-8754. [DOI] [PubMed] [Google Scholar]
- 17.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 19.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301-311. [DOI] [PubMed] [Google Scholar]
- 21.Manzur, K. L., A. Farooq, L. Zeng, O. Plotnikova, A. W. Koch, Sachchidanand, and M. M. Zhou. 2003. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat. Struct. Biol. 10:187-196. [DOI] [PubMed] [Google Scholar]
- 22.Min, J., X. Zhang, X. Cheng, S. I. Grewal, and R. M. Xu. 2002. Structure of the SET domain histone lysine methyltransferase Clr4. Nat. Struct. Biol. 9:828-832. [DOI] [PubMed] [Google Scholar]
- 23.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 25.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 27.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 29.Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib, J. Blechman, T. Nakamura, C. M. Croce, A. Mazo, and E. Canaani. 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 31.Seed, B., and J. Y. Sheen. 1988. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene 67:271-277. [DOI] [PubMed] [Google Scholar]
- 32.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 33.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 35.Trievel, R. C., B. M. Beach, L. M. Dirk, R. L. Houtz, and J. H. Hurley. 2002. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111:91-103. [DOI] [PubMed] [Google Scholar]
- 36.van Holde, K. E., D. E. Lohr, and C. Robert. 1992. What happens to nucleosomes during transcription? J. Biol. Chem. 267:2837-2840. [PubMed] [Google Scholar]
- 37.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, J. R., C. Jing, P. A. Walker, S. R. Martin, S. A. Howell, G. M. Blackburn, S. J. Gamblin, and B. Xiao. 2002. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111:105-115. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe, A. P. 1992. Chromatin: structure and function. Academic Press, London, United Kingdom.
- 40.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, L., L. Xia, D. Y. Wu, H. Wang, H. A. Chansky, W. H. Schubach, D. D. Hickstein, and Y. Zhang. 2002. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21:148-152. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, X., H. Tamaru, S. I. Khan, J. R. Horton, L. J. Keefe, E. U. Selker, and X. Cheng. 2002. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]