Abstract
Both viral effect and immune-mediated mechanism are involved in the pathogenesis of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infection. In this study, we showed that in SARS patient sera there were autoantibodies (autoAbs) that reacted with A549 cells, the type-2 pneumocytes, and that these autoAbs were mainly IgG. The autoAbs were detectable 20 days after fever onset. Tests of non-SARS-pneumonia patients did not show the same autoAb production as in SARS patients. After sera IgG bound to A549 cells, cytotoxicity was induced. Cell cytotoxicity and the anti-epithelial cell IgG level were positively correlated. Preabsorption and binding assays indicated the existence of cross-reactive epitopes on SARS-CoV spike protein domain 2 (S2). Furthermore, treatment of A549 cells with anti-S2 Abs and IFN-γ resulted in an increase in the adherence of human peripheral blood mononuclear cells to these epithelial cells. Taken together, we have demonstrated that the anti-S2 Abs in SARS patient sera cause cytotoxic injury as well as enhance immune cell adhesion to epithelial cells. The onset of autoimmune responses in SARS-CoV infection may be implicated in SARS pathogenesis.
Keywords: autoantibody, cytotoxicity, lung epithelial cell, SARS, spike protein
Introduction
Severe acute respiratory syndrome (SARS), an atypical pneumonia disease caused by a human coronavirus (CoV), is a new global public health problem [1–9]. Major outbreaks of SARS infection occurred in China, Hong Kong, Singapore, Vietnam, Taiwan and Canada. Over 30 countries have reported suspected or probable cases. SARS-CoV is a mutant human CoV that may acquire new virulence factors. Although the temporal progression of the clinical, radiological, and virological changes of SARS has been extensively studied and several treatments have been proposed [10–13], there are as yet no effective strategies to prevent SARS, because its pathogenic mechanisms are still unresolved.
In SARS pathology, the onset of respiratory symptoms is suggested to be a result of multiple factors on respiratory epithelium disruption, including viral cytotoxicity and host factors [12]. Genetic variations of SARS-CoV and viral load seem to be responsible for the severity of SARS. In addition, abnormal immune responses to SARS-CoV infection, such as the unbalance of immune cells and the production and dysregulation of cytokines and chemokines, may also underlie the pathogenesis of disease [9, 12, 14–16]. The immune-mediated mechanisms involved in SARS pathogenesis, however, are not fully understood [9, 17, 18].
The onset of autoimmunity has been related to viral infections. An influenza viral infection-induced autoimmune disease called Goodpasture's syndrome shows the existence of autoantibodies (autoAbs) against the alveolar and glomerular basement membrane [19–21], but there is no report regarding SARS-CoV-induced autoAb production. Nevertheless, murine CoV infection induces autoreactive T cells, B cell polyclonal activation, and autoAb production [22–24]. The immune-mediated pathology related to SARS-CoV infection thus merits further examination. In the present study, the generation and pathogenic role of autoAbs in SARS patients were investigated.
Materials and methods
Patients
SARS patient sera were collected by the Centre for Disease Control, Department of Health, Taiwan (CDC-Taiwan), from March to June 2003. Diagnosis of SARS was based on the clinical criteria established by the WHO. Patients with SARS-CoV were confirmed by laboratory methods, including viral antigen detection, RT-PCR, and serologic methods [25]. The epidemiological characteristics of age and gender and clinical information such as symptoms, underlying diseases, outcomes including death and hospital length-of-stay, as well as laboratory findings were obtained from CDC-Taiwan [25,26, and unpublished observation]. SARS patient sera collected from the early (80 patients, < 20 days after fever onset) and the late (41 patients, ≥ 20 days) stages were included in this study. Sera of some patients were collected two or three times at the late stage; in all, 60 serum samples from 41 patients were tested. Eight serum samples from patients diagnosed with pneumonia from non-SARS aetiology (obtained from Dr T. R. Hsiue, Internal Medicine, National Cheng Kung University Hospital, Tainan) and 10 serum samples from healthy individuals were used as controls.
Cell binding assay
Human lung adenocarcinoma cell line A549, hepatoma cell line Hep3B, and lung fibroblast MRC5 were grown in DMEM, and human lung epithelial cell line HL was grown in MEM, both supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamate, and 50 ng/ml gentamycin. Human microvascular endothelial cell line-1 (HMEC-1) was passed in culture plates containing endothelial cell growth medium (Clonetics, Walkersville, MD, USA) composed of 2% FCS, 1 µg/ml hydrocortisone, 10 ng/ml epidermal growth factor, and antibiotics. Cells were incubated in a CO2 incubator at 37°C and 5% CO2 in a humidified atmosphere. For microscopic observation, monolayers of A549 cells were cultured on sterile glass slides before the experiment. For flow cytometric analysis, cell suspensions were prepared by trypsinization of cell cultures. Cells were washed briefly in phosphate-buffered saline (PBS), fixed with 1% paraformaldehyde in PBS at room temperature for 10 min, and washed again with PBS. Patient serum samples and mouse anti-SARS spike protein (anti-S), spike protein domain 1 (anti-S1), and spike protein domain 2 (anti-S2) Abs were then incubated with cells at 4°C for 1 h. After being washed three times with PBS, cells were incubated with 1 µl of 1 µg/ml FITC-conjugated mouse anti-human IgG, IgM, or IgA and goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA), respectively, at 4°C for 1 h and washed again with PBS. The cell binding activity of sera Abs was analysed by fluorescent microscopy and by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA) with excitation set at 488 nm.
Cell cytotoxicity assay
For the cytotoxicity assay, test sera were pretreated at 56°C for 30 min for complement inactivation. Monolayers of A549 cells were cultured in a 96-well plate before the experiment. Cells were washed briefly in sterile serum-free culture medium, and then treated with serum samples for 72 h. Supernatant was collected and the cytotoxicity was determined using lactate dehydrogenase (LDH) release with a kit (In Situ Cell Cytotoxicity kit; Roche Molecular Biochemicals, Indianapolis, IN, USA) according to the manufacturer's instructions. The percentage of cytotoxicity was calculated as
Preparation of SARS S2 protein
To construct the S2 expression vector, the cDNA of the SARS S2 coding region was amplified by PCR carried out using pcDNA3-Spike-EGFP (a kind gift from Dr C. J. Huang, Academia Sinica, Taipei, Taiwan) as a template, a forward primer (5′-GCGGAATTCGTCGACACTTCTTAT-3′) containing an EcoRI restriction site (in italics), and a reverse primer (5′-GCGTCTAGATTAACTAGTCATGCAACA-3′) containing a stop codon (underlined) and an XbaI restriction site (in italics). The amplified DNA fragment was cloned into the EcoRI and XbaI sites of the pMAL-c2E vector (New England Biolabs, Beverly, MA, USA) after the maltose binding protein (MBP) gene to produce pMBP-TWsp-S2. After ligation, transformation of DH5α was performed using electroporation (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All constructs were verified by DNA sequencing performed using a kit (ABI-Prism dye terminator cycle sequencing kit; PerkinElmer Applied Biosystems, Foster City, CA, USA).
For expression and purification of the SARS S2 fusion proteins, the E. coli DH5α bearing the construct pMBP-TWsp-S2 for expression was grown in Luria-Bertani medium with 100 µg/ml ampicillin. When the cell culture reached an OD of 0·7–1·0 at 600 nm, protein expression was induced by the addition of 0·5 mM IPTG for 3 h at 37°C. To verify expression, cells were collected by centrifugation and disrupted directly in SDS-PAGE sample loading buffer. For large-scale purification, cells were harvested by centrifugation and suspended in binding buffer (20 mM Tris-HCl (pH 7·4) containing 200 mM NaCl and 1 mM EDTA). Cells were lysed with sonication on ice. The cell lysate was clarified by centrifugation at 20 000 × g for 20 min, and then the clear supernatant was loaded onto a column containing amylose resin (New England Biolabs), equilibrated with the binding buffer. The column was washed with three volumes of binding buffer, and then the fusion proteins were eluted by the same buffer containing 10 mM maltose. Finally, the purified fusion protein MBP-S2 was concentrated using a filter unit (Centricon YM-10 Centrifugal Filter Unit; Millipore Corp., Bedford, MA, USA). Protein concentration of various fractions was determined by the Bradford spectrophotometric method (Bio-Rad) in duplicate, and average values were calculated.
Spike peptide prediction and synthesis
Publicly available human and CoV genome sequences at the National Center for Biotechnology Information, USA, were used for in-silico prediction. Algorithms for immunogenicity, second-structure prediction, protein topology analysis, and hydrophobicity were applied to design the tested peptides. The protein sequence of spike protein was obtained from GenBank (accession number AY274119). Immunogenic viral peptides were calculated based on the algorithm developed by Kolaskar and Tongaonkar [27]. In-silico secondary structural analyses of spike protein were performed based on two algorithms: PHD and PREDATOR. Protein topology prediction was based on the algorithm developed by TMHMM. Hydrophobic moment of the peptides was calculated based on the algorithm HMOMENT. Similarity searches were performed between spike protein and human genome databases using the NCBI Blastp program. For blastp analysis, the default database collection of all nonredundant GenBank cDNA sequence translations, PDB, SwissProt, PIR and PRF entries was used, with the species restricted to human. Finally, expert curation was applied for refinement on peptide design. Multiple antigen peptides were synthesized by CytoMol Corp. (Mountain View, CA, USA). Several synthetic peptides were used in this study: D02 (residues 658–669, N-ASYHTVSLLRSTSQK-C), D03 (residues 733–744, N-EEGNLLLQYGSFCTQ-C), D07 (residues 927–937, N-GLGKLQDVVNQNGE-C), and D08 (residues 942–951, N-ALNTLVKQLSSN-C). Extra amino acid residues were added at either N- or C-terminus, as indicated by italic letters, to increase the hydrophobicity.
Pre-absorption and binding assays
Preabsorption of patient sera against SARS-CoV S2 and the four synthetic peptides we used was done using a solid-phase capture technique and individual peptide-coated plates. An ELISA plate was coated with or without peptides in a 10 µg/well and blocked by 5% bovine serum albumin (BSA) in coating buffer (15 mM of sodium carbonate and 35 mM of sodium bicarbonate, pH 9·6). Test serum samples were 1 : 20 diluted and added to the plate at 4°C overnight for absorption. Supernatant was collected from each well and incubated with A549 cells for a binding assay as described above. To detect Ab titres in sera, an ELISA plate was washed with 0·05% PBS-Tween 20 (PBS-T) five times, whereafter HRP-conjugated anti-human IgG was added (0·5 mg/ml; Jackson ImmunoResearch Laboratories) in 1/5000 dilution (200 µl/well). After washing with PBS-T, ABTS peroxidase substrate (Trinity Biotech Plc, Bray, Co Wicklow, Ireland) was added, and the absorbance was measured using a microplate reader (Emax; Molecular Devices Corp., Sunnyvale, CA, USA) at 405 nm.
Adhesion assay
A549 cells (5 × 104 cells/well) were plated into 8-well glass chamber slides (Nalge Nunc International, Naperville, IL, USA). When monolayers were confluent, the cells were treated with anti-S2 hyperimmune sera in serum-free culture medium. After 1 h of incubation, the cells were washed once with medium and incubated for 1 h at 37 °C with isolated healthy human peripheral blood mononuclear cells (PBMC) (1 × 105 cells/well) in a total volume of 250 µl/well. At the end of the incubation period, the nonadherent cells were removed by washing twice with 0·1% BSA in PBS. Adherent cells were stained with Liu's stain (TONYAR Biotech, Taipei, Taiwan) and viewed with light microscopy. The adherent cells were counted on three consecutive microscopic fields [28,29].
Statistical analysis
Comparisons between various treatments were performed using Student's t-test with SigmaPlot version 8·0 for Windows (Cytel Software Corp., Cambridge, MD, USA). Non-paired Student's t-tests were used for the data analyses in Table 1 and Fig. 2, and paired Student's t-tests were used for the data analyses in Figs 3 and 5. Values were considered statistically significant at P < 0·05.
Table 1.
Anti-A549 cell IgG, IgM and IgA levels in SARS patient sera.
| % A549 cell binding (mean ± SD) | |||
|---|---|---|---|
| IgG | IgM | IgA | |
| Healthy controls (n = 10) | 4·5 ± 1·5 | 5·2 ± 3·7 | 4·9 ± 2·3 |
| SARS patients (Fever onset days) | |||
| Early (< 20 days)(n = 80) | 5·8 ± 0·6 | 2·3 ± 1·1 | 2·6 ± 0·8 |
| Late (≥ 20 days)(n = 41) | 24·8 ± 15·5*† | 6·4 ± 9·4 | 10·3 ± 9·2 |
| Late | |||
| 20–40 days (n = 18) | 22·7 ± 15·6*†‡ | 8·0 ± 8·3 | 13·6 ± 13·3 |
| 41–61 days (n = 23) | 22·6 ± 15·5*† | 6·8 ± 12·4 | 9·1 ± 10·6 |
| 62–82 days (n = 14) | 19·8 ± 10·2*† | 4·2 ± 6·7 | 8·4 ± 4·9 |
| > 82 days (n = 5) | 16·9 ± 5·8*† | 5·0 ± 1·8 | 6·5 ± 4·8 |
| Pneumonia patients (n = 8) | 8·9 ± 2·7 | 4·7 ± 2·3 | 2·2 ± 1·7 |
P < 0·001 versus healthy control
P < 0·001 versus early;
P < 0·05 versus fever onset > 82 days.
Fig. 2.
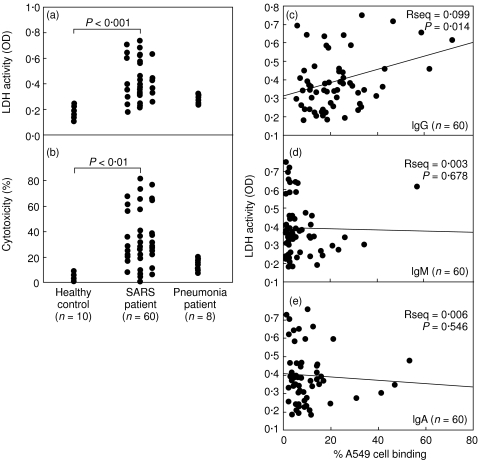
Epithelial cell cytotoxicity caused by SARS patient sera. Culture supernatants were obtained from A549 cells incubated with a 1 : 20 dilution of patient sera for 72 h. The levels of A549 cell cytotoxicity were determined by the lactate dehydrogenase (LDH) activity, and (a) the relative OD and (b) the percentage of cytotoxicity are shown. Relationship between anti-A549 cell IgG (c), IgM (d), or IgA (e) levels of SARS patients as determined by percentage cell binding and cell cytotoxicity as determined by LDH activity.
Fig. 3.
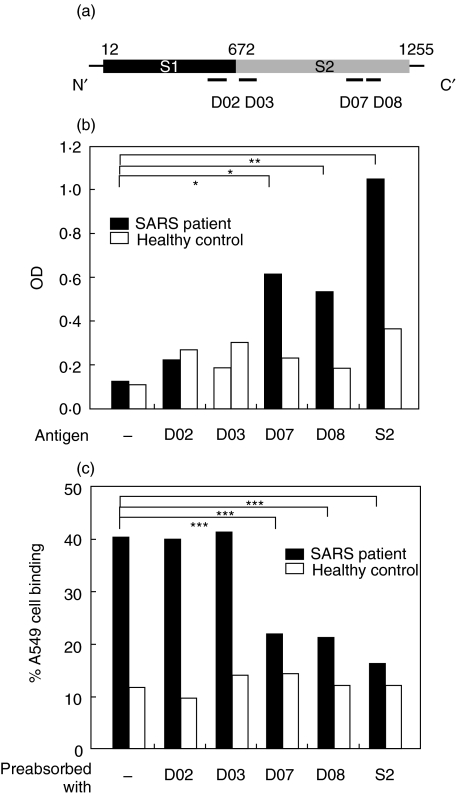
(a) The locations of four synthetic peptides on SARS-CoV spike protein sharing sequence homology with human proteins are indicated. (b) Binding ability of SARS patient sera to spike peptides. SARS patient sera (from a pool of five patients) or healthy control sera (from a pool of five healthy controls) were 1 : 20 diluted and added to the wells coated with spike protein S2 domain, D02, D03, D07, or D08 peptides. ELISA was performed for the detection of the binding activity of patient or healthy control sera to these peptides. Experiments were carried out in duplicate, and the averages are shown. (c) Blockage of A549 cell binding ability by spike peptides. SARS patient sera or healthy control sera were preabsorbed with 10 µg S2, D02, D03, D07, or D08 peptides overnight at 4 °C before incubated with A549 cells. The binding activity of patient or healthy control sera to A549 cells was determined by flow cytometry. Experiments were carried out in duplicate, and the averages are shown. *P < 0·05, **P < 0·01, ***P < 0·001.
Fig. 5.
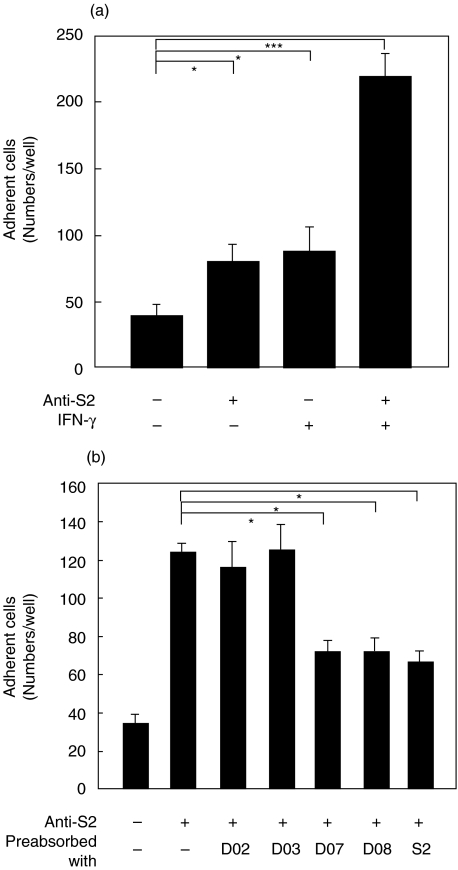
(a) Effect of anti-S2 Abs and IFN-γ on PBMC adherence to epithelial cells. A549 cells were cultured in 8-well glass chamber slides and treated with 25 ng of IFN-γ for 24 h. Cells were washed with PBS, and then cocultured with fresh human PBMC for 1 h in the presence or absence of 1 : 50 dilution of anti-S2 hyperimmune sera. Cell adhesion was observed using Liu's stain. The numbers of adherent cells were quantified in each tested culture. (b) Blockage of PBMC adherence to epithelial cells by spike peptides. Anti-S2 hyperimmune sera were preabsorbed with 10 µg S2, D02, D03, D07, or D08 peptides at room temperature for 2 h before incubated with A549 cells. After 1 h at 37 °C, fresh human PBMC were added and the rest of procedures were followed as described in (a). Experiments were carried out in triplicate, and the averages are shown. *P < 0·05, ***P < 0·001.
Results
SARS patients produced Abs cross-reactive with A549 cells
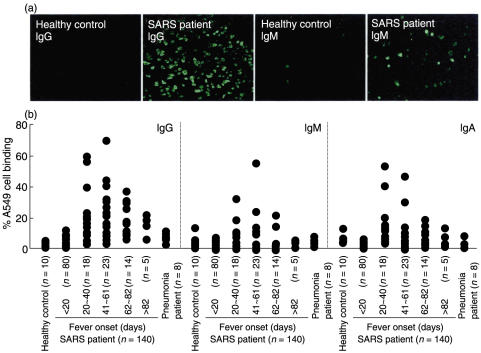
Pulmonary defects are clinical features of SARS-CoV infection. In an attempt to investigate the role played by SARS patient sera, the binding activity of patient sera with human A549 epithelial cells was determined. Fluorescent microscopic observation showed that Abs present in SARS patient sera reacted with uninfected A549 epithelial cells, while sera from healthy controls did not (Fig. 1a). By flow cytometric analysis, the average IgG level was significantly higher at the late stage (≥ 20 days after fever onset) compared to the early stage (< 20 days after fever onset) and the healthy controls (Fig. 1b and Table 1). Although some of the patient sera IgM and IgA at the late stage exhibited high A549 cell binding activity (Fig. 1b), the average levels were not significantly different from those of the early stage and the healthy controls (Table 1). The epithelial cell binding activity of Abs was not significantly elevated in non-SARS-pneumonia patient sera (Fig. 1b and Table 1). The autoAb levels started to increase by day 20, reached the highest levels around day 40, and declined gradually thereafter (60 samples in 41 patients; Fig. 1b and Table 1).
Fig. 1.
Anti-A549 cell autoAbs in SARS patient sera. Human A549 epithelial cells were incubated with a 1 : 20 dilution of sera from SARS patients with different days of fever onset (n = 140), non-SARS-pneumonia patients (n = 8), or healthy controls (n = 10), followed by FITC-conjugated anti-human IgG, IgM, or IgA, and then observed by fluorescent microscopy (a) or analysed by flow cytometry (b). The percentages of A549 cells that reacted with patient or control sera are shown.
Cytotoxic effect of cross-reactive Abs on A549 cells
The consequences of serum binding to A549 cells were next assessed. A549 cell cytotoxicity induced by patient sera was measured using an LDH activity assay. Sera from the late phase were used and results showed that SARS patient sera induced A549 cytotoxicity (average OD = 0·43 ± 0·15, n = 60) compared to sera from healthy controls (average OD = 0·19 ± 0·04, n = 10) and non-SARS-pneumonia patient sera (average OD = 0·29 ± 0·03, n = 8) (Fig. 2a). The percentage of cytotoxicity showed a pattern similar to that of OD values (Fig. 2b). Preabsorption of the IgG fraction by protein G resulted in a reduction of A549 cytotoxicity to levels similar to those of healthy controls, indicating that the cytotoxic effect was caused by the IgG present in the SARS patient sera (data not shown). Complement inactivation of patient sera was performed before experiments. Therefore, epithelial cell injury must have been mediated by patient IgG via a complement-independent pathway. However, the possibility that complement-mediated cytotoxicity may play a role cannot be excluded. Further analysis showed a statistically significant correlation between the levels of anti-A549 IgG, but not IgM or IgA, and the magnitudes of epithelial cell cytotoxicity (Fig. 2c–e).
In addition to A549 cells, we examined whether autoAbs present in SARS patient sera also reacted with other cell types. Some of the SARS patient sera detected bound to the human endothelial cell line HMEC-1 and to the human hepatoma cell line Hep3B, and, to a lesser extent, to the lung fibroblast MRC5 cells, as demonstrated by flow cytometric analysis (data not shown).
Cross-reactive epitopes on SARS-CoV spike protein
Several synthetic peptides were designed from the viral epitopes sharing sequence homology with human proteins (Fig. 3a). After a panel of screening proceeded, two spike-protein peptides, designated D07 and D08, appeared to be bound by SARS patient sera (Fig. 3b). These two peptides are located at the S2 domain of the spike protein. Binding of patient sera to S2 was also confirmed (Fig. 3b). To further validate the epitopes shared between viral- and self-antigens, SARS patient sera were preabsorbed with various peptides, and A549 binding activity was determined. Results indicated that A549 cell binding activity of patient sera was reduced by preabsorption with S2, D07, and D08 (Fig. 3c). The D02 and D03 peptides were not bound by patient sera (Fig. 3b) and, accordingly, did not cause an inhibition (Fig. 3c). The existence of cross-reactive epitopes on SARS-CoV S2 shared homology with host cell proteins was therefore demonstrated.
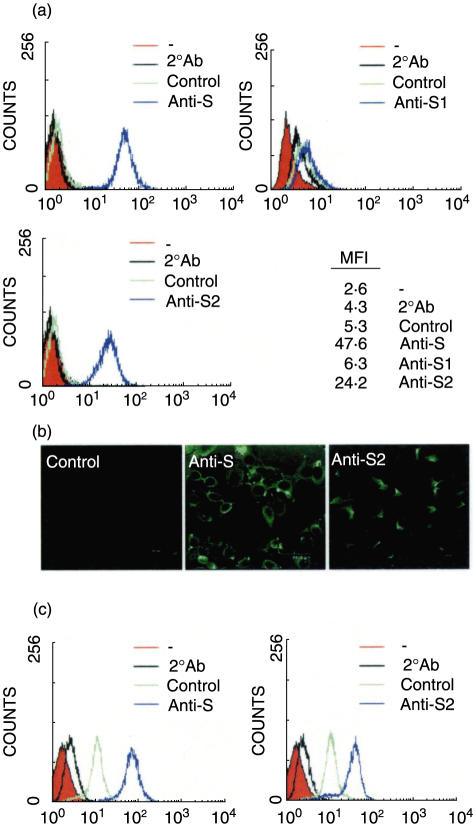
To further characterize the epithelial cell cross-reactivity of anti-S Abs, mouse Abs directed against S, S1, and S2 were tested for their binding activities with A549 cells. Using flow cytometry analysis, there were higher levels of cell binding ability by anti-S and anti-S2, but not anti-S1, Abs (Fig. 4a). The binding ability of anti-S and anti-S2 Abs on the surface of epithelial cells was also confirmed by confocal microscopy (Fig. 4b). Further study using human lung epithelial cell line HL for the binding activity indicated that anti-S and anti-S2 Abs could also bind to these lung epithelial cells (Fig. 4c).
Fig. 4.
The epithelial cell cross-reactivity of anti-spike Abs. A549 cells (a, b) and HL cells (c) were incubated with a 1 : 50 dilution of anti-S, S1, or S2 mouse hyperimmune sera, followed by FITC-conjugated anti-mouse IgG, and then analysed by flow cytometry (a, c) or viewed with confocal microscopy (b). The normal mouse sera were used as the negative control. MFI: mean fluorescence intensity.
Effect of anti-S2 Abs on immune cell adhesion to A549 cells
To further explore the effects of anti-S2 Abs binding on epithelial cells, the adherence of PBMC to A549 cells was investigated. Because a cytokine storm such as the increased production of IFN-γ was previously shown [26], the adhesion of PBMC to IFN-γ-treated A549 cells in the presence or absence of anti-S2 Abs was also assessed. Results showed that higher levels of PBMC adherence could be observed in cells treated with IFN-γ for 24 h compared to the untreated group (Fig. 5a). Interestingly, anti-S2 promoted the adhesion of PBMC to A549 cells and this was greatly enhanced in combination with IFN-γ treatment (Fig. 5a). Therefore, in addition to cell injury after cross-linking, anti-S2 Abs can also up-regulate the immune cell adhesion to epithelial cells. Study using immunostaining indicated that monocytes in PBMC were the major cell population with preferential binding to the anti-S2-treated A549 cells (data not shown). This is in accordance with the finding that macrophages are the prominent leucocyte in the alveoli of SARS patients [12]. In a competition binding assay, synthetic peptides (D07 and D08) and S2 protein inhibited the anti-S2-mediated adhesion of PBMCs to A549 cells (Fig. 5b).
Discussion
A novel human CoV may cause SARS, which is characterized by fever, myalgia, dry cough, and lymphopenia. SARS patients develop an atypical form of pneumonia. Among the changes observed in the lungs of SARS patients are epithelial cell proliferation and desquamation, hyaline membrane formation along alveolar walls, and immune cell infiltration during the early stage of the disease, and increased fibrosis and multinucleated epithelial giant cell formation during a later stage. The increasing viral load suggests an effect caused by viral replication in the early phase [11]. In addition, proinflammatory cytokine production and dysregulation may be involved in the pathogenesis of SARS [12, 15, 16, 26]. One study documented IgG seroconversion in 93% of the patients at a mean of 20 days. The timing of IgG seroconversion, which started on day 10, correlated with falls in viral load, which occurred between days 10–15, when several clinical worsenings, which cannot be explained by uncontrolled viral replication, occurred. The lung damage at this phase is therefore related to immune-mediated pathological effects [11]. In the present study, we showed that autoAb production is also involved in SARS-CoV infection. The autoAbs, mainly IgG, started to appear on day 20.
Viral infections have been associated with the development of autoimmune diseases [30]. Most autoimmune disorders are chronic diseases, but there are several acute autoimmune responses that are initiated shortly after infection. Structural similarities between viral proteins and self-antigens have long been proposed as targets for immune cross-reactivity associated with the initiation of autoimmunity. We have reported this phenomenon in dengue virus infection and hypothesized its role in the immunopathogenesis of dengue virus-induced dengue haemorrhagic fever and dengue shock syndrome [31–34]. Our finding that antiepithelial cell autoAbs are produced after SARS-CoV infection is another example of acute viral infection-induced autoimmunity. Both viruses have some characteristics in common: both are lymphotrophic; the infections cause high fever, lymphopenia, thrombocytopenia, haemorrhage, and mild hepatitis; cytokine storm occurs in the acute phase; and molecular mimicry exists between virus proteins and self-antigens. Mouse CoV infection induces B cell polyclonal activation and the generation of autoAbs [23,24]. Although autoAb production in SARS patients causes cytotoxicity to A549 cells (type-2 pneumocytes), whether the autoimmune response may act as a pathological factor in SARS disease needs further investigation. Interestingly, the elevated Ab titre was associated with the aggravation of respiratory failure [35]. In addition to Ab-mediated cytotoxicity, the release of chemokines and cytokines by A549 cells has been detected. Our preliminary results indicated an increase in the production of IL-6, MCP-1, MIP-1α and MIP-1β after anti-S2 stimulation (unpublished data).
Recent reports [36,37] have shown that the SARS-CoV spike protein might elicit the protective Ab responses in mice. The neutralizing effect of anti-SARS-CoV spike Abs was demonstrated. Based on our results by sequence comparison from the NCBI protein database, however, several regions of SARS-CoV spike protein show sequence homology to self-antigens expressed in human cells. We have identified several candidate proteins recognized by SARS patient sera and anti-SARS-CoV spike Abs, including annexin II, glyceraldehyde-3-phosphate dehydrogenase, albumin, α1-antitrypsin, aldo-keto reductase, and transferrin (manuscript in preparation). The local alignment by Java-European Molecular Biology Open Software Suite (JEMBOSS)-Water analysis showed that these proteins have a high degree of homology with the spike peptide sequences of D07 and D08 located in S2 portion. Recombinant S2 protein or the spike peptides D07 and D08 blocked the binding of SARS patient sera to A549 cells. Furthermore, murine anti-S2, but not S1, antisera can bind A549 cells. These results suggest that anti-SARS-CoV Abs can recognize the cross-reactive epitope on human lung epithelial A549-cell proteins by molecular mimicry between spike proteins and self-antigens. The epitope for neutralizing Abs is in the S1 domain [38,39], while the cross-reactive Abs recognize the epitope on the S2 domain of spike protein. Analysis of Ab expression profile from SARS patient sera used in this study indicated that anti-S2 IgG started to appear around day 20, while antinucleocapsid (N) IgG could be detected around day 10 [40]. This is consistent with the previous report that IgG seroconversion started on day 10 [11] and with our findings that the autoAbs, which are mainly anti-S2 IgG, appeared on day 20.
The onset of autoimmune responses in SARS-CoV infection may have important implications. It is still not clear why intense lung inflammation develops even after the viral load has dropped two weeks after the onset of fever. The virus itself and virus-induced cytokine production might be responsible for the early stage of lung epithelial cell damage, while the late autoAb-mediated or infiltrating cell-mediated inflammation causes continued and sustained alveolar damage and fibrosis. In addition to A549 cells, other cell types, including endothelial cells, hepatocytes, and fibroblasts, were also bound by autoAbs in SARS patients. The autoAb production caused by the molecular mimicry between self-antigens and the spike protein of SARS-CoV might relate to the systemic effects and the sequelae of SARS disease [41]. Furthermore, the binding of immune cells with lung epithelial cells, which is accelerated by the combination of IFN-γ and anti-S2, may contribute to the inflammatory responses associated with SARS pathogenesis. This hypothesis needs to be further tested. Studies on the role of autoAbs will be crucial to gain an insight into SARS immunopathologic mechanism.
Acknowledgments
We thank everyone from the Division of Disease Surveillance and Investigation, CDC-Taiwan, involved in combating the SARS outbreak in Taiwan, and the experts from the CDC-USA and WHO who helped us in this SARS storm. We are indebted to the laboratory investigation team members who conducted the examinations of SARS-CoV RT-PCR and the serological analyses. We thank Bill Franke for editorial assistance. This work was supported by SARS Research Project grants NSC92-2751-B006-007, -008 and -009 from the National Science Council, Taiwan.
References
- 1.Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuiken T, Fouchier RAM, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai MMC. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.So LKY, Lau ACW, Yam LYC, Cheung TMT, Poon E, Yung RWH, Yuen KY. Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet. 2003;361:1615–1617. doi: 10.1016/S0140-6736(03)13265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris JSM, Chu CM, Cheng VCC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholls JM, Poon LLM, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JT, Sheng WH, Fang CT, Chen YC, Wang JL, Yu CJ, Chang SC, Yang PC. Clinical manifestations, laboratory findings, and treatment outcomes of SARS patients. Emerg Infect Dis. 2004;10:818–824. doi: 10.3201/eid1005.030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Qiu Z, Zhang L, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong CK, Lam CWK, Wu AKL, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:195–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones BM, Ma ESK, Peiris JSM, et al. Prolonged disturbance of in vitro cytokine production in patients with severe acute respiratory syndrome (SARS) treated with ribavirin and steroids. Clin Exp Immunol. 2004;135:467–473. doi: 10.1111/j.1365-2249.2003.02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholls J, Dong XP, Jiang G, Peiris M. SARS clinical virology and pathogenesis. Respirology. 2003;8:S6–S8. doi: 10.1046/j.1440-1843.2003.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Openshaw PJM. What does the peripheral blood tell you in SARS? Clin Exp Immunol. 2004;136:11–12. doi: 10.1111/j.1365-2249.2004.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunnarsson A, Hellmark T, Wieslander J. Molecular properties of the Goodpasture epitope. J Biol Chem. 2000;275:30844–30848. doi: 10.1074/jbc.M004717200. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds J, Moss J, Duda MA, et al. The evolution of crescentic nephritis and alveolar haemorrhage following induction of autoimmunity to glomerular basement membrane in an experimental model of Goodpasture's disease. J Pathol. 2003;200:118–129. doi: 10.1002/path.1336. [DOI] [PubMed] [Google Scholar]
- 21.DePaso WJ, Winterbauer RH. Interstitial lung disease. Dis Mon. 1991;37:61–133. [PubMed] [Google Scholar]
- 22.Kyuwa S, Yamaguchi K, Toyoda Y, Fujiwara K. Induction of self-reactive T cells after murine coronavirus infection. J Virol. 1991;65:1789–1795. doi: 10.1128/jvi.65.4.1789-1795.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooks JJ, Percopo C, Wang Y, Detrick B. Retina and retinal pigment epithelial cell autoantibodies are produced during murine coronavirus retinopathy. J Immunol. 1993;151:3381–3389. [PubMed] [Google Scholar]
- 24.Mathieu PA, Gomez KA, Coutelier JP, Retegui LA. Identification of two liver proteins recognized by autoantibodies elicited in mice infected with mouse hepatitis virus A59. Eur J Immunol. 2001;31:1447–1455. doi: 10.1002/1521-4141(200105)31:5<1447::AID-IMMU1447>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HS, Chiu SC, Tseng TC, et al. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg Infect Dis. 2004;10:304–310. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang KJ, Su IJ, Theron M, Wu YC, Lai SK, Liu CC, Lei HY. An interferon-γ-related cytokine storm in SARS patients. J Med Virol. 2004;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 28.Frostegard J, Nilsson J, Haegerstrand A, Hamsten A, Wigzell H, Gidlund M. Oxidized low density lipoprotein induces differentiation and adhesion of human monocytes and the monocytic cell line U937. Proc Natl Acad Sci USA. 1990;87:904–908. doi: 10.1073/pnas.87.3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams MR, Jessup W, Hailstones D, Celermajer DS. L-Arginine reduces human monocyte adhesion to vascular endothelium and endothelial expression of cell adhesion molecules. Circulation. 1997;95:662–668. doi: 10.1161/01.cir.95.3.662. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz MS, Sarvetnick N. Viruses, host responses, and autoimmunity. Immunol Rev. 1999;169:241–253. doi: 10.1111/j.1600-065X.1999.tb01319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei HY, Yeh TM, Liu HS, Lin YS, Chen SH, Liu CC. Immunopathogenesis of dengue virus infection. J Biomed Sci. 2001;8:377–388. doi: 10.1007/BF02255946. [DOI] [PubMed] [Google Scholar]
- 32.Lin CF, Lei HY, Shiau AL, et al. Endothelial cell apoptosis induced by antibodies against dengue virus nonstructural protein 1 via production of nitric oxide. J Immunol. 2002;169:657–664. doi: 10.4049/jimmunol.169.2.657. [DOI] [PubMed] [Google Scholar]
- 33.Lin CF, Lei HY, Shiau AL, et al. Antibodies from dengue patient sera cross-react with endothelial cells and induce damage. J Med Virol. 2003;69:82–90. doi: 10.1002/jmv.10261. [DOI] [PubMed] [Google Scholar]
- 34.Lin YS, Lin CF, Lei HY, et al. Antibody-mediated endothelial cell damage via nitric oxide. Curr Pharm Design. 2004;10:213–221. doi: 10.2174/1381612043453469. [DOI] [PubMed] [Google Scholar]
- 35.Hsueh PR, Hsiao CH, Yeh SH, et al. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg Infect Dis. 2003;9:1163–1167. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subbarao K, McAuliffe J, Vogel L, et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babcock GJ, Esshaki DJ, Jr, Thomas WD, Ambrosino DM. Amino acids 270–510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu HS, Hsieh YC, Su IJ, et al. Early detection of antibodies against various structural proteins of the SARS-associated coronavirus in SARS patients. J Biomed Sci. 2004;11:117–126. doi: 10.1007/BF02256554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan KS, Zheng JP, Mok YW, Li YM, Liu YN, Chu CM, Ip MS. SARS: prognosis, outcome and sequelae. Respirology. 2003;8:S36–S40. doi: 10.1046/j.1440-1843.2003.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]