Abstract
Mast cells have recently been found to be a major player in the host defence against bacterial infection through secretion of potent mediators. Identification of bacteria-induced mast cell mediators and intracellular signalling molecules involved during bacterial infection remains a major area of investigation. Recently we found an active interaction between mast cells and Pseudomonas aeruginosa bacteria. To further characterize specific genes in mast cells modulated by P. aeruginosa, we used a new approach for the study of mast cell–bacteria interaction; the suppression subtractive hybridization (SSH). SSH approach does not require a prerequisite knowledge of target genes and does not rely on the availability of the assay reagents for the specific genes. Using SSH, 94 clones were randomly selected from the subtracted cDNA library for differential screening leading to the identification of 14 P. aeruginosa–up-regulated transcripts. Sequence analysis revealed that expression of IL-1, IL-8 and CCL4 was increased by human mast cells after P. aeruginosa infection. Increased production of IL-1, IL-8 and CCL4 was confirmed at the protein levels. In addition, sequence analysis of the clones also suggests that ribosomal protein S3 and cytochrome b as well as additional 4 uncharacterized genes may potentially be involved in P. aeruginosa pathogenesis. Thus, SSH is an effective approach by identifying potential molecular targets for the study of mechanisms involved in P. aeruginosa and mast cell interaction.
Keywords: Pseudomonas aeruginosa, mast cell, suppression subtractive hybridization, cytokine, chemokine
Introduction
Mast cells play a critical role in host defence against bacterial infection through secretion of mast cell mediators [1–3]. Recently we demonstrated that human mast cells produce several cytokines and chemokines after stimulation by Pseudomonas aeruginosa as determined by traditional technical approaches such as enzyme-linked immunosorbent assay (ELISA) or reverse transcription-polymerase chain reaction (RT-PCR) [4–6]. In addition, several intracellular signalling molecules such as protein kinase Cα, protein phosphatase 2 A, caspase 3 and caspase 8 have been found to be involved in mast cell–P. aeruginosa interaction by Western blotting [4,7]. Since a better understanding of molecular mechanisms involved in mast cell–P. aeruginosa interaction largely relies on the identification of specific genes modulated during this interaction, characterization of P. aeruginosa-induced genes remains to be an area of active investigation. Recently, several high throughput assays such as cDNA microarrays have been used and generated valuable information [8,9]. Here we employed an alternative approach – the suppression subtractive hybridization (SSH). This method does not require a prerequisite knowledge of the target gene and allows us to identify P. aeruginosa-induced ‘novel’ target in human mast cells for further study.
SSH is a powerful technique that combines the traditional subtractive hybridization method with suppression polymerase chain reaction [10,11]. It has been used to compare two populations of mRNA and to obtain clones of genes that are expressed in one population but are lower or not expressed in the other [12]. Based on this feature it is well suited for identifying genes in mammalian cells that are modulated by pathogen infection [11].
Here we report that using SSH several P. aeruginosa-induced genes were identified. Expression of IL-1, IL-8 (CXCL8) and CCL4 was confirmed by Northern blot analysis and the production of IL-1, IL-8 and CCL4 protein was confirmed by ELISA. The finding of IL-1 production by P. aeruginosa is consistent with our previous studies, while IL-8 and CCL4 production by human mast cells in response to P. aeruginosa has not been reported previously. Since both IL-8 and CCL4 are potent chemoattractants for leucocyte migration [13], the finding of P. aeruginosa-induced CCL-4 and IL-8 production supports the conceptual model that mast cells function as a sentinel in host defense against bacterial infection through recruiting other immune cells such as neutrophils [1–3].
Materials and methods
Mast cells and Pseudomonas aeruginosa treatment
Highly purified human cord blood-derived mast cells were obtained by long-term culture of cord blood progenitor cells as previously described [6]. Briefly, heparinized cord blood was centrifuged over a Ficoll separating solution (Seromed, Berlin, Germany). Light-density cells including the progenitors were cultured at 37 °C in a humidified atmosphere containing 5% CO2 at a starting density of 1 × 106 cells/ml in RPMI 1640 medium supplemented with l-glutamine, penicillin, streptomycin, 10% fetal bovine serum (all from Invitrogen-Life Technologies, Burlington, ON, USA), 1% (w/v) bovine serum albumin (BSA; Sigma St Louis, MO, USA), 50 µM 2-mercaptoethanol (Sigma), 100 ng/ml human recombinant stem cell factor (SCF; PeptroTech, Inc., Sharpsburg, MD, USA) and 20% CCL-204 (ATCC) normal human fibroblast supernatant as a source of IL-6. The medium was renewed every 7 days. The percentage of mast cells in the cultures was assessed by toluidine blue staining (pH 1·0) of cytocentrifuged samples. After >8 weeks in culture, mature MC (>95% purity) were identified by their morphological features and the presence of metachromatic granules (toluidine blue staining). These cells were >98% positive for c-kit when stained by antic-kit Ab (Exalpha Biologicals, Boston, MA, USA) and analysed by flow-cytometry. Experimental procedures were approved by the Ethical Committee of Issac Walton Killam Health Center, Dalhousie University.
For RNA isolation, cord blood-derived mast cells at the concentration of 5 × 105 cells/ml were treated with P. aeruginosa strain 8821 for 3 h at the mast cell-bacteria ratio of 1 : 50. In other experiments, cord blood-derived mast cells (5 × 105 cells/ml) were treated by P. aeruginosa for 24 h, cell free supernatants were collected for the determination of CCL4 and IL-8 levels by ELISA.
RNA isolation
Total RNA was isolated from mast cells stimulated with P. aeruginosa or untreated mast cells using TRIzol® reagent (Invitrogen) according to the manufacture's instructions. To remove the genomic DNA, the RNA samples were treated with DNase I (Promega, Madison, WI, USA) for 15 min at 37 °C, then extracted with phenol: chloroform: isopropyl alcohol (25 : 24 : 1), and concentrated by ethanol precipitation. The amount and quality of the total RNA obtained was determined by spectrophotometry (OD = 260/280) and electrophoresis on a 1% agarose gel containing formaldehyde, respectively. The RNA was then stored at −70 °C.
Suppression subtractive hybridization (SSH)
SSH was performed using the PCR-select cDNA subtraction system in combination with the Super SMART PCR cDNA Synthesis and Advantage cDNA PCR Systems (BD Biosciences Clontech, Palo Alto, CA, USA). In brief, total RNAs from the P. aeruginosa stimulated (tester) or untreated mast cells (driver) were synthesized into double-stranded cDNAs according to the manufacturer's instructions. The first strand cDNA synthesis reaction included 1 µg total RNA, the 3′-SMART CDS primer, the SMART II A Oligonucleotide, dNTP and PowerScript reverse transcriptase. The product was purified with Chromaspin columns (BD Biosciences Clontech) according to the manufacture's instruction. A 80 µl diluted ssDNA (1 : 16) aliquot was mixed with Advantage 2 Polymerase Mix, dNTP and 5′ PCR primer II A to synthesize the double-strand cDNA. The PCR condition was 20 cycles of 95 °C for 15 s, 65 °C for 30 s, and 68 °C for 3 min on Perkin-Elmer GeneAmp PCR system (PE 480). Both ‘tester’ and ‘driver’ PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN Inc., Valencia, CA, USA). The purified double stranded cDNAs were digested with Rsa I and purified with QIAquick PCR Purification Kit (QIAGEN). The tester cDNA (cDNA from P. aeruginosa-treated mast cells) then was subdivided into two portions, and each was ligated with a different adaptor (adaptor 1 or adaptor 2), provided by the manufacturer. After the ligation, the resulting cDNAs (‘tester’ cDNAs ligated with adaptors) were divided into two populations: one for subtraction study and the other for nonsubtraction, that allow evaluation of subtraction efficiency.
For subtraction study, the adaptor-ligated cDNAs were used for two hybridizations with ‘driver’ cDNAs from un-stimulated mast cells. In the first round, an excess of driver was added to each sample (cDNA-adaptor 1 and cDNA-adaptor 2) of tester, leading to the enrichment of differentially expressed sequences. During the second round of hybridization, the two primary hybridization samples (cDNA-adaptor 1 and cDNA-adaptor 2) were mixed together to form new double-stranded hybrids with different ends. Freshly denatured driver cDNA was added again to further enrich differentially expressed sequences. After first and second hybridizations were performed, the resulting annealed material was used as the PCR template. PCR amplifications were performed using the primers that matched the different adaptors to the 5′- and 3′- ends. In the first amplification, only double strand cDNAs with different adaptor sequences on each end are exponentially amplified. In the second PCR, nested PCR is used to further reduce background and to enrich the differentially expressed sequences. The hybridization mixture was diluted with 200 µl dilution buffer, and 1 µl diluted mixture was amplified for 27 primary PCR cycles. As a control for subtraction efficiency, the ‘Un-subtracted’ cDNA population without hybridization process was also amplified for 27 primary PCR cycles. The condition of the primary PCR was: 94 °C for 25 s followed by 27 cycles of 94 °C for 10 s, 66 °C for 30 s and 72 °C for 90 s. One µl of one-tenth diluted primary PCR products from both ‘Subtracted’ and ‘Un-subtracted’ samples were used for a second round of PCR. The secondary PCR condition was 10 cycles of 94 °C for 10 s, 68 °C for 30 s, and 72 °C for 90 s. These cDNAs were then used for evaluation of subtraction efficiency as described below.
The PCR products from ‘Subtracted’ population were ligated into the PCR 2·1 TA cloning vector (Invitrogen), and transformed into DH5α competent cells. These samples were then used for differential hybridization and cloning as described in the following ‘differential hybridization’ section.
Evaluation of subtraction efficiency
The resulting cDNAs from above ‘Subtracted’ and ‘Un-subtracted’ population were used for evaluation of subtraction efficiency by determining the relative amount of glyceraldehye-3-phosphate dehydrogenase (GAPDH). GAPDH cDNA present in the subtracted and unsubtracted cDNA populations after SSH was amplified by PCR using the GAPDH 5′-and 3′ primers provided in the PCR-Select™ cDNA Subtraction System (BD Biosciences, Clontech). Various PCR cycles (13–33) were performed. Successful subtraction is reflected by the delayed appearance of GAPDH after various PCR cycles.
Differential hybridization
The transformed DH5α competent cells were grown overnight at 37 °C on Luria-Bertani (LB) agar plates containing ampicillin, X-gal, and isopropyl-β-D-thiogalactopyranoside for white/blue colony selection. White colonies were isolated and grown individually in 200 µl of LB broth containing ampicillin for overnight at 37 °C. The insert of each colony was PCR amplified using a 1-µl aliquot of the bacteria growth while rest was kept in 50% glycerol at −80 °C. The PCR amplifications were done using 0·1 µl of Taq Gold polymerase (PE Applied Biosystems) and Nested primer 1 and nested primer 2R from supplier (BD Biosciences, Clontech) in an iCycler (Bio-Rad). The amplification profile comprised an initiating cycle of 30 s at 94 °C followed by 23 cycles as follows: 95 °C for 30 s, 68 °C for 3 min. The final cycle included a further 30 s at 68 °C. A 7-µl aliquot of each PCR product was denatured by adding equal amount of 0·6 N NaOH, and 2·5 µl of the mixture was dotted onto Tropilon-plus membrane (Tropix Inc., Bedford MA, USA). Two identical membranes were done for 94 cDNA inserts and two negative controls provided with the PCR-Select Differential Screening Kit (BD Biosciences, Clontech). The cDNA from the P. aeruginosa stimulated or untreated mast cells were labelled using DIG HIGH Prime DNA labelling kit (Roche Diagnostics Canada, Quebec, Canada). Membranes were prehybridized for 2 h in a solution of 1 mM ethylenediaminetetraacetic acid (EDTA), 7% sodium dodecyl sulphate (SDS), 0·25 M sodium phosphate at 70 °C. Membranes were then hybridized in the same solution with Dig-High Prime labelled probe at 70 °C overnight. The membranes were washed with 2 × 15 min at room temperature with 2x SSC/1%SDS (1 × SSC = 150 mM NaCl and 15 mM sodium citrate, pH 7·4), 2 × 15 min at 68 °C with 0·1 × SSC/1% SDS, and 1 × 10 min at 68 °C with 1 × SSC. The signals were detected on X-ray film using a Southern-Star biotin-labelled DNA detection kit (Tropix).
DNA sequencing and analysis
DNA sequencing was performed using an ABI 3700 DNA Analyser in the Dalhousie University. Sequence homologies were determined at the NCBI (National Center for Biotechnology Information, Bethesda, MD, USA) using the BLAST (Basic Local Alignment Search Tool) program.
Virtual Northern blot analysis
A 20 µl of the synthesized double stranded cDNA of the P. aeruginosa stimulated and untreated mast cells were electrophoresed on 1% agarose gel. DNA was transferred onto a Tropilon-plus membrane (Tropix) in 20× SSC overnight and corss-linked to the membrane for 2 min using a UV transilluminator. Based on the result of sequencing analysis, the differential clones that represented various genes were used as the probes. The probes were amplified by PCR using M13 forward and reverse primers. PCR conditions were: 94 °C for 2 min; 35 cycles of 94 °C for 1 min, 50 °C for 1 min, 72 °C for 2 min; final extension at 72 °C for 10 min. Amplicons were labelled using Dig-High Prime (Roche Diagnostics Canada). Probes were denatured at 95 °C for 5 min and added to nylon membranes that had been prehybridized in a solution of 1 mM EDTA, 7% SDS, 0·25 M Sodium Phosphate at 65 °C for 2 h. Membranes were then hybridized at 65°C overnight in the same solution and washed twice with 2× SSC/1%SDS (5 min each) at room temperature followed by three washes with 0·5× SSC/0·1% SDS at 65 °C (10 min each). The signals were detected on X-ray film using a Southern-Star biotin-labelled DNA detection kit (Tropix).
IL-8 and CCL4 assays
Human IL-8 and CCL4 levels in supernatants were measured using Abs from R & D Systems (Minneapolis, MN, USA) by ELISA. The detection limit was 10 pg/ml.
Results
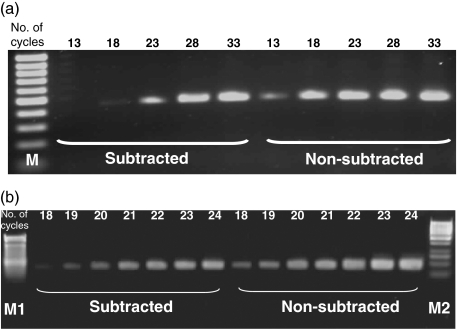
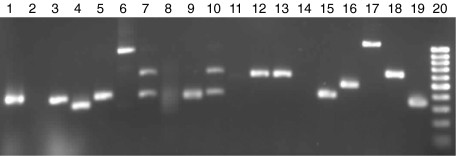
Total RNAs from the P. aeruginosa stimulated or untreated mast cells were used to generate double strand cDNAs using SMART PCR cDNA Synthesis kit. The Advantage™ cDNA PCR kit was used for the cDNA amplification. Based on the results of preliminary experiment, the optimal cycle number of PCR reaction was 20 cycles. A subtracted cDNA library was constructed to enrich the transcripts that were differentially expressed by mast cells after P. aeruginosa stimulation. The subtraction efficiency was evaluated by detecting the housekeeping gene, GAPDH in both the subtracted and nonsubtracted cDNA populations by PCR amplification. The PCR product of GAPDH gene appeared to be detectable on agarose gel after amplification for 18 cycles and 13 cycles in the subtracted and the nonsubtracted cDNA populations, respectively (Fig. 1a). The difference of band density of GAPDH gene is visible between the subtracted and the nonsubtracted cDNA populations in the PCR less than 20 cycles (Fig. 1b). The PCR products of the subtraction were cloned and transferred into bacteria that were grown on agar plates. Each isolated colony containing a plasmid with a cDNA insert was PCR amplified. Figure 2 shows the example of insert amplification of the isolated bacterial colony. The size of the amplified 96 inserts ranged from 200 bp up to 1·2 kb. In Fig. 2, lanes 7 and 10 show several bands suggesting some bacteria colonies had more than one plasmid.
Fig. 1.
Evaluation of the subtraction efficiency by amplifying the cDNA of GAPDH in both the subtracted and nonsubtracted cDNA pools by PCR. (a). The products amplified for different cycles were electrophoresized on the 1·5% agarose gel. The numerical number indicated the number of PCR cycles. Lane M: 100 bp molecular mass markers. Note that the GAPDH band appeared at the 13rd cycle in the nonsubtracted pool, while it appeared at the 18th cycle in the subtracted pool. (b). The cDNA of GAPDH in both the subtracted and nonsubtracted cDNA pools were amplified for 18–24 cycles by PCR. The numerical number indicated the number of PCR cycles. Lane M1: 100 bp molecular markers, lane M2: 1kb molecular markers. The difference in GAPDH band density between the subtracted and nonsubtracted pool was detected in the PCR less than 20 cycles.
Fig. 2.
PCR assays to verify the insert of each colony. The PCR amplified subtracted cNDA was ligated into the PCR 2·1 TA cloning vector, and transformed into DH5α competent cells. The transformed DH5α competent cells were grown overnight at 37 °C on LB agar plates containing ampicillin, X-gal, and isopropyl-β-D-thiogalactopyranoside for white/blue colony selection. White colonies were isolated and grown individually overnight at 37 °C in 200 µl of LB broth containing ampicillin. The insert of each colony was PCR amplified using a 1-µl aliquot of the bacteria growth. The size of inserts showing in this example varied from about 350 base pair (bp) to about 1100 bp. Lanes 7 and 10 showed that bacteria colonies had more than one plasmid. Lanes 2 and 14 showed no insert in these colonies. Lane 20 is 100 bp DNA markers.
Before characterization of the clones, a differential hybridization procedure was performed to identify the false positives. Ninety-four clones were differentially screened resulting in the identification of 14 P. aeruginosa-up-regulated clones. All the positive clones were analysed by DNA sequencing and then compared against the NCBI databases using the online BLAST program. The sequences of clones psa12B and psa10C are identical, and match with Homo sapiens chemokine (C-C motif) ligand 4 (CCL4). The clone psa8C shows homologous to human interleukin 1. The sequences of clone psa1G and clone psa1D are the same, and match with human interleukin 8. Interestingly, two clones have the sequence homology with the cell structural proteins, ribosomal protein S3 and cytochrome b, respectively. Other three clones also match with different human DNA sequences in GenBank. No matches were found in GenBank for four clones (Table 1).
Table 1. Analysis of positive clone sequences from subtracted library.
| Clone | Insert size (bp) | Best BALST hit | Identity |
|---|---|---|---|
| psa1C | 870 | Homo sapiens chromosome 5 clone (AC136632) | 97% (578/591) |
| psa1D & psa1G | 565 | Homo sapiens interleukin 8 (NM_000584) | 98% (557/565) |
| psa1E | 436 | Human DNA sequence from clone RP13—37K15 on chromosome 9 (AL772267) | 99% (273/274) |
| psa1F | 503 | Homo sapiens heterogeneous nuclear ribonucleoprotein d-like, mRNA (BC007392) | 98% (495/503) |
| psa1H | 846 | Homo sapiens ribosomal protein S3 (NM 001005) | 97% (747/763) |
| psa2D | 460 | No significant hits | |
| psa8B | 909 | No significant hits | |
| psa8C | 513 | Homo sapiens interleukin 1 (NM_000576) | 99% (459/460) |
| psa12C | 709 | Homo sapiens cytochrome b (AY195792) | 98% (676/687) |
| psa10C & psa12B | 331 | Homo sapiens chemokine (C-C motif) ligand 4 precursor (NP_002984) | 99% (330/331) |
| psa10G | 818 | No significant hits | |
| psa12D | 380 | No significant hits |
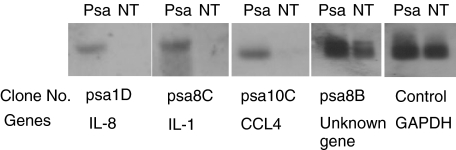
Reverse Northern blot analysis was used to further evaluate the relevance of 4 positives clones found with differential screening. The expression pattern of these 4 clones is presented in Fig. 3. The clone psa1D, psa8C and psa10C that matched with interleukin 8, interleukin 1 and CCL4, respectively, showed elevated expressions following P. aeruginosa treatment, whereas psa8B, the function of which is unknown was enhanced after P. aeruginosa stimulation.
Fig. 3.
Virtual Northern assay of gene expression. The SMART double strand cDNAs from P. aeruginosa stimulated mast cells (lane Psa), and untreated mast cells (lane NT) were hybridized with the probes prepared from the positive clone psa1D, psa8C, psa10C, psa8B and GAPDH (control).
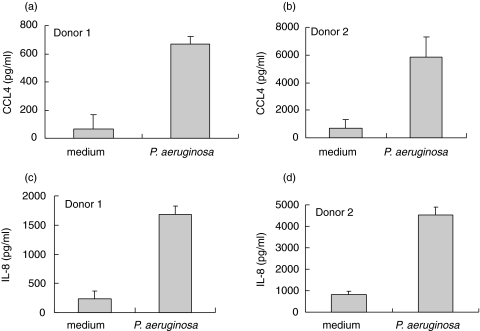
To further confirm the expression of CCL4, IL-8 and IL-1 by human mast cells, human cord blood-derived mast cells (5 × 105 cells/ml) from two individual donors were treated with P. aeruginosa strain 8821 (1/50 cell-bacteria ratio) for 24 h. Cell free supernatants were collected for the determination by ELISA. P. aeruginosa treatment strongly stimulated human mast cells to produce CCL4 and IL-8 (Fig. 4). Increased IL-1α and IL-1β production from these samples following P. aeruginosa treatment was also observed (data not shown), which is consistent with our recent report [6]. These results support the above SSH data that CCL4, IL-8 and IL-1 are up-regulated in human mast cells following P. aeruginosa stimulation.
Fig. 4.
CCL4 and IL-8 production by human cord blood-derived mast cells following P. aeruginosa stimulation. Human cord blood-derived mast cells (5 × 105 cells/ml) from two individual donors were treated with P. aeruginosa (1/50 cell-bacteria ratio) for 24 h. Cell free supernatants were collected for the determination by ELISA. Treatment of mast cells with P. aeruginosa induced increased CCL4 and IL-8 production. Error bars represent replicate.
Discussion
Our study provides evidence that SSH technique can be used to identify genes differentially expressed in P. aeruginosa stimulated and unstimulated mast cells. P. aeruginosa is the predominant pathogen in cystic fibrosis and colonizes almost all cystic fibrosis patients at some time during the disease process [14]. P. aeruginosa is also a common cause of nosocomial pneumonia [15]. The host response to this bacterium is a multifaceted process involving various cell populations such as epithelial cells, macrophages, neutrophils, endothelial cells and mast cells. Recently we demonstrated that toll-like receptor (TLR) 2, TLR4 and a TLR adaptor protein, MyD88, are involved in the host defence against P. aeruginosa lung infection in mice in vivo[16]. Since human mast cells express various members of TLR family [17–19], it is possible that members of TLR family may be involved in P. aeruginosa–mast cell interaction.
A role of mast cells in P. aeruginosa infection has only recently been reported by our laboratory based on the conceptual model that mast cells function as a sentinel in the host defense against bacterial infection through recruiting other immune cells. Based on this concept, several cytokines and chemokines in mast cells that are potentially important for other immune cell recruitment have been selected for further study. These selected targets include IL-1α, IL-1β, TNF, GM-CSF, CCL20 and IL-6 [5–7]. The selection of target genes for further study requires prerequisite background knowledge of the specific gene and its potential biological roles in infection. This limitation prevents the finding of ‘unexpected’ genes that are important for the interaction between mast cells and P. aeruginosa. To overcome this limitation, we reported here that SSH is a useful tool for the identification of mast cell genes that are regulated during P. aeruginosa infection without bias of the prerequisite knowledge of a specific gene.
This approach is based on the principle of subtraction of two gene populations; P. aeruginosa-stimulated mast cell population minus unstimulated mast cell population, to identify P. aeruginosa-induced differentially expressed genes. The transcripts that were differentially expressed in mast cells after P. aeruginosa stimulation were enriched by constructing a subtracted cDNA library. The efficiency of the subtraction was verified by testing level of GAPDH transcript by PCR. Transcripts from this subtracted cDNA library were randomly selected for cloning. After differential hybridization to exclude the false positives from 94 clones, 14 P. aeruginosa-up-regulated clones were identified. All these positive clones were sequenced and compared against the NCBI databases for sequence match. As illustrated in Table 1, these genes include IL-1, IL-8, CCL4, ribosomal protein S3, cytochrome b and several others including 4 sequences with no matches.
Among these newly identified genes, P. aeruginosa-induced IL-1 production has been reported previously [6]. We demonstrated previously that human mast cells after P. aeruginosa stimulation secrete biologically active IL-1α and IL-1β which stimulate endothelium to promote human neutrophil transmigration [6]. Thus, it appears that SSH-generated information is consistent with our previous data using traditional approaches such as ELISA. Production of IL-8 and CCL4 by human mast cells in response to P. aeruginosa has not been reported previously. We confirmed the production of these two chemokines at the protein level using ELISA. Human IL-8 is a potent neutrophil chemoattractant [20]. Since neutrophils infiltration is one of the major features of P. aeruginosa infection [21], mast cell-derived IL-8 likely contributes to the neutrophil recruitment during P. aeruginosa infection.
CCL4 is a potent chemoattractant for a number of cell populations including lymphocytes, immature dendritic cells, neutrophils, NK cells and monocytes [22]. Up-regulation of CCL4 production by human mast cells in response to P. aeruginosa suggests that mast cells may have additional previous un-recognized roles in the initiation of immune response to P. aeruginosa infection. Further studies are required to clarify the biological significance of CCL4 production by mast cells during P. aeruginosa infection. Since both IL-8 and CCL4 are potent chemoattractants and play a major role in the recruitment of leucocytes to sites of infection, P. aeruginosa-induced increased production of IL-8 and CCL4 by mast cells as identified by this study, supports the conceptual model that mast cells likely function as one the sentinels in host defense against P. aeruginosa infection.
Interestingly, SSH also identified that several intracellular proteins including ribosomal protein S3 and cytochrome b were modulated by P. aeruginosa stimulation. Recently, a novel role for ribosomal protein S3 was found in caspase-8/caspase-3-mediated apoptosis [23]. Cytochrome b together with other mitochondrial membrane proteins are important components for the reduction of cytochrome c in mitochondrial respiratory chains [24] which also play an essential role in apoptosis [25]. We recently reported that P. aeruginosa exotoxin A induces human mast cell apoptosis through caspase-8/caspase-3 pathway [4] and induces loss of mitochondrial membrane potential (C. Jenkins and T.J. Lin, unpublished observation). Treatment of human mast cells with P. aeruginosa (24 h) induced mast cell apoptosis (56%) as determined by single stand DNA formation (data not shown). A role for ribosomal protein S3 and cytochromine b in P. aeruginosa-induced human mast cell apoptosis will be further investigated. It is also noteworthy that no matches were found in GenBank for four clones. These may represent P. aeruginosa-induced novel genes yet to be further cloned and characterized.
In summary, we have employed SSH in the characterization of P. aeruginosa-induced genes in human mast cells. A cDNA library containing differentially expressed genes in P. aeruginosa stimulated human mast cells were constructed. Ninety four clones were randomly selected and were differentially screened resulting in the identification of 14 P. aeruginosa-up-regulated transcripts. Sequencing analysis of these clones revealed novel information such as up-regulation of IL-8 and CCL4 that have not been associated with P. aeruginosa in human mast cells previously. While other information such as P. aeruginosa-induced IL-1 production is consistent with our previous report [6]. In addition, sequencing analysis also suggests that several intracellular molecules such as ribosomal protein S3 and cytochrome b as well as other uncharacterized genes may potentially be involved in P. aeruginosa pathogenesis. Thus, SSH is an effective approach for the characterization of molecular mechanisms involved in P. aeruginosa–mast cell interaction.
References
- 1.Echtenacher B, Mannel DN, Hultner L. Critical protective role of mast cells in a model of acute septic peritonitis. Nature. 1996;381:75–7. doi: 10.1038/381075a0. [DOI] [PubMed] [Google Scholar]
- 2.Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381:77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 3.Prodeus AP, Zhou X, Maurer M, Galli SJ, Carroll MC. Impaired mast cell-dependent natural immunity in complement C3-deficient mice. Nature. 1997;390:172–5. doi: 10.1038/36586. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins CE, Swiatoniowski A, Issekutz AC, Lin TJ. Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase-8 and -3-dependent mechanism. J Biol Chem. 2004;279:37201–7. doi: 10.1074/jbc.M405594200. [DOI] [PubMed] [Google Scholar]
- 5.Lin TJ, Maher LH, Gomi K, McCurdy JD, Garduno R, Marshall JS. Selective early production of CCL20, or macrophage inflammatory protein 3alpha, by human mast cells in response to Pseudomonas aeruginosa. Infect Immun. 2003;71:365–73. doi: 10.1128/IAI.71.1.365-373.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin TJ, Garduno R, Boudreau RT, Issekutz AC. Pseudomonas aeruginosa activates human mast cells to induce neutrophil transendothelial migration via mast cell-derived IL-1 alpha and beta. J Immunol. 2002;169:4522–30. doi: 10.4049/jimmunol.169.8.4522. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau RT, Garduno R, Lin TJ. Protein phosphatase 2A and protein kinase Calpha are physically associated and are involved in Pseudomonas aeruginosa-induced interleukin 6 production by mast cells. J Biol Chem. 2002;277:5322–9. doi: 10.1074/jbc.M108623200. [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa JK, Norris A, Bangera MG, Geiss GK, van 't Wout AB, Bumgarner RE, Lory S. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc Natl Acad Sci USA. 2000;97:9659–64. doi: 10.1073/pnas.160140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMorran B, Town L, Costelloe E, Palmer J, Engel J, Hume D, Wainwright B. Effector ExoU from the type III secretion system is an important modulator of gene expression in lung epithelial cells in response to Pseudomonas aeruginosa infection. Infect Immun. 2003;71:6035–44. doi: 10.1128/IAI.71.10.6035-6044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanders ED, Goulden MG, Kennedy TC, Kempsell KE. Analysis of immune system gene expression in small rheumatoid arthritis biopsies using a combination of subtractive hybridization and high-density cDNA arrays. J Immunol Meth. 2000;233:131–40. doi: 10.1016/s0022-1759(99)00126-x. [DOI] [PubMed] [Google Scholar]
- 11.Rebrikov DV, Desai SM, Siebert PD, Lukyanov SA. Suppression subtractive hybridization. Meth Mol Biol. 2004;258:107–34. doi: 10.1385/1-59259-751-3:107. [DOI] [PubMed] [Google Scholar]
- 12.Parsons YN, Panagea S, Smart CH, Walshaw MJ, Hart CA, Winstanley C. Use of subtractive hybridization to identify a diagnostic probe for a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J Clin Microbiol. 2002;40:4607–11. doi: 10.1128/JCM.40.12.4607-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennouna S, Bliss SK, Curiel TJ, Denkers EY. Cross-talk in the innate immune system: neutrophils instruct recruitment and activation of dendritic cells during microbial infection. J Immunol. 2003;171:6052–8. doi: 10.4049/jimmunol.171.11.6052. [DOI] [PubMed] [Google Scholar]
- 14.Koch C, Hoiby N. Pathogenesis of cystic fibrosis. Lancet. 1993;341:1065–9. doi: 10.1016/0140-6736(93)92422-p. [DOI] [PubMed] [Google Scholar]
- 15.Cross A, Allen JR, Burke J, et al. Nosocomial infections due to Pseudomonas aeruginosa: review of recent trends. Rev Infect Dis. 1983;5(Suppl. 5):S837–45. doi: 10.1093/clinids/5.supplement_5.s837. [DOI] [PubMed] [Google Scholar]
- 16.Power MR, Peng Y, Maydanski E, Marshall JS, Lin TJ. The development of early host response to Pseudomonas aeruginosa lung infection is critically dependent on myeloid differentiation factor 88 in mice. J Biol Chem. 2004;279:49315–22. doi: 10.1074/jbc.M402111200. [DOI] [PubMed] [Google Scholar]
- 17.Kulka M, Alexopoulou L, Flavell RA, Metcalfe DD. Activation of mast cells by double-stranded RNA. evidence for activation through Toll-like receptor 3. J Allergy Clin Immunol. 2004;114:174–82. doi: 10.1016/j.jaci.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 18.McCurdy JD, Olynych TJ, Maher LH, Marshall JS. Cutting edge: distinct Toll-like receptor 2 activators selectively induce different classes of mediator production from human mast cells. J Immunol. 2003;170:1625–9. doi: 10.4049/jimmunol.170.4.1625. [DOI] [PubMed] [Google Scholar]
- 19.Varadaradjalou S, Feger F, Thieblemont N, Hamouda NB, Pleau JM, Dy M, Arock M. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human mast cells. Eur J Immunol. 2003;33:899–906. doi: 10.1002/eji.200323830. [DOI] [PubMed] [Google Scholar]
- 20.Murphy PM. Neutrophil receptors for interleukin-8 and related CXC chemokines. Semin Hematol. 1997;34:311–8. [PubMed] [Google Scholar]
- 21.Cripps AW, Dunkley ML, Clancy RL, Kyd J. Pulmonary immunity to Pseudomonas aeruginosa. Immunol Cell Biol. 1995;73:418–24. doi: 10.1038/icb.1995.65. [DOI] [PubMed] [Google Scholar]
- 22.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 23.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560(1–3):81–5. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 24.Crofts AR. The cytochrome bc1 complex. function in the context of structure. Annu Rev Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 25.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–12. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]