Abstract
India is at the epicentre of the global HIV/AIDS epidemic in South-east Asia, predominated by subtype C infections. It is important to characterize HIV-1-specific T cell responses in this particular population with the aim of identifying protective correlates of immunity to control HIV-1 infection. In this study, we performed a comprehensive analysis of the breadth and magnitude of T cell responses directed at HIV-1 subtype C Gag, one of the most conserved HIV-1 proteins. The study population consisted of antiretroviral naive, chronic HIV-1 subtype C-infected individuals at various stages of infection. We used recent advanced techniques such as enzyme-linked immunospot (ELISPOT) assay and intracellular cytokine staining to quantify the total CD4+ and CD8+ T cell response to HIV-1 gag at single peptide level, regardless of HLA haplotype of the infected individual. The p24-Gag was identified as the most frequently recognized subunit protein with the greatest magnitude of CD4+ and CD8+ T cell responses. Stronger and broader CD8 T cell responses were recognized, contrasting with the weaker and narrower CD4 T cell responses with regard to Gag protein subunits. The magnitude of the HIV-specific interferon (IFN)-γ responses was observed to be higher than the corresponding interleukin (IL)-2 response, indicating the persistence of antigenic load in chronically infected Indian population due to the probable dysfunction of HIV-specific, IFN-γ-secreting CD8 T cells in absence of IL-2 help.
Keywords: CD4, CD8, Gag, HIV-1-specific, India
Introduction
It is estimated that 39·4 million adults and children are living with HIV-1/AIDS, with 4·3 million new infections detected as at the end of 2004. South-east Asia accounts for approximately 6·4 million of the global HIV-infected population, with India alone accounting for 5·1 million HIV-1 subtype C infections [1]. Understanding the correlates of protective immunity is the first logical step in the development of immune-system-based approaches such as vaccines to control the HIV-1 infection. Most HIV-infected patients initially develop HIV-specific immune responses comprising HIV-specific CD8 cell, CD4 cell and B cell activity. However, these responses are suboptimal and ultimately fail in the vast majority of patients, primarily because of viral mechanisms.
HIV-specific CD8 cell responses are generated early in acute infection followed by a rapid decline in HIV-1 plasma viraemia, reflecting the strong antiviral activities of these cells [2,3]. Increasing evidence also indicates the critical role of virus-specific CTL responses in control of SIV viraemia in animal models [4,5]. However, broadly diversified CD8 responses persist into chronic phases of HIV disease but fail to control viral replication [6–8]. HIV-specific CD4 cell responses, while being generated in the acute stages of HIV disease, are absent or severely impaired in the majority of patients with chronic HIV disease, thus also leading to the waning of HIV-specific CD8 cell responses that require help from CD4 responses [2,9–16].
The identification of the optimal and dominant HIV-1 specific responses are important for defining the property of immunogenecity in HIV-1 vaccine trials and are a desired outcome of vaccine-induced T cell immunity. In addition to the identification and definition of significant epitopes, it is also imperative to define the qualitative nature of anti-HIV T cell immunity. Recent studies have comprehensively analysed total HIV-specific CD4 and CD8 T cell responses to the entire HIV-1 genome [6,7,17–19]. The most frequent and robust responses have been found to be directed against immunodominant regions within the Gag and Nef proteins [9]. However, most of the HIV-1-specific CTL and T helper immune response studies performed until now have been based on HIV-1 B, found mainly in the Caucasian population. Only a limited number of studies have targeted non-B subtypes in non-Caucasian ethnic groups, which illustrate lacunae in the information relevant to the worst affected epicentres of global AIDS epidemic, such as India [17–19].
Hence, with the aim of identifying HIV-1 C Gag-specific responses, we investigated the magnitude and frequency of T cell responses against overlapping subtype-C-based peptides corresponding to the HIV-1 Gag region. We used an interferon (IFN)-γ enzyme-linked immunospot (ELISPOT) assay, that has been used successfully as an effective tool to map immunodominant regions, for the rapid, effective and preliminary screening of a large number of HIV-infected individuals against a wide spectrum of synthetic peptides. Intracellular cytokine staining, having considerable advantages over other techniques, was used to detect low-frequency CD4 or CD8 T cell responses at a single peptide level. It also helps to examine the response to all potential HIV epitopes, regardless of the HLA haplotype of the infected individual. The functional and phenotypic patterns of CD4+ and CD8+ T cell responses characteristic of chronic HIV infection were observed in the HIV-1 subtype C-infected Indian population.
Materials and methods
Study population
Twenty-five HIV-1-infected individuals at different stages of infection were recruited from the AIDS clinic at the Department of Microbiology at the All India Institute of Medical Sciences, New Delhi. The study group consisted of chronically infected individuals with confirmed serodiagnosis of HIV and having no prior history of antiretroviral therapy. The characteristics of the study population are detailed in Table 1. The appropriate Institutional Ethics Committee approved the studies and all the patients gave written and informed consent prior to entry in the study. We also included five individuals seronegative for HIV-1/2 and for hepatitis B and C as healthy controls, who were screened for acute illness and infection requiring medication. Pregnant females and patients with blood dyscrasias were excluded from the study.
Table 1. Characteristics of the study patient cohort.
| Patient no. | Laboratory ID | Age | Sex | CD4 counts | CD8 counts | CDC stage | SFC/10–6 (range)* | No. of recognized peptides† |
|---|---|---|---|---|---|---|---|---|
| 1 | 110976 | 35 | Female | 264 | 2852 | C2 | 230–2670 | 24 |
| 2 | 110977 | 28 | Male | 97 | 1207 | C3 | 120–260 | 12 |
| 3 | 110983 | 33 | Female | 89 | 1244 | C3 | 200–500 | 10 |
| 4 | 111000 | 38 | Male | 698 | 652 | A1 | 140–160 | 4 |
| 5 | 111002 | 30 | Female | 368 | 1250 | B2 | 170–500 | 18 |
| 6 | 111052 | 30 | Female | 344 | 1068 | C2 | 110–440 | 8 |
| 7 | 111053 | 26 | Male | 556 | 1112 | B1 | 280–850 | 19 |
| 8 | 111054 | 32 | Female | 361 | 1394 | A2 | 320–900 | 9 |
| 9 | 111086 | 55 | Female | 403 | 1198 | B2 | 100–220 | 34 |
| 10 | 111102 | 31 | Female | 566 | 4099 | B1 | 160–660 | 24 |
| 11 | 111104 | 27 | Male | 527 | 1032 | B1 | 160–310 | 10 |
| 12 | 111118 | 36 | Male | 249 | 1221 | A2 | 100–330 | 16 |
| 13 | 111154 | 26 | Male | 483 | 1544 | A2 | 100–1590 | 20 |
| 14 | 111181 | 27 | Female | 572 | 971 | A1 | 250–810 | 20 |
| 15 | 111188 | 28 | Male | 970 | 1469 | A1 | 180–2120 | 46 |
| 16 | 111190 | 27 | Female | 347 | 679 | A2 | 260–420 | 6 |
| 17 | 111221 | 30 | Male | 314 | 1172 | A2 | 210–660 | 20 |
| 18 | 111231 | 32 | Male | 611 | 1025 | A1 | 390–430 | 1 |
| 19 | 111246 | 28 | Female | 389 | 1009 | A2 | 120–350 | 2 |
| 20 | 111288 | 21 | Female | 280 | 715 | A2 | 129–142 | 2 |
| 21 | 111292 | 24 | Female | 155 | 1452 | C3 | 120–340 | 4 |
| 22 | 111296 | 20 | Female | 295 | 544 | A2 | 60–260 | 6 |
| 23 | 111428 | 31 | Male | 108 | 1076 | B3 | 140–330 | 4 |
| 24 | 111434 | 32 | Female | 620 | 505 | B1 | 160–180 | 1 |
| 25 | 111440 | 30 | Male | 173 | 434 | B3 | 110–120 | 1 |
| Median | 30 | 360 | 1112 | 10 |
Range of SFC/106 peripheral blood mononuclear cells (PBMC) in enzyme-linked immunospot (ELISPOT) assay (lowest and highest positive response obtained in each patient.
Number of positive peptides recognized after matrix analysis in ELISPOT assay. SFC: spot-forming cells.
Peripheral blood mononuclear cells (PBMC) isolation and CD4 and CD8 counts
Fresh blood was processed within 4 h of phlebotomy. PBMC were isolated from ethylenediamine tetraacetic acid (EDTA)-treated and heparinized blood on a Ficoll-Hypaque density gradient (Sigma, St Louis, MO, USA) and cryopreserved [90% fetal calf serum (FCS), 10% dimethylsulphoxide (DMSO)] at −196°C until use. Absolute CD4 and CD8 counts were determined on a FACSCalibur by the CellQuest software (Becton Dickinson, San José, CA, USA) using a set of criteria for quality control [20,21].
Synthetic HIV-1 peptides
A set of 125 overlapping peptides corresponding to the consensus Indian HIV-1 subtype-C gag (510 amino acids) sequence was synthesized commercially (Bio-Synthesis Incorporated, TX, USA). This sequence was based on the consensus of subtype C sequences and previously reported Indian clade C isolates from our laboratory (GenBank numbers AF533118–AF533141). Each peptide was 15 ± 1 amino acids (aa) in length, overlapping the next peptide by 11 aa. The lyophilized peptides were dissolved in DMSO at a concentration of 100 mg/ml (stock) and stored in aliquots at −70°C until use.
ELISPOT assays
Screening for CD4 and CD8 T cell responses was performed by IFN-γ ELISPOT using a strategy based on a matrix of peptide pools. The 125 peptides spanning the gag region of HIV-1 were divided into rows and columns such that 22 pools of 11 peptides each were formed, the last two pools having 15 peptides. Each peptide was included in two different pools; the positive responses to both the pools indicated a positive response to that specific peptide, which was then retested individually. Cryopreserved PBMCs were thawed rapidly and cultured overnight at 37°C, 5% CO2 atmosphere in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% FCS, 10 m M HEPES buffer, 2 mMl-glutamine and 50 U of penicillin-streptomycin. The 96-well polyvinylidene plates (Millipore, Bedford, MA, USA) were coated overnight with anti-IFN-γ antibody (R&D Systems, Minneapolis, MN, USA) at a concentration of 0·5 µg/ml in phosphate-buffered saline (PBS). The plates were washed and blocked as per manufacturer's instructions. PBMC were added at a concentration of 100 000 per well in 200 µl of RPMI medium. The gag peptide pools were plated in duplicates and the final concentration of each peptide in the pool was 2 µg/ml. Negative control wells (cells and medium only) were run in quadruplicate for each patient sample along with duplicate positive control wells (cells and phytohaemagglutinin at 2 µg/ml) to ensure that the cells were responsive. The plates were incubated for 18–20 h at 37°C and 5% CO2 and then washed six times with PBS 0·05% Tween 20. The plates were then incubated at room temperature for 2 h with the corresponding biotinylated secondary antibody (R&D Systems), followed by washes and a 1-h incubation with a streptavidin–alkaline phosphatase conjugate. After the plates were washed, 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium substrates (BCIP-NBT) were added for colour development. The number of spots per well was determined using an automated ELISPOT plate reader (Carl Zeiss Vision GmbH, Hallbergmoos, Germany). The results were expressed as spot-forming cells (SFC) per million PBMC. The negative response was < 30 SFC/106 PBMC in all cases. The response was considered positive if > 50 SFC/106 PBMC were detected and they were at least 3 standard deviations (s.d.) above background. Responses to two adjacent overlapping peptides were evaluated as responses to one epitopic region, because some T cell epitopes can be located in the overlapping region of two adjacent peptides, resulting in responses to both overlapping peptides. The matrix was analysed for each patient, and candidate positive peptides were retested individually by intracellular cytokine (ICS) detection.
ICS detection by flow cytometry
The positive peptides by IFN-γ ELISPOT assay were confirmed individually by ICS. Cryopreserved PBMCs were thawed and rested overnight at 37°C, 5% CO2 atmosphere in complete RPMI-1640/10% FCS medium. For each patient sample, 0·5–1 × 106 PBMCs were distributed in 96-well round-bottomed plates (Costar, Cambridge, MA, USA) and stimulated with anti-CD28 and anti-CD49d co-stimulatory antibodies (1 µg/ml each; BD Biosciences) and individual peptide (2 µg/ml). A negative control (cells and co-stimulatory antibodies) was included to assess spontaneous production of IFN-γ or interleukin (IL)-2. Positive controls (Staphylococcus enterotoxin B at 1 µg/ml, Sigma-Aldrich) and (CMV lysate at 1% stock solution; BioWhittaker, Walkersville, MD, USA) were included to ensure the cells were responsive. The cultures were incubated for 2 h at 37°C in a 5% CO2 incubator, followed by additional 4-h incubation in the presence of cytokine secretion inhibitor Brefeldin A (10 µg/ml; Sigma-Aldrich). The cells were placed at 4°C overnight and stained the next day.
The stimulated cells were treated with 2 mM EDTA for 15 min and with 1× FACS lysing solution (BD Biosciences) for 10 min at room temperature. The cells were washed once (400 × g for 5 min) with FACS buffer (PBS, 0·5% bovine serum albumin, 0·1% sodium azide) and permeabilized with 1× permeabilizing solution II (BD Biosciences). After 10 min incubation at room temperature, the cells were washed twice in FACS buffer (400 × g for 5 min) and stained for 30–45 min with a cocktail of fluorochrome conjugated antibodies specific for surface markers: CD3/CD4/CD8/CD69 and intracellular cytokines: IFN-γ and IL-2. The cells were washed and resuspended in PBS 1% paraformaldehyde. For each analysis 100 000 events were acquired on a FACSCalibur flow cytometer (Becton Dickinson). Data were analysed using FlowJo software (Tree Star, CA, USA). The lymphocyte gate (G1) was defined on forward scatter (FSC) and side scatter (SSC) for each of the stimulation in the sample. CD8+ and CD4+ T cells were defined by the second gate (G2) as the CD3+ CD8+ and CD3+ CD4+ subsets in the staining panels IFN-γ/CD69/CD8/CD3 and IL-2/CD69/CD4/CD3, respectively. Alternatively, the CD8+ and CD4+ T cells were defined as the CD3+ CD4− and CD3+ CD8− subsets in the IL-2/CD69/CD4/CD3 and IFN-γ/CD69/CD8/CD3 staining panels, respectively [22]. Comparative analysis of responding T cell populations showed that the responses were directly equivalent even if responses are measured in this manner (data not shown). The statistics of IFN-γ+ CD69+ and IL-2+ CD69+ subsets on the double gate (G3 = G1 + G2) were then considered to be the response frequency. The net percentage of cytokine positive cells within the CD4 and CD8 T cell compartments was determined by subtracting frequencies of cells stimulated with co-stimulatory antibodies without antigen (not exceeding 0·10%) from the statistics of IFN-γ+ CD69+ and IL-2+ CD69+ subsets of cells stimulated with antigen. In five HIV seronegative healthy controls, no IFN-γ and IL-2 responses to the Gag peptides were observed.
T cell depletions
The peptides confirmed to be positive for CD4 T cell responses by ICS were also confirmed by CD4+ T cell ELISPOT. Briefly, anti-CD8 monoclonal antibody (mAb)-coated magnetic beads (Miltenyi Biotech, Germany) were used to selectively deplete effector CTLs (CD8+ T cells) from PBMC, as described in the manufacturer's instructions. After negative depletion the supernatant, containing ≥ 98% CD4+ T cells, was washed twice with RPMI-1640/10% FCS and used directly in an ELISPOT assay as described above.
Results
Patients' profiles
The study group consisted of 25 chronically HIV-infected individuals with a median age of 30 years. The median CD4 counts were 360/µl (range: 89–970/µl) and CD8 counts were 1112/µl (range: 434–4099/µl) as detailed in Table 1.
HIV-specific T cell responses by ELISPOT assay using overlapping peptide mixtures
IFN-γ ELISPOT assays were performed using mixtures of 15-mer peptides in a matrix format such that each peptide was present in two pools. The pattern of positive wells in the matrix allowed the prediction of individual positive peptides. The total magnitude of positive responses against the entire Gag region of HIV-1 subtype C ranged from 60 to 2670 SFC/106 PBMC in the study group (Table 1). All the subjects recognized at least one peptide, and in people with more than one detectable response there was a broad range in the number of responses to individual peptides targeted (range: 1–46; median: 10). In order to characterize the function and phenotype of T cell responses at the single peptide level, the peptides positive by ELISPOT assay were retested and confirmed by intracellular cytokine staining.
Quantification of HIV-specific CD8+ T cell responses to Gag protein
The dominant target in terms of magnitude and breadth of responses was observed to be the p24 subunit of Gag protein (Table 2). Fifteen peptides located in p24-Gag were targeted by 64% of the subjects. Six peptides in p17-Gag and 7 peptides in p15-Gag were recognized by 28% and 36% individuals, respectively. Two patients recognized the peptide sequence Gag133–143: GKKVSQNYPIV at the p17–p24 junction region (Table 3). Analysis of intracellular IFN-γ and IL-2 production in all the patients demonstrates a high frequency of IFN-γ+ HIV-specific CD8+ T cells (median: 0·17; range: 0·11–1·49; Table 2,Fig. 1), but no IL-2-producing CD8+ T cell subset was observed in any of the patients.
Table 2.
.Cytokine production by HIV peptide-specific CD8+ T cells in chronic individuals.
| Patient no. | Laboratory ID | CDC stage | Peptide no. | Region of protein | Amino acid sequence | Parental protein | CD69+ IL-2+ (%) | CD69+ IFN-γ+ (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 111000 | A1 | 67–68 | Gag264–274 | PVGDIYKRWII | p24 | < 0·01 | 0·2 |
| 2 | 111181 | A1 | 89–90 | Gag347–357 | EEMMTACQGVG | p24 | < 0·01 | 0·18 |
| 3 | 111188 | A1 | 20–21 | Gag82–92 | SLFNTVATLY | p17 | < 0·01 | 0·3 |
| 22–23 | Gag92–103 | YCVHAGIEVRD | p17 | < 0·01 | 0·51 | |||
| 55–56 | Gag216–226 | AAEWDRLHPVH | p24 | < 0·01 | 0·56 | |||
| 109–110 | Gag426–436 | DCTERQANFLG | p15 | < 0·01 | 0·1 | |||
| 4 | 111231 | A1 | 79–80 | Gag308–318 | RAEQATQDVKN | p24 | < 0·01 | 0·41 |
| 5 | 111054 | A2 | 46–47 | Gag176–186 | ALSEGATPQDL | p24 | < 0·01 | 1·13 |
| 6 | 111118 | A2 | 51–52 | Gag195–205 | GGHQAAMQMLKD | p24 | < 0·01 | 0·11 |
| 7 | 111154 | A2 | 5, 6 | Gag25–35 | RLRPGGKKHYM | p17 | < 0·01 | 0·2 |
| 33–34 | Gag133–143 | GKKVSQNYPIV | p17–p24 | < 0·01 | 0·45 | |||
| 8 | 111190 | A2 | 20–21 | Gag82–92 | SLFNTVATLY | p17 | < 0·01 | 0·25 |
| 70–71 | Gag275–285 | LGLNKIVRMYS | p24 | < 0·01 | 0·1 | |||
| 9 | 111221 | A2 | 5, 6 | Gag25–35 | RLRPGGKKHYM | p17 | < 0·01 | 0·1 |
| 7, 8 | Gag33–43 | HYMLKHLVWAS | p17 | < 0·01 | 0·1 | |||
| 33–34 | Gag133–143 | GKKVSQNYPIV | p17–p24 | < 0·01 | 0·14 | |||
| 99–100 | Gag386–396 | SKRIVKCFNCG | p15 | < 0·01 | 0·15 | |||
| 10 | 111246 | A2 | 49–51 | Gag188–201 | NTMLNTVGGHQAAM | p24 | < 0·01 | 0·16 |
| 11 | 111288 | A2 | 43–44 | Gag171–181 | AFSPEVIPMFT | p24 | < 0·01 | 1·49 |
| 12 | 111296 | A2 | 124–125 | Gag486–496 | PLTSLKSLFGS | p15 | < 0·01 | 0·11 |
| 13 | 111053 | B1 | 22–23 | Gag93–103 | YCVHAGIEVRD | p17 | < 0·01 | 0·13 |
| 109–110 | Gag426–436 | DCTERQANFLG | p15 | < 0·01 | 0·19 | |||
| 124–125 | Gag486–496 | PLTSLKSLFGS | p15 | < 0·01 | 0·14 | |||
| 14 | 111102 | B1 | 21–22 | Gag87–97 | VATLYCVHAGI | p17 | < 0·01 | 0·3 |
| 45–46 | Gag178–188 | PMFTALSEGAT | p24 | < 0·01 | 0·17 | |||
| 15 | 111104 | B1 | 5, 6 | Gag25–35 | RLRPGGKKHYM | p17 | < 0·01 | 0·18 |
| 103–104 | Gag402–412 | IAKNCRAPRKK | p15 | < 0·01 | 0·14 | |||
| 16 | 111434 | B1 | 51–52 | Gag195–205 | GGHQAAMQMLK | p24 | < 0·01 | 0·24 |
| 17 | 111002 | B2 | 55–56 | Gag212–222 | AAEWDRLHPVH | p24 | < 0·01 | 0·64 |
| 87–88 | Gag343–353 | ALGPGASLEEM | p24 | < 0·01 | 0·6 | |||
| 18 | 111086 | B2 | 99–100 | Gag386–396 | SKRIVKCFNCG | p15 | < 0·01 | 0·14 |
| 109–110 | Gag426–436 | DCTERQANFLG | p15 | < 0·01 | 0·15 | |||
| 124–125 | Gag486–496 | PLTSLKSLFGS | p15 | < 0·01 | 0·11 | |||
| 19 | 111428 | B3 | 79–80 | Gag308–318 | RAEQATQDVKN | p24 | < 0·01 | 0·12 |
| 87–88 | Gag343–353 | ALGPGASLEEM | p24 | < 0·01 | 0·14 | |||
| 20 | 111440 | B3 | 1, 2 | Gag5–15 | ASILRGGKLDK | p17 | < 0·01 | 0·11 |
| 21 | 110976 | C2 | 56–57 | Gag217–227 | RLHPVHAGPIA | p24 | < 0·01 | 0·45 |
| 59–60 | Gag229–239 | GQMREPRGSDIA | p24 | < 0·01 | 0·2 | |||
| 110–111 | Gag429–439 | ERQANFLGKIW | p15 | < 0·01 | 0·4 | |||
| 22 | 111052 | C2 | 70–71 | Gag275–285 | LGLNKIVRMYS | p24 | < 0·01 | 0·15 |
| 23 | 110977 | C3 | 45–46 | Gag178–188 | PMFTALSEGAT | p24 | < 0·01 | 0·12 |
| 24 | 110983 | C3 | 59–60 | Gag229–239 | GQMREPRGSDIA | p24 | < 0·01 | 0·17 |
| 25 | 111292 | C3 | 56–57 | Gag217–227 | RLHPVHAGPIA | p24 | < 0·01 | 0·15 |
Table 3.
Summary of frequently recognized peptides in the study cohort.
| Peptide no. | Region of protein | Amino acid sequence | Subtype B epitopes in subtype C peptide sequence | Parental protein | Study subjects with response (%) |
|---|---|---|---|---|---|
| CD8+ T cell responses | |||||
| 5, 6 | Gag20–30 | RLRPGGKKHYM | WEKIRLRPGGKKKYK, KIRLPGGK, KIRLPGGKKKYKLIRLRPGGKK, RLRPGGKKKY | p17 | 12% |
| 20–23 | Gag78–96 | SLFNTVATLYCVHAGIEVRD | RSLYNTVATLY, SLYNTVATL, NTVATLYCV, TLYCVHQR, TLYCVHQRI, YCVHQRIEIKDTKEAL | p17 | 16% |
| 33–34 | Gag128–138 | GKKVSQNYPIV | QVSQNYPIV, NYPIVQNL | p17–p24 | 8% |
| 45–46 | Gag173–183 | PMFTALSEGAT | AFSPEVIPMFSALSEGATPQ, EVIPMFASALSEGATP, IPMFSALSEGATPQDL | p24 | 8% |
| 51–52 | Gag196–206 | GGHQAAMQMLKD | GHQAAMQMLKE | p24 | 8% |
| 55–57 | Gag212–227 | AAEWDRLHPVHAGPIA | LKETINEEAAEWDRVHPV, AEWDRVHPV, VHPVHAGPIA, HPVHAGPIA | p24 | 16% |
| 59–60 | Gag229–240 | GQMREPRGSDIA | HAGPIAPGQMREPRG, IAPGQMREPRGSDIAGTTST | p24 | 8% |
| 70–71 | Gag271–281 | LGLNKIVRMYS | IYKLWIILGLNKIVRMYSPT, KRWIILGLNKIVRMYSPTSI | p24 | 8% |
| 79–80 | Gag308–318 | RAEQATQDVKN | FKTLRAEQATQDVKNWMTDT | p24 | 8% |
| 87–88 | Gag339–349 | ALGPGASLEEM | KTILRALGPGATLEEMMTAC | p24 | 8% |
| 99–100 | Gag387–397 | SKRIVKCFNCG | SNFKGNKRMVKCFNCGKEGH | p15 | 8% |
| 109–111 | Gag426–439 | DCTERQANFLGKIW | KEGHQMKDCTERQANF, CTERQANFL, TERQANFL, RQANFLGKIWPSYKG, CTERQANFL, FLGKIWPS | p15 | 16% |
| 124–125 | Gag483–493 | PLTSLKSLFGS | KELYPLTSL, TPLSTLRSLF | p15 | 12% |
| CD4+ T cell responses | |||||
| 43–46 | Gag166–183 | AFSPEVIPMFTALSEGAT | AFSPEVIPMFSALSEC | p24 | 8% |
| 62–63 | Gag241–251 | GTTSTLQEQIA | GSDIAGTTSTLQEQIC | p24 | 8% |
| 64–66 | Gag248–262 | EQIAWMTSNPPVPVG | STLQEQIGWMTNNPPIPVGE | p24 | 8% |
Fig. 1.
Representation of CD8+ CD69+ interferon (IFN)-γ+ cells [20 000 events within the lymphocyte gate (G1)] in the study population following stimulation. (a) Unstimulated, (b) Staphylococcus enterotoxin B stimulation, (c) cytomegalovirus lysate stimulation and (d) Gag peptide stimulation.
Quantification of HIV-specific CD4+ T cell responses to Gag protein
The CD4+ T cell responses were observed in eight of the total 25 individuals (Table 4). The p24-Gag subunit again dominated the detectable responses, with all eight patients recognizing at least one peptide in the p24-Gag region and 24% of the study subjects responding to nine peptides in this subunit (Table 3). p17-Gag responses were observed in four of the patients, while only one patient had CD4+ T cell responses to p15-Gag region. HIV-specific CD4+ T cells in these eight patients produced IFN-γ (median: 0·23; range: 0·11–1·34; Table 4, Fig. 2) when stimulated with individual gag peptides. However, low IL-2 frequencies were observed in only three patients (median: 0·13; range: 0·08–0·38; Table 4, Fig. 3) suggesting the inability of viraemic, chronic HIV-infected patients to produce IL-2.
Table 4.
Cytokine production by HIV peptide-specific CD4+ T cells in chronic individuals.
| Patient no. | Laboratory ID | CDC stage | Peptide no. | Region of protein | Amino acid sequence | Parental protein | CD69+ IL-2+ (%) | CD69+ IFN-γ+ (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 111181 | A1 | 13–14 | Gag53–63 | PGLLETSEGCK | p17 | < 0·01 | 0·1 |
| 62–63 | Gag241–251 | GTTSTLQEQIA | p24 | < 0·01 | 0·12 | |||
| 2 | 111154 | A2 | 5, 6 | Gag25–35 | RLRPGGKKHYM | p17 | 0·14 | 0·21 |
| 59–60 | Gag229–239 | GQMREPRGSDI | p24 | < 0·01 | 0·14 | |||
| 3 | 111221 | A2 | 7, 8 | Gag32–43 | HYMLKHLVWAS | p17 | < 0·01 | 0·23 |
| 83–85 | Gag324–338 | LLVQNANPDCKTILR | p24 | < 0·01 | 0·3 | |||
| 4 | 111246 | A2 | 49–51 | Gag188–201 | NTMLNTVGGHQAAM | p24 | < 0·01 | 0·3 |
| 5 | 111288 | A2 | 43–44 | Gag171–181 | AFSPEVIPMFT | p24 | 0·38 | 0·14 |
| 6 | 111002 | B2 | 64–66 | Gag248–262 | EQIAWMTSNPPVPVG | p24 | < 0·01 | 0·12 |
| 76–77 | Gag296–306 | FRDYVDRFFKT | p24 | < 0·01 | "0·11 | |||
| 7 | 111086 | B2 | 1, 2 | Gag5–15 | ASILRGGKLDKW | p17 | < 0·01 | 0·15 |
| 21–22 | Gag87–97 | VATLYCVHAGI | p17 | 0·13 | 0·26 | |||
| 45–46 | Gag178–188 | PMFTALSEGAT | p24 | < 0·01 | 0·27 | |||
| 54–55 | Gag208–218 | INEEAAEWDRL | p24 | < 0·01 | 0·25 | |||
| 56–57 | Gag217–227 | RLHPVHAGPIA | p24 | < 0·01 | 0·3 | |||
| 65–66 | Gag251–261 | WMTSNPPVPVG | p24 | < 0·01 | 0·35 | |||
| 100–101 | Gag390–400 | VKCFNCGKGEH | p15 | < 0·01 | 0·36 | |||
| 109–110 | Gag426–436 | DCTERQANFLG | p15 | < 0·01 | 1·34 | |||
| 120–121 | Gag466–476 | FEETTPAPPKQ | p15 | 0·08 | 0·25 | |||
| 123–124 | Gag479–489 | KDREPLTSLKS | p15 | < 0·01 | 0·2 | |||
| 8 | 110977 | C3 | 62–63 | Gag241–251 | GTTSTLQEQIA | p24 | < 0·01 | 0·12 |
Fig. 2.
Representation of CD4+ CD69+ interferon (IFN)-γ+ cells [20 000 events within the lymphocyte gate (G1)] in the study population following stimulation. (a) Unstimulated, (b) Staphylococcus enterotoxin B stimulation, (c) cytomegalovirus lysate stimulation and (d) Gag peptide stimulation.
Fig. 3.
Representation of CD4+ CD69+ interleukin (IL)-2+ cells [20 000 events within the lymphocyte gate (G1)] in the study population following stimulation. (a) Unstimulated, (b) Staphylococcus enterotoxin B stimulation, (c) cytomegalovirus lysate stimulation and (d) Gag peptide stimulation.
Discussion
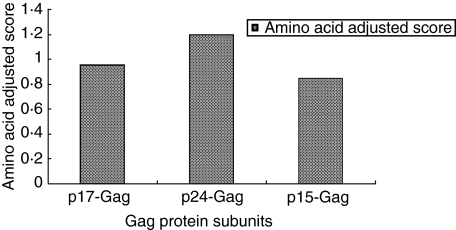
All the individuals studied responded to at least one peptide of Gag protein. The distribution of the individual peptide responses and their frequency of recognition in the total study population of 25 HIV-1 infected individuals across the entire Gag protein is summarized in Fig. 4. The p24-Gag was one of the most frequently recognized subunit proteins and the greatest magnitude of CD4+ and CD8+ T cell responses were directed to this protein when adjusted for the amino acid-adjusted (AA) score (Fig. 5). The AA score is defined as the frequency of recognition of the individual protein divided by the protein length in amino acids [6,19]. The reasons for this high frequency and magnitude of response are due probably to amino acid sequence conservation among this protein subunit. The present study identified several peptide sequences that were recognized as CD8+ and CD4+ T cell responses in more than one study subject (Table 3). One of them overlaps or contains the CD4+ T cell epitope, p24-Gag171–188 (AFSPEVIPMFTALSEGAT). All these confirmed subtype C peptide responses were matched with previously described epitope sequences for Gag derived from subtype B (obtained from hiv-web.lanl.gov). Although some of the subtype B epitope sequences were variant from the subtype C peptide sequences (Table 3), it was apparent that many of the responses targeted were in highly conserved regions of the Gag gene.
Fig. 4.
Frequency of recognition of the individual Gag peptides by the study subjects.
Fig. 5.
Amino acid-adjusted scores for Gag protein subunit recognition.
We observed stronger CD8 responses in terms of the magnitude and number of peptides targeted, as well as broader targeting by CD8 T cells with regard to Gag protein subunits recognized, contrasting with the weaker and less numerous CD4 T cell responses, which were more narrowly directed. Sequence variation of the autologous viral sequence compared to the consensus sequence used for the synthetic peptides may contribute to the pattern of immunodominance of CD8 responses compared to CD4 responses [9]. The variable recognition of HIV-1 peptides by CD4 T cells might also be affected by a differential ability of HLA class II molecules to bind peptides in multiple registers, thus the core region engages the binding grove with additional flanking residues, which is not usually an optimal ligand for T cell responses [9,23]. A smaller burst size than the CD8 response and the disease stage may also influence the magnitude and specificity of the CD4 response [6,22,24,25]. However, we were still able to detect HIV-specific CD4+ T cell responses to Gag protein in IFN-γ ELISPOT and intracellular cytokine staining assays, suggesting that these responses persist at low precursor frequencies, undetectable during acute HIV-1 infection, but can be detected later during chronic HIV-1 infection, as in our study population [26].
The importance of CD4 T cell help for the adequate generation of efficient, protective memory CD8 T cell responses has been documented recently. The loss of CD4 T cells during chronic HIV-1 infection adversely affects the function of antigen-specific CTLs [2,9–16]. In our study, the magnitude of the HIV-specific IFN-γ response was observed to be generally higher in both CD8+ and CD4+ T cell subsets than the corresponding IL-2 response, that was detectable only in CD4+ T cells. These data are consistent with those who have used global stimulation methods to show that progression to disease is associated with the loss of HIV-specific CD4+ T cells that secrete IL-2 but not IFN-γ+ IL-2− CD4+ T cells that secrete only IFN-γ in the absence of IL-2, after stimulation with HIV peptides as detected by ICS [2,22,27–29]. Thus, the effector Th1 cells defined by IFN-γ secretion do not develop into long-term memory cells as efficiently as CD4 memory cells defined by IL-2 secretion, that includes both central memory cells and a subset of effectors that secrete both IL-2 and IFN-γ[30,31]. This may also explain that the patients with both detectable IL-2 and IFN-γ CD4+ T cell responses are able to contain viraemia and belong to CDC stages A2 and B2 (Table 4). However, patients with high IFN-γ CD8+ T cell responses may still belong to the CDC C group, thus progressing towards the later stages of disease. This may be due to the probable dysfunction of HIV-specific, IFN-γ-secreting CD4+ T cells in viraemic patients that lack IL-2 production, thus limiting their ability to express T cell help functions required for establishment and/or maintenance of memory CD4 T cells and CD8 T cell-mediated responses (Table 2). A substantial proportion of these HIV-1-specific CD8+ T cells is also susceptible to apoptosis and, hence, may not manifest effective cytolytic activity [32].
Thus, our results are novel as we have attempted to quantify the total HIV-specific CD4+ and CD8+ T cell responses to the possible epitopes in HIV-1 subtype C Gag protein in chronically infected untreated individuals. The detection of broader HIV-1-specific T cell responses of higher magnitude reflects the higher response to increasing antigenic load during chronic infection [29]. The identification of these responses at a single peptide level across the HIV-1 subtype C-infected Indian population and correlation with data from the Caucasian population may provide useful insight for the design of new immunotherapies and vaccines for the effective control of HIV-1 infection.
References
- 1.World Health Organization. 3. Geneva: World Health Organization; 2004. HIV/AIDS: meeting the challenge. [Google Scholar]
- 2.Lichterfeld M, Kaufmann DE, Yu XG, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med. 2004;200:701–12. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary HIV-1 infection. J Virol. 1994;68:6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 5.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Addo MM, Yu XG, Rathod A, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betts MR, Ambrozak DR, Douek DC, et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;73:11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draenert R, Verrill CL, Tang Y, et al. Persistent recognition of autologous-virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J Virol. 2004;78:630–41. doi: 10.1128/JVI.78.2.630-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufmann DE, Bailey PM, Sidney J, et al. Comprehensive analysis of HIV-1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78:4463–77. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNeil AC, Shupert WL, Iyasure CA, et al. High-level HIV-1 viremia suppresses viral antigen-specific CD4+ T cell proliferation. Proc Natl Acad Sci USA. 2001;98:13878–83. doi: 10.1073/pnas.251539598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer BE, Boritz E, Blyveis N, Wilson CC. Discordance between frequency of HIV-1-specific gamma interferon-producing CD4+ T cells and HIV-1-specific lymphoproliferation in HIV-1 infected subjects with active viral replication. J Virol. 2002;76:5925–36. doi: 10.1128/JVI.76.12.5925-5936.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg ES, Billingsley JM, Caliendo AM, et al. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–50. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 13.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–9. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 14.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–42. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen EM, Lemmens EE, Wolfe T, et al. CD4+ Tcells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–6. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 16.Kaech SM, Ahmed R. CD8 T cells remember with a little help. Science. 2003;300:263–5. doi: 10.1126/science.1084511. [DOI] [PubMed] [Google Scholar]
- 17.Novitsky V, Rybak N, McLane MF, et al. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific ELISPOT-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J Virol. 2001;73:9210–28. doi: 10.1128/JVI.75.19.9210-9228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masemola A, Mashishi T, Khoury G, et al. Hierarchical targeting of Subtype C HIV-1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78:3233–43. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan M, Goepfert PA. Magnitude of functional CD8+ T cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol. 2002;76:2298–305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandy F, Nicholson J, McDougal J. Revised guidelines for performing single-platform absolute CD4+ t-cell determinations with CD45 gating for persons infected with human immunodeficiency virus. MMWR. 2003;52:1–13. [PubMed] [Google Scholar]
- 21.Vithayasai V, Sirisanthana T, Sakonwasun C, et al. Flow cytometric analysis of T lymphocyte subsets in adult Thais. Asian Pacific J Allergy Immunol. 1997;15:141–6. [PubMed] [Google Scholar]
- 22.Boaz MJ, Waters A, Murad S, et al. CD4 responses to conserved HIV-1 T helper epitopes show both negative and positive associations with virus load in chronically infected subjects. Clin Exp Immunol. 2003;134:454–63. doi: 10.1111/j.1365-2249.2003.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bitmansour AD, Douek DC, Maino VC, Picker LJ. Direct ex vivo analysis of human CD4+ memory T cell activation requirements at the single clonotype level. J Immunol. 2002;169:1207–18. doi: 10.4049/jimmunol.169.3.1207. [DOI] [PubMed] [Google Scholar]
- 24.Ahmadzadeh M, Farber DL. Functional plasticity of an antigen-specific memeory CD4 T cell population. Proc Natl Acad Sci USA. 2002;99:11802–7. doi: 10.1073/pnas.192263099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goulder PJ, Altfeld MA, Rosenberg ES, et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–94. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boritz E, Palmer BE, Wilson CC. Human immunodeficiency virus type (HIV-1)-specific CD4+ T cells that proliferate in vitro detected in samples from most viremic subjects and inversely associated with plasma HIV-1 levels. J Virol. 2004;78:12638–46. doi: 10.1128/JVI.78.22.12638-12646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Younes SA, Yassine-Diab B, Dumont AR, et al. HIV-1 viremia prevents the establishment of interleukin 2- producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–22. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boaz MJ, Waters A, Murad S, et al. Presence of HIV-1 Gag-specific IFN-γ+ IL-2+ and CD28+ IL-2+ Cd4 T cell responses is associated with nonprogression in HIV-1 infection. J Immunol. 2002;169:6376–85. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- 29.The Newsletter on International AIDS. Vaccine research. p. 1. IAVI Report December 2004–March 2005.
- 30.Wu CY, Kirman JR, Rotte MJ, et al. Distinct lineages of Th 1 cells have differential capacities for memory cell generation in vivo. Nat Immunol. 2002;3:852–8. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- 31.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 32.Chun TW, Justement JS, Sanford C, et al. Relationship between the frequency of HIV-specific CD8+ T cells and the level of CD38+CD8+ T cells in untreated HIV-infected individuals. Proc Natl Acad Sci USA. 2004;101:2464–9. doi: 10.1073/pnas.0307328101. [DOI] [PMC free article] [PubMed] [Google Scholar]