Abstract
Paramyosin, a Schistosoma mansoni myoprotein associated with human resistance to infection and reinfection, is a candidate antigen to compose a subunit vaccine against schistosomiasis. In this study, 11 paramyosin peptides selected by TEPITOPE algorithm as promiscuous epitopes were produced synthetically and tested in proliferation and in vitro human leucocyte antigen (HLA)-DR binding assays. A differential proliferative response was observed in individuals resistant to reinfection compared to individuals susceptible to reinfection in response to Para (210–226) peptide stimulation. In addition, this peptide was able to bind to all HLA-DR molecules tested in HLA-DR binding assays, confirming its promiscuity. Para (6–22) and Para (355–371) were also shown to be promiscuous peptides, because they were able to bind to the six and eight most prevalent HLA-DR alleles used in HLA-DR binding assays, respectively, and were also recognized by T cells of the individuals studied. These results suggest that these paramyosin peptides are promising antigens to compose an anti-schistosomiasis vaccine.
Keywords: paramyosin, Schistosoma mansoni, synthetic peptides, T cells, vaccine
Introduction
Schistosomiasis is a chronic disease endemic in 76 countries worldwide, causing an estimated 200 000 deaths/year [1]. Mass chemotherapy is the control strategy used nowadays; however, this control strategy has some drawbacks, such as a high frequency of reinfection and the difficulties and costs involving the long-lasting maintenance of these programmes [2]. Parasite drug resistance is also a concern that has to be considered [3,4]. In this context, the development of a defined anti-schistosomiasis vaccine would contribute enormously to the current therapies mainly because immunization provides long-lasting immunity to the disease. Several studies are in progress testing different antigens of the parasite and several vaccination strategies in experimental immunization protocols [5–7].
Six schistosome antigens have been selected by the World Health Organization (WHO) as candidates to compose a subunit vaccine against schistosomiasis [5]. One of these antigens is paramyosin, a filamentous, α-helical, coiled-coil protein of approximately 100 kDa that is present in the muscle of invertebrates, including Schistosoma mansoni [5,8]. Paramyosin was identified as the major antigen recognized by sera of mice vaccinated intradermally with an S. mansoni adult parasite extract [9]. Purified paramyosin used in mice immunization conferred significant resistance against challenge infection [8,10]. The resistance induced by immunization involved interferon (IFN)-γ production by stimulated T cells, suggesting that during challenge of vaccinated hosts paramyosin may elicit a protective T cell response as a result of its release from migrating parasite larvae [8]. It thus appears that both cellular and humoral immune responses are important mechanisms in the protective immunity induced by paramyosin immunization.
Studies of human immune responses to paramyosin support the potential of this protein as a vaccine candidate. S. mansoni paramyosin is strongly recognized by sera of individuals naturally resistant to infection and it also induced high levels of IFN-γ production by peripheral blood mononuclear cells (PBMC) from these individuals [11,12]. Paramyosin also induced significant lymphocyte proliferation and interleukin (IL-2) and IL-5 production in individuals resistant to S. mansoni reinfection [13]. These results suggest the involvement of paramyosin in natural resistance and in resistance to reinfection against Schistosoma mansoni.
The identification of immunodominant epitopes within vaccine candidate antigens is extremely important, not only because it makes immunization possible, excluding cross-reactivity and the accompanying risk of autoimmunity or allergy, but also because it allows the construction of vaccines with relevant peptides from different candidate antigens, improving the chance of achieving high levels of protection.
Here we report the T cell proliferative responses by PBMC from individuals living in endemic areas for schistosomiasis in response to paramyosin epitopes selected by TEPITOPE algorithm. Eleven paramyosin peptides were synthesized and presented as antigens to PBMC of individuals infected with S. mansoni (INF), naturally resistant to S. mansoni infection (NR) resistant (RR) and susceptible (SR) to S. mansoni reinfection after treatment. We observed differential T cell proliferation in response to paramyosin epitopes according to individual status of resistance and susceptibility to S. mansoni reinfection. Among the paramyosin synthetic peptides tested in this study, Para (210–226) was the only epitope recognized preferentially by T cells of individuals resistant to S. mansoni reinfection when compared to the response of cells from infected patients. Further, this peptide, together with Para (6–22) and Para (355–371), were shown to be promiscuous peptides as predicted by TEPITOPE and proliferation assays. Additionally, in vitro human leucocyte antigen (HLA)-DR binding assays showed that Para (6–22), Para (210–226) and Para (355–371) bound to the majority of HLA-DR molecules used in this study. Therefore, the results presented here suggest that these peptides are promising candidates to compose an anti-schistosomiasis vaccine.
Materials and methods
Study population
Heparinized peripheral blood was obtained from individuals with different genetic backgrounds living in three different areas endemic for schistosomiasis (Melquiades, Côrrego do Onça and Caatinga do Moura). The individuals included in this study reflect the racial distribution profile of Brazilian population according to data from the official Brazilian Census (http://www.ibge.gov.br). These individuals were classified into five groups with regard to their infection status, and the selection of subjects was performed based only on the criteria for inclusion and exclusion of each group independently of previous knowledge of immune responses for each individual. Non-infected (NI) individuals are healthy people without any parasite infection or contaminated water contact. Some individuals, despite contact with contaminated water, showed repeated stool-negative examination and no clinical signs of disease for at least 3 consecutive years and were classified as naturally resistant to S. mansoni infection (NR). The water contact exposure was evaluated objectively by observers and the population studied had at least one contact daily. Another group showed stool-negative examination after treatment and was classified as resistant to S. mansoni reinfection (RR). Individuals classified as susceptible to S. mansoni reinfection (SR) showed stool-positive examination following treatment, and individuals grouped as infected individuals (INF) showed stool-positive examination and no treatment history. In spite of having similar water contact levels, these subjects exhibited a high variability in infection levels before and after treatment with praziquantel (40 mg/kg). These patients or their legal guardians gave informed consent after explanation of the protocol that had been approved previously by the Ethical Committee of the Federal University of Minas Gerais. Details regarding sex and age of the individuals included in this study are described in Table 1.
Table 1. Study population.
| Groups | No. of subjects | Age (mean ± s.d.) | Age range | Sex (male/female) |
|---|---|---|---|---|
| NI | 8 | 32·14 ± 12·46 | 21–59 | 3/5 |
| INF | 16 | 27·61 ± 17·46 | 9–58 | 6/10 |
| RR | 23 | 27·21 ± 21·04 | 10–71 | 10/13 |
| SR | 17 | 25·68 ± 16·64 | 9–69 | 7/10 |
| NR | 19 | 49·36 ± 16·35 | 11–73 | 9/10 |
| All | 83 | 32·4 ± 16·79 | 9–73 | 35/48 |
NI: non-infected individuals; INF: infected individuals; RR: individuals resistant to Schistosoma mansoni reinfection; SR: individuals susceptible to S. mansoni reinfection; NR: naturally resistant individuals.
Antigen preparation
SWAP (soluble adult worm antigen preparation) was prepared as described previously [14]. Adult worms were obtained from Swiss outbred mice following 8 weeks of infection. The soluble fraction was obtained after homogenization of worms in a Virtiss homogenizer, and centrifuged at 4000 g for 1 h at 4°C. The supernatant was collected and protein concentration was determined according to Bradford's method [15].
Synthetic peptides
The amino acid sequence of S. mansoni paramyosin (GenBank Accession no. M35499) was scanned using the TEPITOPE algorithm (Vaccinome, http://www.vaccinome.com) that can predict, after scanning all 9 mer windows starting with a hydrophobic residue on a protein sequence, those that have the potential ability to bind one or more of 25 different HLA-DR molecules, by use of 25 virtual matrices that cover most of the HLA-DR specificities in the Caucasian population [16]. We synthesized peptides that correspond to an inner nonamer core selected by TEPITOPE as the HLA-binding motif with flanking amino acids added at both N- and C- teminal ends, to increase the efficiency of in vitro peptide presentation to CD4+ T cells. The complete sequences of the synthetic peptides are shown in Table 2. Eleven 17-mer synthetic peptides containing the paramyosin epitopes selected were purchased from Mimotope (CA, USA). A peptide sequence identity search to other myosin peptides was performed using the computer program PepPepSearch (http://cbrg.inf.ethz.ch/Server/PepPepSearch.html).
Table 2. Peptides from Schistosoma mansoni paramyosin selected by TEPITOPE algorithm.
| Peptide (amino acid position) | Amino acid sequence* | % HLA able to bind peptides according to TEPITOPE prediction | % individuals able to respond to peptide stimulation within the study population |
|---|---|---|---|
| Para (6–22) | TESHVKISRTIYRGVSP | 21 | 20 |
| Para (13–29) | SRTIYRGVSPSTTRLES | 13 | 11·6 |
| Para (151–167) | LDGALKAKQSAESKLEG | 8 | 16·5 |
| Para (168–184) | LDSQLNRLKSLTDDLQR | 17 | 12·6 |
| Para (210–226) | YEAQILNYSKAKSSLES | 21 | 18·5 |
| Para (355–371) | NSELIRRAKAAESLASD | 26 | 14·6 |
| Para (399–415) | RLKSLVNDLTDKNNLLE | 13 | 13·8 |
| Para (584–600) | AENNLQITEHKRLQLAN | 4 | 16 |
| Para (676–692) | NNEVLRLADELRQEQGN | 17 | 19·3 |
| Para (760–776) | FERQYKELQTQAEDDRR | 4 | 8·76 |
| Para (830–846) | AERTVTVRRVGPGGRAV | 13 | 16·5 |
Underlined amino acids correspond to the nonamer peptide predicted by TEPITOPE to bind different human leukocyte antigen (HLA)-DR.
HLA-DR typing
DNA was extracted from blood samples using the Wizard genomic DNA purification kit (Promega Corp., Madison, WI, USA) according to the manufacturer's instructions. Genomic DNA was quantified on agarose gel and by spectrophotometry. HLA-DRB1 (DR1, DR4, DR7, DR8, DR9, DR10, DR11, DR12, DR13, DR14, DR15, DR16, DR17 and DR18) typing was performed using a low-resolution sequence specific primer amplification as described previously [17].
HLA-DR peptide-binding assay
HLA class II molecules were purified from Epstein–Barr virus-transformed homozygous B lymphoblastoid cell lines or transfected fibroblasts by affinity chromatography, as described previously [18]. Peptide binding assays were performed by incubating purified human class II molecules (5–500 n M) with various concentrations of unlabelled peptide and 1–10 n M125I-radiolabelled probe peptide for 48 h in phosphate buffered saline (PBS) containing 0·05% to 0·15% Nonidet P-40 in the presence of a protease inhibitor cocktail [18,19]. Radiolabelled peptides were iodinated using the chloramine-T method [20]. Major histocompatibility complex (MHC) class II peptide complexes were separated from free peptide by gel filtration on TSK200 columns (Tosoh Biosciences LLC, Montgomeryville, PA, USA) and the fraction of bound peptide calculated [21]. Alternatively, the percentage of radioactivity from peptides bound to MHC was determined by capturing MHC/peptide complexes on LB3·1 antibody coated Lumitrac 600 plates (Greiner Bio-one, Fickenhousen, Germany), and determining bound counts per minute (CPM) using the Top-Count (Packard Instrument Co. Meriden, CT, USA) microscintillation counter. The radiolabelled probe peptides utilized were HÁ Y307-319 (sequence YPKYVKQNTLKLAT; DRB1*0101), an analogue of TT Y828-843 (sequence YATFFIKANSKTE; DRB5*0101, DRB1*1101, DRB1*0701), MBP Y85-100 (sequence PVVHFFKNIVTPRTPPY; DRB1*1501, DRB1*0401, DRB1*0405), and an analogue of TT830-843 (sequence QYIKANAKTE; DRB1*1302) as described previously [19]. Previous experiments have shown that peptides showing IC50 percentage values below 1000 were recognized much more frequently by sensitized individuals bearing that HLA allele than peptides showing higher values [22].
Cellular immune response to paramyosin synthetic peptides
PBMC were isolated from peripheral blood by Ficoll-Hypaque density gradient centrifugation (d = 1·077), washed three times with sterile saline and incubated in RPMI-1640 medium (Gibco, St. Louis, MO, USA) supplemented with 10% normal human AB serum (Sigma, Carlsbad, CA, USA), 100 U/ml penicillin G sodium, 100 µg/ml streptomycin sulphate and 250 ng/ml amphotericin B. In the proliferation assay, cell cultures from RR, SR, INF, NR and NI individuals were carried out in triplicate in 96-well flat-bottomed culture plates (105 cells/well; final volume 0·2 ml) with SWAP (25 µg/ml) and paramyosin peptides (10 µ M). Phytohaemaglutinim (PHA) (2 µg/ml) was used as positive control and unstimulated culture was used as negative control. The antigens used were titred to determine optimal concentrations for proliferation assay. Plates were incubated in 5% CO2 at 37°C for 5 days and cultures were pulsed with 0·5 µCi/well [3H]-thymidine (Amersham, Sydney, Australia) for 18 h. Cells were collected in glass fibre filters by Filtermate Harvester (Packard, IL, USA). [3H]-thymidine incorporation was measured at the Beta Counter apparatus (Packard).
Statistical analysis
The Mann–Whitney rank sum test was used to compare continuous variables in the proliferation assay. Fisher's χ2 test was used to compare frequency values of HLA-DR distribution.
Results
Peptides selection
The entire sequence of S. mansoni paramyosin was scanned by TEPITOPE algorithm at 3% threshold, which led to the identification of regions predicted to bind promiscuously with high affinity to several different HLA-DR molecules. To each of the 9-mer windows along the scanned sequence, the algorithm provided an HLA binding score for each of the 25 HLA-DR molecules analysed. Nonamers attaining a score above threshold (e.g. 3% of the best binding peptides for high affinity) for a given HLA-DR molecule were selected and are indicated in Table 2. The algorithm also allows the selection of sequences predicted to bind simultaneously, thus promiscuously, to several HLA-DR molecules. Peptides predicted to bind to the largest number of HLA-DR molecules at the 3% threshold within paramyosin were selected to be used in this study. TEPITOPE scanning of paramyosin resulted in the selection of 11 paramyosin epitopes (Table 2) that were synthesized by Mimotope (San Diego, CA, USA) and used as antigens in the in vitro assays. Paramyosin epitopes selected did not show identity with human peptides; however, all peptides presented high levels of identity to S. japonicum and Taenia solium myoprotein peptides (ranging from 82·4% to 100% and 58·8% to 94·1%, respectively) (data not shown). According to TEPIPTOPE analysis, Para (355–371) was predicted to be the most promiscuous peptide able to bind to 26% of the HLA-DR molecules tested (Table 2).
Cellular immune response to paramyosin synthetic peptides
To test the proliferative response of individuals living in areas endemic for schistosomiasis, the PBMC of these individuals were stimulated with paramyosin peptides or SWAP. To evaluate the proliferative response to antigen stimulation, a ‘cut-off’ was determined by the mean proliferation index of PBMC from non-infected (NI) individuals stimulated with paramyosin peptides plus two standard deviations. The proliferation index was calculated for each individuals by the formula: mean cpm of stimulated cells/mean cpm of non-stimulated cells. Therefore, individuals with a proliferation index (PI) equal to or higher than 2·0 were considered individual responders to antigen stimulation. NI individuals showed a proliferation index of ≤2·0 and were considered non-responders. PBMC of 14% of individuals in this study did not proliferate in response to paramyosin peptides (data not shown). Para (6–22) was the most promiscuous peptide according to the proliferative responses. PBMC from 20% of the individuals tested proliferated in response to this epitope (Table 2). With regard to the proliferative response to SWAP, 89% of PBMC from the individuals tested in this study proliferated in response to SWAP stimulation (data not shown).
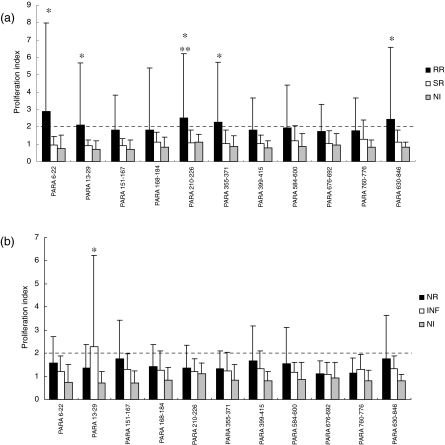
In order to identify peptides that could be recognized by resistant versus susceptible individuals, we compared the proliferation index of individuals who responded to paramyosin epitopes in the NR group to INF individuals and RR patients compared to the SR group. Before praziquantel treatment, a lower proliferation index was observed in response to paramyosin peptides in both resistant and susceptible individuals (Fig. 1b). Only Para (13–29) was recognized by INF patients when compared to NI individuals above the ‘cut-off’ level. Additionally, no statistically significant difference in the proliferation index was observed between the INF and NR groups (Fig. 1b). After praziquantel treatment an improvement in the proliferation index was observed in individuals resistant to S. mansoni reinfection. Para (6–22), Para (13–29), Para (210–226), Para (355–371) and Para (830–846) induced a significant proliferation index in RR individuals compared to NI individuals (PI > 2·0); however, only Para (210–226) showed significant T cell activation in the RR group compared to the SR group (Fig. 1a). A statistically significant difference in T cell proliferation in response to SWAP was observed in the RR (154 ± 87) group compared to the SR group (101·7 ± 127); however, no statistically significant difference was observed in the proliferation index by both RR (120 ± 96) and SR (195 ± 93·3) PBMC stimulated with PHA, a polyclonal stimulus. With regard to NR and INF groups, no statistically significant difference was observed between these groups even when PBMC of these individuals were stimulated with SWAP (NR −135 ± 81 and INF −46·5 ± 54·7) or PHA (NR −124 ± 135 and INF −41 ± 41).
Fig. 1.
Proliferative response to paramyosin peptides by peripheral blood mononuclear cells (PBMC) of individuals resistant to Schistosoma mansoni reinfection (RR) and susceptible to reinfection (SR) (a), and of individuals naturally resistant to S. mansoni infection (NR) and infected with the parasite (INF) (b). Results are expressed as mean of proliferation index plus standard deviation. Statistically significant differences determined by Mann–Whitney test analysis (P < 0·05) among all tested groups compared to non-infected individuals are denoted by an asterisk, and between RR versus SR are denoted by two asterisks. Dashed lines represent the cut-off.
Paramyosin peptide binding to MHC class II molecules
An MHC class II/paramyosin peptide binding assay was performed with the eight most prevalent HLA-DR molecules in the population studied. This assay showed that Para (13–29), Para (210–226) and Para (355–371) were able to bind to 100% (8/8) of the HLA-DR molecules used in this assay, followed by Para (6–22) that bound to 75% (6/8) of them (Table 3). The other peptides studied bound to less than six of the HLA-DR molecules analysed.
Table 3. Human leucocyte antigen (HLA)-DR binding capacity of each paramyosin peptide selected by TEPITOPE.
| Binding capacity (IC 50 nM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Position | DR1 DRB1*0101 | DR4w4 DRB1*0401 | DR4w15 DRB1*0405 | DR7 DRB1*0701 | DR5w11 DRB1*1101 | DR6w19 DRB1*1302 | DR2w2 B1 DRB1*1501 | DRw2B2 DRB5*0101 | Molecules bound |
| TESHVKISRTIYRGVSP | 6–22 | 870 | 3739 | 2399 | 288 | 198 | 457 | 387 | 28 | 6 |
| SRTIYRGVSPSTTRLES | 13–29 | 7·5 | 437 | 86 | 53 | 737 | 596 | 334 | 43 | 8 |
| LDGALKAKQSAESKLEG | 151–167 | 1406 | 6055 | 22267 | 21959 | 3039 | 23034 | 884 | 412 | 2 |
| LDSQLNRLKSLTDDLQR | 168–184 | 1280 | 34 | 14 | 8824 | 1985 | 9529 | 5599 | 314 | 3 |
| YEAQILNYSKAKSSLES | 210–226 | 49 | 861 | 12 | 10 | 23 | 255 | 42 | 28 | 8 |
| NSELIRRAKAAESLASD | 355–371 | 4·4 | 11 | 820 | 377 | 75 | 279 | 151 | 19 | 8 |
| RLKSLVNDLTDKNNLLE | 399–415 | 294 | 784 | 2630 | – | – | 84 | – | – | 3 |
| AENNLQITEHKRLQLAN | 584–600 | 1780 | – | – | 30 | 43 | 3721 | 59 | 18110 | 3 |
| NNEVLRLADELRQEQGN | 676–692 | 2804 | 7690 | 24506 | – | 4940 | – | 42159 | 1079 | 0 |
| FERQYKELQTQAEDDRR | 760–776 | 7·4 | 305 | 16 | 22380 | 89 | – | 5401 | 3098 | 4 |
| AERTVTVRRVGPGGRAV | 830–846 | 7·7 | – | – | 7855 | 9153 | – | 27 | 2·4 | 3 |
Significant affinity threshold < 1000, shown in bold type. Dash (–) means IC50% > 30 000.
HLA-DR distribution in the population studied
To evaluate the distribution of HLA-DR allele groups in the population studied, genomic DNA was obtained from blood samples and HLA-DR allele groups identified by low-resolution sequence-specific primer amplification. The frequency of HLA-DR allele groups in the study population is shown in Table 4. No predominance of a particular HLA-DR allele expression was observed within this population, not even when the frequency of HLA-DR expression was evaluated separately by each patient group (NR, INF, RR or SR) Table 4.
Table 4. Human leucocyte antigen (HLA)-DR distribution in the study population.
| HLA-DR frequency (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groups | HLA-DR1 | HLA-DR4 | HLA-DR7 | HLA-DR8 | HLA-DR9 | HLA-DR10 | HLA-DR11 | HLA-DR12 | HLA-DR13 | HLA-DR14 | HLA-DR15 | HLA-DR16 | HLA-DR17 | HLA-DR18 |
| NR | 11 | 22 | 11 | 0 | 11 | 11 | 22 | 0 | 22 | 22 | 44 | 22 | 11 | 0 |
| INF | 57·1 | 28·5 | 0 | 14·3 | 0 | 0 | 28·6 | 0 | 14·3 | 0 | 0 | 28·6 | 28·6 | 0 |
| RR | 20 | 30 | 30 | 0 | 0 | 20 | 20 | 10 | 10 | 0 | 20 | 20 | 20 | 0 |
| SR | 11 | 0 | 33 | 0 | 11 | 11 | 22 | 0 | 33 | 22 | 0 | 0 | 11 | 22 |
| All | 23·6 | 18·4 | 18·4 | 7·9 | 5·25 | 7·9 | 21 | 2·6 | 18·4 | 10·5 | 18·4 | 15·8 | 15·7 | 5·3 |
Discussion
Peptide vaccines present some advantages compared to subunit and live attenuated formulations. An advantage to the use of peptide vaccines is the possibility of excluding from vaccine formulation immunosuppressive epitopes and epitopes with the risk of eliciting autoimmunity. However, this type of vaccine presents a limitation in terms of obtaining an effective immune response in a population with high genetic variability. The choice of epitopes to compose a peptide vaccine should take into account not only their ability to elicit the desirable immune response but also their ability to be presented by a wide range of HLA molecules. Therefore, human epitope mapping is an important tool towards the development of epitope-based vaccines. The TEPITOPE algorithm eases the identification of relevant epitopes in a protein sequence, as it indicates the probability of an epitope to bind to a given HLA allele. Epitopes able to bind to several HLA molecules can be selected for further study, reducing the number of peptides to be tested in in vitro assays. Several in vitro studies have confirmed the TEPITOPE prediction of peptides to different HLA molecules [22,23].
In this study, 11 paramyosin epitopes were selected by TEPITOPE by their ability to bind to several HLA-DR molecules. Peptides promiscuity was confirmed by an in vitro proliferative response of PBMC from individuals living in areas endemic for schistosomiasis and in vitro HLA-binding assays. Although a correlation between TEPITOPE prediction and in vitro proliferative response was observed for the majority of paramyosin peptides, for some peptides such as Para (151–167), Para (355–371) and Para (584–600) no correlation was detected. One possible explanation for this TEPITOPE analysis discrepancy could be the fact that the algorithm predicts HLA binding and not necessarily T cell recognition. Additionally, the effective response depends not only on the ability of a given peptide to bind to different HLA molecules, but also on the individual T cell receptor repertoire. Para (151–167) and Para (584–600), for which TEPITOPE analysis predicted an ability to bind to only two and one HLA-DR molecules, respectively; were able to induce a proliferative response in a greater number of individuals. This contrasting result may be explained by the usage of HLA-DR alleles in TEPITOPE analysis being distinct, to some extent, from those observed in the studied population. For example, HLA-DR 14, 16, 17 and 18 molecules are present in the individuals studied here and these alleles are not used in the TEPITOPE algorithm.
With regard to the HLA-binding assay, it confirmed the ability of Para (6–22), Para (210–226) and Para (355–371) to bind to a great number of HLA-DR molecules. These paramyosin epitopes were able to bind to eight [Para (210–226) and Para (355–371)] and six [Para (6–22)] of the eight most prevalent HLA-DR used in the binding assay. Even though Para (13–29) was able to bind to eight HLA-DR molecules, showing its promiscuity, we decided to not select this peptide for further vaccine studies because it was recognized preferentially by T cells of infected individuals (Fig. 1b). With regard to HLA frequency within the population, no predominance of a particular HLA-DR allele expression was observed. HLA-DR expression in each individual, correlated with the response to a given peptide, could not be performed due to a low numbers of HLA-typed patients.
Herein, Para (210–226) showed some relevant characteristics as a potential candidate to compose a peptide vaccine against schistosomiasis. This peptide was recognized by 18·5% of the individuals tested, and Para (210–226) was also able to bind to all the HLA-DR molecules used in the peptide-HLA binding assay. These results confirm Para (210–226) promiscuity, but what makes this peptide interesting in composing an anti-schistosome vaccine is its ability to induce cellular responses in individuals resistant to S. mansoni reinfection compared to infected patients. In a preliminary study performed by our group using far fewer subjects, we also demonstrated the ability of Para (210–226) peptide to induce proliferative responses in individuals resistant to reinfection [24]. A recent study performed with mice infected with S. mansoni has shown that chronic infection is related to IL-10 production, which can be responsible for the Th2 type of immune response observed after egg deposition [25]. IL-10 mediates IFN-γ suppression in human schistosomiasis, and therefore T cell proliferative responses to S. mansoni antigens in infected individuals are reduced [26]. However, we believe that preferential recognition of Para (210–226) by the RR group compared to the SR group cannot be attributed to immunomodulation mediated by IL-10 produced by the SR group, as no statistically significant difference was observed between both groups regarding proliferative responses to PHA. Because the human immune response to paramyosin has been associated with resistance to S. mansoni infection and reinfection [11–13], this peptide might represent one possible component responsible to induce resistance in individuals living in areas endemic for schistosomiasis. Additionally, Para (6–22) and Para (355–371) peptides induced T cell responses in 20% and 14·6% of tested individuals, respectively, and bound to six and eight of the HLA alleles studied in the in vitro HLA-binding assay. These results indicate the immunological relevance of these epitopes as potential vaccine candidates. Furthermore, Para (6–22) possesses only three amino acids that are different from the Taenia solium paramyosin B cell epitope identified in a phage display library [27], and may suggest Para (6–22) as a potential B cell epitope.
The relative ease and low cost of preparation, safety and prolonged shelf life and the ability to focus on defined epitopes has encouraged the use of synthetic peptide vaccines against animal and human disease [28]. The amino acid sequence of the most schistosome vaccine antigens shows several regions with relevant identity to the mammalian counterpart. Paramyosin synthetic peptides used in this study did not present identity with mammalian peptides, but present high identity with peptides from S. japonicum and other parasites such as T. solium. Para (6–22), Para (210–226) and Para (355–371), the most promiscuous peptides from paramyosin, presented 100%, 100% and 82·4% identity with S. japonicum myoprotein peptides, respectively, and 85·7%, 58·8% and 0% identity with T. solium. The identity with peptides from S. japonicum and T. solium open the possibility for use of these peptides as candidates to compose a multivalent vaccine.
Several immunodominant epitopes from other S. mansoni antigens have also been identified and used successfully as antigens in immunization protocols. A B cell epitope of 9B antigen, peptides from SG3PDH, Sm14 or from Sm-37-GAPDH, together with Sm-10-DLC, were able to induce partial protective immunity in the murine model [29–34]. With regard to parasite paramyosin T cell immunodominant epitopes, a peptide in the 176–192 position of the T. solium paramyosin was identified using the murine model [34]. Also, T. solium paramyosin B and T cell epitope mapping demonstrated that B cell immunodominant epitopes are present in the C-terminal region of this protein, while T cell epitopes are located in the N-terminus and central region of T. solium paramyosin [35].
Herein, Para (6–22), Para (210–226) and Para (355–371), the most promiscuous paramyosin peptides identified by this study, are located in the N-terminus and central region of the S. mansoni paramyosin amino acid sequence. These peptides were able to induce a PBMC proliferative response in a great number of individuals living in areas endemic for schistosomiasis; however, T cell recognition cannot be attributed to HLA-DR molecules’ predominance in these individuals, as HLA-DR low-resolution typing demonstrated a broad distribution of HLA-DR alleles in the population studied. Para (6–22), Para (210–226) and Para (355–371) immunological features demonstrated here suggest these peptides as potential candidates in the composition of an anti-schistosomiasis vaccine. However, our understanding is that an efficient anti-schistosomiasis vaccine should contain peptides from other S. mansoni antigens, because 14·6% of the individuals included in this study did not respond to any paramyosin peptide.
Acknowledgments
This work was supported by FAPEMIG, CNPq and PADCT/CNPq.
References
- 1.Chitsulo L, Loverde P, Engels D. Schistosomiasis. Nat Rev Microbiol. 2004;2:12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- 2.Boros DL. Immunopathology of Schistosoma mansoni infection. Clin Microbiol Rev. 1989;2:250–69. doi: 10.1128/cmr.2.3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coles GC, Bruce JI, Kinoti GK, Mutahi WT, Dias EP, Katz N. Drug resistance in schistosomiasis. Trans R Soc Trop Med Hyg. 1986;80:347–51. doi: 10.1016/0035-9203(86)90055-6. [DOI] [PubMed] [Google Scholar]
- 4.Fallon PG, Doenhoff MJ. Drug-resistant schistosomiasis: resistance to praziquantel and oxaminiquine induced in Schistosoma mansoni in mice is drug specific. Am J Trop Med Hyg. 1994;51:83–8. doi: 10.4269/ajtmh.1994.51.83. [DOI] [PubMed] [Google Scholar]
- 5.Bergquist NR. Schistosomiasis vaccine development: approaches and prospects. Mem Inst Oswaldo Cruz. 1995;90:221–7. doi: 10.1590/s0074-02761995000200017. [DOI] [PubMed] [Google Scholar]
- 6.Capron A, Riveau G, Capron M, Trottein F. Schistosomes: the road from host–parasite interactions to vaccines in clinical trials. Trends Parasitol. 2005;21:143–9. doi: 10.1016/j.pt.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 7.McManus DP, Bartley PB. A vaccine against Asian schistosomiasis. Parasitol Int. 2004;53:163–73. doi: 10.1016/j.parint.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Pearce EJ, James SL, Hieny S, Lanar DE, Sher A. Induction of protective immunity against Schistosoma mansoni by vaccination with schistosome Paramyosin (Sm97), a nonsurface parasite antigen. Proc Natl Acad Sci USA. 1988;85:5678–82. doi: 10.1073/pnas.85.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanar DE, Pearce EJ, James SL, Sher A. Identification of Paramyosin as schistosome antigen recognized by intradermally vaccinated mice. Science. 1986;234:593–6. doi: 10.1126/science.3094144. [DOI] [PubMed] [Google Scholar]
- 10.Flanigan TP, King CH, Lett RR, Nanduri J, Mahmoud AA. Induction of resistance to Schistosoma mansoni infection in mice by purified parasite Paramyosin. J Clin Invest. 1989;83:1010–14. doi: 10.1172/JCI113942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeiro de Jesus A, Araujo I, Bacellar O, et al. Human immune responses to Schistosoma mansoni vaccine candidate antigens. Infect Immun. 2000;68:2797–803. doi: 10.1128/iai.68.5.2797-2803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correa-Oliveira R, Pearce EJ, Oliveira GC, et al. The human immune response to defined immunogens of Schistosoma mansoni: elevated antibody levels to Paramyosin in stool-negative individuals from two endemic areas in Brazil. Trans R Soc Trop Med Hyg. 1989;83:798–804. doi: 10.1016/0035-9203(89)90334-9. [DOI] [PubMed] [Google Scholar]
- 13.Al-Sherbiny M, Osman A, Barakat R, El Morshedy H, Bergquist R, Olds R. In vitro cellular and humoral responses to Schistosoma mansoni vaccine candidate antigens. Acta Trop. 2003;88:117–30. doi: 10.1016/s0001-706x(03)00195-5. [DOI] [PubMed] [Google Scholar]
- 14.Colley DG, Cook JA, Freeman GL, Jr, Bartholomew RK, Jordan P. Immune responses during human Schistosomiasis mansoni. I. In vitro lymphocyte blastogenic responses to heterogeneous antigenic preparations from schistosome eggs, worms and cercariae. Int Arch Allergy Appl Immunol. 1977;53:420–33. [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 16.Sturniolo T, Bono E, Ding J, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–61. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 17.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39:225–35. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 18.Sette A, Sidney J, del Guercio MF, et al. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994;31:813–22. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 19.Southwood S, Sidney J, Kondo A, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–73. [PubMed] [Google Scholar]
- 20.Buus S, Sette A, Colon SM, Miles C, Grey HM. The relation between major histocompatibility complex (MHC) restriction and the capacity of Ia to bind immunogenic peptides. Science. 1987;235:1353–8. doi: 10.1126/science.2435001. [DOI] [PubMed] [Google Scholar]
- 21.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. The measurement of MHC/peptide interactions by gel infiltration. Curr Protocols Immunol. 1998;18:1–19. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 22.Iwai LK, Yoshida M, Sidney J, et al. In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasileinses frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals. Mol Med. 2003;9:1–12. [PMC free article] [PubMed] [Google Scholar]
- 23.Panigada M, Sturniolo T, Besozzi G, et al. Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect Immun. 2002;70:79–85. doi: 10.1128/IAI.70.1.79-85.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fonseca CT, Cunha-Neto E, Kalil J, et al. Identification of immunodominant epitopes of Schistosoma mansoni vaccine candidate antigens using human T cells. Mem Inst Oswaldo Cruz. 2004;99(Suppl. 1):63–6. doi: 10.1590/s0074-02762004000900011. [DOI] [PubMed] [Google Scholar]
- 25.Pearce EJ, MacDonald AS. The immunobiology of schistosomiasis. Nat Rev Immunol. 2002;2:499–511. doi: 10.1038/nri843. [DOI] [PubMed] [Google Scholar]
- 26.Correa-Oliveira R, Malaquias LC, Falcao PL, et al. Cytokines as determinants of resistance and pathology in human Schistosoma mansoni infection. Braz J Med Biol Res. 1998;31:171–7. doi: 10.1590/s0100-879x1998000100024. [DOI] [PubMed] [Google Scholar]
- 27.Gazarian KG, Gazarian TG, Solís CF, Hernández R, Shoemarker CB, Laclet JP. Epitope mapping on N-terminal region of Taenia solium paramyosin. Immunol Lett. 2000;72:191–5. doi: 10.1016/s0165-2478(00)00189-9. [DOI] [PubMed] [Google Scholar]
- 28.Meloen RH, Langeveld JP, Schaaper WM, Slootstra JW. Synthetic peptide vaccines: unexpected fulfillment of discarded hope? Biologicals. 2001;29:233–6. doi: 10.1006/biol.2001.0298. [DOI] [PubMed] [Google Scholar]
- 29.Agiro L, Henri S, Dessein H, Kouriba B, Dessein AJ, Bourgois A. Induction of protection against S. mansoni with a MAP containing epitopes of Sm37-GAPDH and Sm10-DLC. Effect of coadsorbtion with GM-CSF on alum. Vaccine. 2000;18:2033–8. doi: 10.1016/s0264-410x(99)00523-x. [DOI] [PubMed] [Google Scholar]
- 30.Tallima H, Montash M, Veprek P, Velek J, Jezek J, El Ridi R. Differences in immunogenicity and vaccine potential of peptides from Schistosoma mansoni glyceraldehyde 3-phosphate dehydrogenase. Vaccine. 2003;21:3290–300. doi: 10.1016/s0264-410x(03)00180-4. [DOI] [PubMed] [Google Scholar]
- 31.Tarrab-Hazdai R, Schechtman D, Arnon R. Synthesis and characterization of a protective peptide-based vaccine against Schistosoma mansoni. Infect Immun. 1998;66:4526–30. doi: 10.1128/iai.66.9.4526-4530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nara T, Tanabe K, Mahakunkijcharoen Y, Osada Y, Matsumoto N, Kita K, Kojima S. The B-cell epitope of Paramyosin recognized by a protective monoclonal IgE antibody to Schistosoma japomicum. Vaccine. 1997;15:79–84. doi: 10.1016/s0264-410x(96)00100-4. [DOI] [PubMed] [Google Scholar]
- 33.Vilar MM, Barrientos F, Almeida M, et al. An experimental bivalent peptide vaccine against schistosomiasis and fascioliasis. Vaccine. 2003;22:137–44. doi: 10.1016/s0264-410x(03)00300-1. [DOI] [PubMed] [Google Scholar]
- 34.López-Moreno HS, Correa D, Laclette JP, Ortiz-Navarrete VF. Identification of CD4+ T cell epitopes of Taenia solium Paramyosin. Parasite Immunol. 2003;25:513–16. doi: 10.1111/j.1365-3024.2003.00658.x. [DOI] [PubMed] [Google Scholar]
- 35.Vázquez-Talavera J, Solís CF, Medina-Escutia E, et al. Human T and B cell epitope mapping of Taenia solium Paramyosin. Parasite Immunol. 2001;23:575–9. doi: 10.1046/j.1365-3024.2001.00416.x. [DOI] [PubMed] [Google Scholar]