Abstract
The synthesis of 60S ribosomal subunits in Saccharomyces cerevisiae requires Tif6p, the yeast homologue of mammalian eukaryotic translation initiation factor 6 (eIF6). In the present work, we have isolated a protein kinase from rabbit reticulocyte lysates on the basis of its ability to phosphorylate recombinant human eIF6. Mass spectrometric analysis as well as antigenic properties of the purified kinase identified it as casein kinase I. The site of in vitro phosphorylation, which is highly conserved from yeast to mammals, was identified as the serine residues at positions 174 (major site) and 175 (minor site). The homologous yeast protein Tif6p was also phosphorylated in vivo in yeast cells. Mutation of Tif6p at serine-174 to alanine reduced phosphorylation drastically and caused loss of cell growth and viability. When both Ser-174 and Ser-175 were mutated to alanine, phosphorylation of Tif6p was completely abolished. Furthermore, while wild-type Tif6p was distributed both in nuclei and the cytoplasm of yeast cells, the mutant Tif6p (with Ser174Ala and Ser175Ala) became a constitutively nuclear protein. These results suggest that phosphorylatable Ser-174 and Ser-175 play a critical role in the nuclear export of Tif6p.
Eukaryotic translation initiation factor 6 (eIF6), a monomeric protein of about 26 kDa, was originally isolated from the postribosomal supernatant of both wheat germ and mammalian cell extracts based on an assay that measured the ability of the protein to bind to the cytoplasmic 60S ribosomal subunit and prevent its association with the 40S ribosomal subunit (18, 21, 22, 25, 27). Because of this ribosomal subunit antiassociation property, eIF6 was thought to provide a pool of free ribosomal subunits required for translation initiation in eukaryotic cells. The protein was therefore classified as an eIF (16), although its role in translation of mRNAs was not defined in these original studies. More recently, the cloning of the human cDNA (25) and then of the Saccharomyces cerevisiae gene encoding eIF6 has allowed detailed characterization of the function of eIF6 in yeast cells (23, 26, 30). Molecular genetic analysis has now provided compelling evidence that at least in yeast cells, eIF6, encoded by the essential gene, TIF6, does not function as a canonical translation initiation factor (26). Rather, eIF6, designated Tif6p in yeast, is necessary for the formation of 60S ribosomal subunits. Lack of Tif6p prevents the processing of pre-rRNA to form the mature 25S and 5.8S rRNAs, the constituents of the 60S ribosomal particle (2). In agreement with this observation, it was observed that Tif6p, in addition to its localization in the cytoplasm, is also present in the nuclei of yeast cells (2). Furthermore, recent proteomic analysis of yeast nucleolar 60S preribosomal particles (also called 66S preribosomes), which are the intermediates in the synthesis and assembly of 60S ribosomal subunits, has shown that Tif6p is part of a multiprotein assembly complex associated with the 60S preribosomal particles (4, 10).
During the course of purification of eIF6 from mammalian and yeast cells, we observed that in cruder protein fractions, eIF6 migrated as a doublet as judged by Western blot analysis. Treatment of these protein fractions with calf intestinal alkaline phosphatase resulted in the migration of eIF6 as a single polypeptide band. These results suggested that eIF6 may be phosphorylated in eukaryotic cells. In view of the well-established notion that phosphorylation regulates the biological activity of many proteins, we purified and characterized the mammalian protein kinase responsible for the phosphorylation of eIF6.
In this paper, we describe the purification and the characterization of the mammalian protein kinase from rabbit reticulocyte lysates that is responsible for phosphorylation of mammalian eIF6 in vitro. We identify the protein kinase as casein kinase I (CK I). The site of phosphorylation, which is highly conserved between yeast and mammals, has been identified as the serine residues at positions 174 and 175. We further show that the yeast protein Tif6p is also phosphorylated in vivo in yeast cells, and mutation of Ser-174 and Ser-175 of Tif6p to alanine completely abolishes phosphorylation of Tif6p and causes loss of yeast cell growth and viability. Furthermore, unphosphorylated Tif6p was localized exclusively in the nuclei of yeast cells, suggesting that phosphorylation of Tif6p is involved in passage of Tif6p to the cytoplasm.
MATERIALS AND METHODS
Strains, plasmids, and genetic techniques.
The S. cerevisiae haploid strain KSY606, containing the chromosomal copy of the TIF6 gene inactivated by insertion of a HIS3 marker gene and harboring a centromeric (CEN) expression plasmid that expresses Tif6p from its natural promoter, was constructed from the diploid yeast strain W303a/α as described previously (2). All other yeast strains in this work were derived from KSY606 and are described in Table 1. Yeast cells were grown at 30°C either in rich YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] dextrose, 2% [wt/vol] Bacto Peptone) or in YPGal medium, in which 2% [wt/vol] galactose replaced dextrose as the carbon source. Where indicated, haploid yeast cells were also grown in synthetic complete medium (0.67% Bacto yeast nitrogen base without amino acids, 0.2% amino acid mixture supplemented with nutrients required for auxotrophic deficiencies) containing either 2% galactose (SGal medium) or 2% dextrose (SD medium) as the carbon source. Yeast genetic methods were carried out by following standard protocols (19).
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference(s) or source |
|---|---|---|
| W303a/α | MATa/α leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 | 19 |
| KSY601 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3/TIF6 | 26 |
| KSY603 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6] | 2, 26 |
| KSY605 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[TRP1 TIF6-Myc] | 2 |
| KSY606 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 TIF6] | 2 |
| KSY607 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[LEU2 TIF6-HA] | 2 |
| UBY608 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura 3-1 can1-100 tif6::HIS3 p[TRP1 TIF6-Myc] p[LEU2 tif6-HA (S174A, S175A)] | This work |
| UBY609 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6] p[LEU2 tif6-GFP(S174A, S175A)] | This work |
| UBY610 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[TRP1 TIF6-Myc] p[LEU2 TIF6-HA] | This work |
| UBY611 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6] p[LEU2 TIF6-GFP] | This work |
| UBY612 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 TIF6] p[LEU2 GAL10::TIF6-GFP] | This work |
| UBY613 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 TIF6] p[LEU2 GAL10::tif6-GFP(S174A, S175A)] | This work |
Purification of eIF6 kinase from rabbit reticulocyte lysate.
The preparation of crude ribosomal 0.5 M KCl wash proteins from 300 ml of rabbit reticulocyte lysates was as described previously for the isolation of eIF5 (3). The dialyzed 0.5 M KCl wash proteins were loaded onto a DEAE cellulose column (60-ml bed volume), previously equilibrated in buffer A (20 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol [DTT], 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], and 5% glycerol) containing 100 mM KCl. The column was washed with the same equilibration buffer. The unretarded protein fraction was then applied to a 60-ml-bed-volume phosphocellulose column equilibrated in a buffer A-100 mM KCl. After the column was washed with buffer A-100 mM KCl, the bound proteins were eluted stepwise with buffer A-300 mM KCl followed by buffer A-750 mM KCl. The eIF6 kinase activity was detected in the 750 mM KCl eluate. Fractions containing the bulk of the kinase activity were pooled, and the proteins present in the pooled fractions were precipitated by adding solid ammonium sulfate to 70% saturation. The precipitated proteins were dissolved in 1.5 ml of buffer A-500 mM KCl, dialyzed for about 2 h against the same buffer, and then concentrated to about 0.7 ml by Centricon-10 filtration. The concentrated protein sample (5 mg of protein) was then applied to a 35-ml-bed-volume Sephadex G-75 superfine column (1.4 by 89 cm) equilibrated in buffer A-500 mM KCl. The column was developed with the same buffer, and fractions of 1 ml were collected. Fractions 17 to 20 containing eIF6 kinase activity (500 μg of protein) were pooled, dialyzed against buffer A-100 mM KCl to reduce the ionic strength of the pooled fraction to that of buffer A-100 mM KCl, and then purified by gradient elution (buffer A-100 mM KCl to buffer A-500 mM KCl) from a fast- performance liquid chromatography (FPLC) Mono S column (1-ml bed volume). eIF6 kinase activity eluted at about 320 mM KCl. Active fractions were pooled and stored in small aliquots at −70°C.
Assay for eIF6 kinase activity.
Phosphorylation of eIF6 was carried out in reaction mixtures (10 μl each) containing 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 100 mM KCl, 1 mM DTT, 1 mM [γ-32P]ATP (2,000 cpm/pmol), and 50 to 200 ng of eIF6 kinase-containing protein fraction. Following incubation for 15 min at 30°C, the reactions were terminated by the addition of 10 μl of an electrophoresis loading buffer (250 mM Tris-HCl [pH 6.8], 570 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], and 0.01% bromophenol blue) and heating of the mixtures at 100°C for 3 min. The reaction mixtures were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (15% gel), and the dried gel was analyzed by using autoradiography.
Isolation of maximally phosphorylated eIF6 for mass spectrometric analysis.
Two reaction mixtures (50 μl each), one containing [γ-32P]ATP (100 cpm/pmol) and the other without ATP, were prepared as described above in “Assay for eIF6 kinase activity” except that each reaction mixture contained 100 pmol of recombinant mammalian eIF6 and 0.5 μg of purified eIF6 kinase. After incubation at 30°C for 30 min, each reaction mixture was subjected to SDS-PAGE, the polypeptide band corresponding to eIF6 was excised and destained, and enzymatic digestion with trypsin was performed in-gel. Resultant peptides were extracted and analyzed by mass spectrometry by using a PerSeptive Biosystems matrix-assisted laser desorption ionization-time of flight voyager DE-STR mass spectrometer in the laboratory for macromolecular analysis at the Albert Einstein College of Medicine of Yeshiva University.
Purification of recombinant His-tagged wild-type mammalian eIF6.
The recombinant mammalian eIF6 expression plasmid pRSET-A-eIF6 was generated by inserting PCR-amplified human eIF6-encoding sequences (25) at the EcoRI/BamHI sites of the expression vector pRSET-A. The construct was then used to transform Escherichia coli BL21(DE3) cells, and the expression of His-tagged human eIF6 was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to 4 liters of an exponentially growing culture of these bacterial cells carrying the expression plasmid pRSET-A-eIF6. Cells (7 g) were harvested by centrifugation at 3 h postinduction, suspended in a solution containing 20 ml of 20 mM potassium phosphate (pH 7.8), 100 mM NaCl, 0.5 mM PMSF, and 3 mM potassium imidazole, treated with 3 mg of lysozyme for 30 min at 4°C, and then disrupted by sonication. After the cell debris was removed by centrifugation, a cocktail of protease inhibitors was added to the supernatant and the mixture was treated with 40 μg of pancreatic DNase, incubated at 0°C for 30 min, and then centrifuged at 48,000 rpm for 150 min in a 50 Ti rotor. The postribosomal supernatant was adjusted to 0.5 M NaCl by the addition of 2 M NaCl and then loaded onto a 4-ml-bed-volume Ni-nitrilotriacetic acid-agarose column preequilibrated in buffer N (20 mM potassium phosphate [pH 7.8], 500 mM NaCl, and 0.5 mM PMSF) containing 3 mM potassium imidazole. After the column was washed first with buffer N-3 mM potassium imidazole and then with buffer N-30 mM imidazole, His6-tagged eIF6 was eluted from the column by using buffer N-300 mM potassium imidazole. The eluate was dialyzed against buffer B (20 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, and 1 mM DTT) containing 80 mM KCl for about 3 h and then purified by gradient elution (buffer B-100 mM KCl to buffer B-500 mM KCl) from an FPLC Mono Q column (1-ml bed volume; Pharmacia Biotech). eIF6 activity, assayed by Western blotting by using polyclonal anti-eIF6 antibodies, eluted at about 340 mM KCl. Active fractions were pooled and stored in small aliquots at −70°C.
Site-directed mutagenesis of mammalian and yeast eIF6-encoding sequences and expression of mutant eIF6 proteins in yeast cells.
Point mutations within the sequences coding for eIF6 (Tif6p) present in the yeast LEU2-based CEN plasmid pRS315-TIF6(-HA) or the bacterial recombinant expression plasmid pRSET-A-eIF6 were constructed by one-stage PCR by using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. We designed appropriate 26- to 30-mer mutagenic oligonucleotide primers to create the desired serine-to-alanine or -aspartate mutations at position 174 or 175 of either mammalian eIF6 or yeast Tif6p by maintaining the reading frame of TIF6. All the mutated open reading frames (ORFs) were sequenced to confirm the desired mutations and ensure error-free DNA synthesis in other regions of the ORF. Recombinant pRSET-A plasmids containing the mutant eIF6 coding sequences were used to transform E. coli BL21(DE3) cells, and the mutant proteins were expressed and purified from bacterial cells by using a procedure similar to that described above for the purification of the wild-type protein. It should be noted that the derivation of the Stokes' radius by Sephadex G-75 gel filtration showed that both the wild-type eIF6 and the Ala mutant eIF6 have the same values as that obtained for native eIF6 isolated from rabbit reticulocyte lysates (18, 25).
Metabolic labeling of Tif6p in yeast cells and isolation of labeled Tif6p by immunoprecipitation.
Exponentially growing yeast cells (25 ml) in low-phosphate medium were labeled with 32P by using 5 mCi of carrier-free [32P]orthophosphate as described by Warner (28). The harvested cells were washed twice with ice-cold water and then resuspended in 200 μl of lysis buffer (20 mM HEPES [pH 7.9], 10 mM EDTA, 1 mM DTT, 100 mM ammonium sulfate, 0.5 mg of bovine serum albumin/ml, and 10% glycerol) containing a cocktail of protease inhibitors as described previously (14).
Cells were disrupted by vortexing with glass beads and centrifuged. The supernatant (250 μl) was treated with 100 mg of solid ammonium sulfate, and the precipitated proteins were recovered by centrifugation. The pellet was dissolved in 600 μl of lysis buffer containing 1% Triton X-100, 0.1% SDS, and 5% nonfat milk and incubated with 10 μl of agarose-conjugated anti-hemagglutinin (anti-HA) antibody (Santa Cruz, Berkeley, Calif.) for about 4 h with gentle mixing in a rotator. Immunocomplexes bound to agarose beads were isolated by centrifugation and washed sequentially with (i) lysis buffer containing 0.4 M ammonium sulfate (twice) and (ii) lysis buffer containing 50 mM ammonium sulfate (once). Proteins bound to the immunocomplexes were eluted by suspending the beads in 50 μl of 125 mM Tris-HCl (pH 6.8), 2% SDS, and 10% glycerol and incubating the suspension in a boiling water bath for 5 min. After centrifugation, the supernatant containing 32P-labeled Tif6p was treated with 100 mM DTT, cooled, and then subjected to SDS-PAGE (10% gel). The dried gel was analyzed by using autoradiography.
Other methods.
Exponentially growing yeast cells were fractionated into nuclear and cytosolic fractions by an adaptation of the procedure of Aris and Blobel (1). Fractionation of HeLa cell extracts into nuclear and postnuclear supernatant fractions was carried out by using standard protocols, and these fractions were kind gifts of Danny Reinberg of Robert Wood Johnson Medical School, Piscataway, N.J. Where indicated, the nuclear and the cytosolic fractions of HeLa and yeast cells were preincubated with calf intestinal phosphatase in a buffer containing 20 mM HEPES (pH 7.9), 20 mM MgCl2, and 1 mM PMSF prior to SDS-PAGE. Recombinant mammalian His6-tagged CK Iα was purified from BL21(DE3) cells carrying the sequence coding for bovine CK Iα in the plasmid pRSET-A by following a procedure similar to that described above for the purification of His6-tagged eIF6, except that after affinity purification from a Ni+-nitrilotriacetic acid-agarose column, the kinase was further purified by phosphocellulose chromatography and finally by gradient elution from an FPLC Mono S column. Recombinant mammalian CK Iδ was purchased from New England Biolabs. Polyclonal anti-mammalian eIF6 antibodies were prepared as described previously (25). These antibodies were specific for denatured conformation of eIF6 and did not immunoprecipitate native eIF6.
The procedures used for (i) pulse-chase labeling of pre-rRNA with [3H]uracil and subsequent analysis of RNA by gel electrophoresis and (ii) indirect immunofluorescence studies to detect localization of HA- and Myc-tagged Tif6p in yeast cells were as described previously (2). The subcellular localization of wild-type and Ala mutant Tif6p proteins was also analyzed by monitoring the fluorescent signal produced by Tif6p in which green fluorescent protein (GFP) was fused at the carboxy terminus of the protein. The fusion protein was expressed from a pRS315 plasmid under the control of a GAL10 promoter in KSY606. The intensity of the gel bands was quantitated by using ImageQuant (version 1.2).
RESULTS
eIF6 appears to be phosphorylated in mammalian and yeast cells.
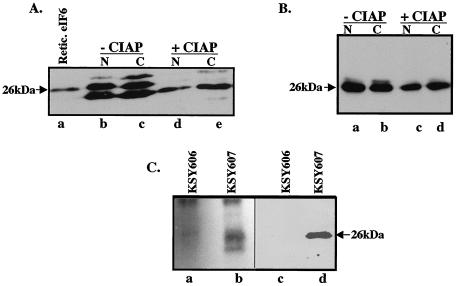
Our initial experiments were to determine whether eIF6 is phosphorylated in eukaryotic cells. HeLa cell extracts were fractionated into nuclear and cytoplasmic fractions. Western blot analysis of these fractions showed that eIF6 migrated as multiple immunoreactive bands. In contrast, purified mammalian eIF6 from rabbit reticulocyte lysates migrated as a single band (Fig. 1A, compare lanes b and c with lane a). When these extracts were preincubated with calf intestinal phosphatase prior to Western blot analysis (Fig. 1A, lanes d and e), a single immunoreactive polypeptide corresponding to the faster-migrating polypeptide was observed. These results suggested that in HeLa cell extracts, a fraction of eIF6 is phosphorylated. When extracts of KSY607 yeast cells, expressing HA-tagged Tif6p (Table 1), were fractionated into nuclear and cytoplasmic fractions and then subjected to Western blot analysis, two immunoreactive polypeptide bands were observed, although the faster-migrating band was much more intense than the slower-migrating polypeptide (Fig. 1B, lanes a and b). Here again, treatment of these fractions with calf intestinal phosphatase prior to Western blot analysis resulted in the appearance of only one immunoreactive polypeptide band corresponding to the faster-migrating band of Tif6p (Fig. 1B, lanes c and d).
FIG. 1.
eIF6 is phosphorylated in HeLa and yeast cells. (A) Cell extracts of HeLa cells were fractionated into nuclear (N) and cytoplasmic (C) fractions. Where indicated, aliquots of each fraction were preincubated with calf intestinal alkaline phosphatase (CIAP). These fractions (lanes d and e) and untreated controls (lanes b and c) along with purified rabbit reticulocyte eIF6 (Retic. eIF6; lane a) were subjected to Western blot analysis by using anti-mammalian eIF6 antibodies as probes. (B) Nuclear and cytoplasmic fractions derived from extracts of KSY607 yeast cells, expressing HA-tagged Tif6p, were preincubated either with calf intestinal alkaline phosphatase (lanes c and d) or with buffer (lanes a and b) and then subjected to Western blot analysis by using anti-HA antibodies as probes. (C) Cell extracts of 32P-labeled yeast cells expressing either HA-Tif6p (strain KSY607) or untagged Tif6p (strain KSY606) were immunoprecipitated with anti-HA antibodies. The washed immunocomplexes were subjected to SDS-PAGE followed by electrophoretic transfer onto a polyvinylidene difluoride membrane. The blot was first subjected to autoradiography (lanes a and b) and then analyzed by immunoblotting by using anti-HA antibodies (lanes c and d). The faster-migrating band in lane b may represent a degradation product of Tif6p.
To confirm that Tif6p is indeed phosphorylated in vivo in yeast cells, we labeled KSY607 cells with [32P]orthophosphate, and Tif6p present in cell lysates was immunoprecipitated by using monoclonal anti-HA antibodies. The immunocomplexes were resolved by SDS-PAGE followed by autoradiography of the dried gel. A major 32P-labeled polypeptide, which migrated with the same mobility as HA-Tif6p isolated from KSY607 cells, was observed (Fig. 1C, compare lanes b and d). No radioactive polypeptide band corresponding to Tif6p was immunoprecipitated from 32P-labeled cell extracts of an isogenic yeast strain KSY606, whose Tif6p was not tagged (Fig. 1C, lanes a and c). These results suggest that Tif6p is phosphorylated in yeast cells.
Isolation and initial characterization of an eIF6 protein kinase from rabbit reticulocyte lysate.
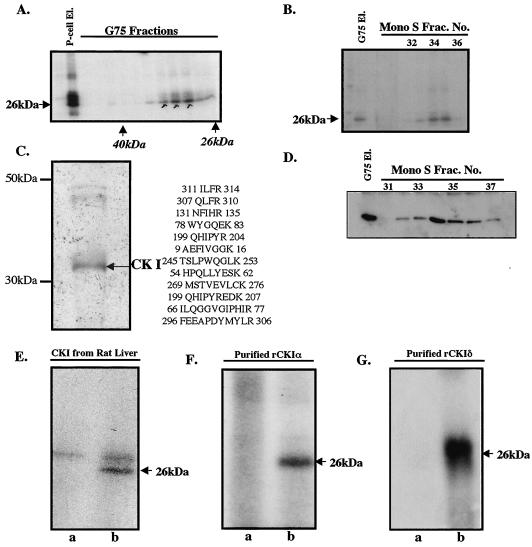
We made the initial observation that rabbit reticulocyte lysates contain a protein kinase activity that phosphorylates bacterially expressed recombinant human eIF6 by using [γ-32P]ATP as the phosphoryl donor (data not shown). Fractionation of the crude lysate into ribosomes and postribosomal supernatant showed that eIF6 kinase activity was associated mainly with ribosomes, from which it can be eluted by treatment of the ribosomes with 0.5 M KCl (data not shown). The purification of the eIF6 kinase from 0.5 M KCl ribosomal wash fraction is outlined in Materials and Methods. The eIF6 kinase activity was unretarded in the DEAE cellulose column but bound strongly to a phosphocellulose column from which the kinase activity was eluted with a buffer containing 0.75 M KCl. When the concentrated phosphocellulose eluate was purified by using a Sephadex G-75 gel filtration column, the eIF6 kinase activity eluted as a protein of about 30 to 35 kDa (Fig. 2A). The pooled Sephadex fractions were further purified by FPLC on a Mono S column (Fig. 2B). Analysis of the purified Mono S fraction by SDS-PAGE followed by silver staining showed one major polypeptide band of apparent molecular mass of about 35 kDa and two minor higher-molecular-mass bands of about 45 to 48 kDa (Fig. 2C). Tryptic peptides from the major species were fractionated by high-performance liquid chromatography and analyzed by mass spectrometry and Edman degradation sequencing. The derived peptide sequences were identified to be those of CK Iα (20). In agreement with these results, we observed that when the pooled Sephadex G-75 fractions as well as eluates from the Mono S column were subjected to Western blot analysis by using polyclonal anti-human CK Iα antibodies (Santa Cruz) as probes (Fig. 2D), immunoreactive polypeptide bands corresponding to mammalian CK Iα were found to comigrate with the eIF6 kinase activity. These results suggested that eIF6 kinase present in rabbit reticulocyte lysates is CK Iα. Further confirmation for this conclusion came from the observation that recombinant eIF6 was efficiently phosphorylated by purified CK I from rat liver (Fig. 2E) as well as by purified bacterially expressed recombinant mammalian CK Iα and CK Iδ isoforms (Fig. 2F and G, respectively).
FIG. 2.
Purification and characterization of eIF6 kinase from rabbit reticulocyte lysates. (A) A partially purified eIF6 kinase preparation (dialyzed ammonium sulfate fraction derived from the phosphocellulose eluate [P-cell El.]) was subjected to Sephadex G-75 gel filtration as described in Materials and Methods. Aliquots of the protein fractions eluted from the column were assayed for eIF6 kinase activity. (B) The pooled Sephadex G-75 eluate [G75 El.] of eIF6 kinase was subjected to FPLC on a Mono S column. Aliquots of each fraction as well as the pooled G-75 fraction that was loaded onto the Mono S column were assayed for eIF6 kinase activity. Frac. No., fraction number. (C) The pooled Mono S fraction was analyzed by SDS-PAGE followed by silver staining. The major polypeptide band was digested with trypsin and analyzed by mass spectrometry. The sequenced peptides that correspond to CK Iα are indicated. (D) Fractions eluting from the FPLC Mono S column that contained eIF6 kinase activity were analyzed by Western blotting by using anti-mammalian CK Iα antibodies. (E, F, and G) Recombinant human eIF6 (3 pmol) was phosphorylated by using [γ-32P]ATP and either purified rat liver CK I (E), a purified recombinant mammalian CK Iα isoform (purified rCKIα) (F), or purified recombinant mammalian CK Iδ (purified rCKIδ) (G). In each case, the reaction mixture in lane a did not contain eIF6 while the reaction mixture in lane b was the complete system containing eIF6. In panel E, the slower-migrating bands present in both lanes a and b represent autophosphorylated CK I.
Identification of the amino acid residues of eIF6 phosphorylated by purified eIF6 kinase.
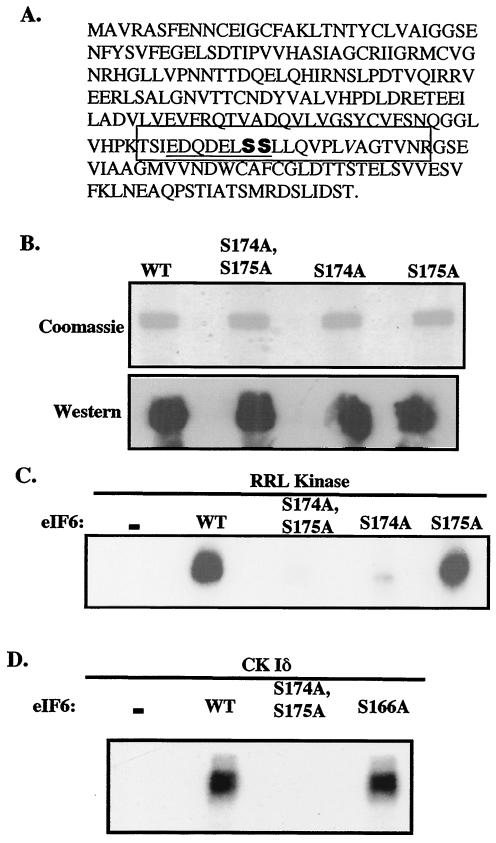
Phosphoamino acid analysis of mammalian eIF6 phosphorylated in vitro by purified eIF6 kinase from rabbit reticulocyte lysates showed that serine was the only amino acid residue phosphorylated (data not shown). To identify the serine residue(s) in mammalian eIF6 that is phosphorylated in vitro, eIF6 was maximally phosphorylated in vitro by the purified kinase, and phosphorylated eIF6 was isolated free of the unreactive components by SDS-PAGE as described in Materials and Methods. Both the maximally phosphorylated and the unphosphorylated eIF6 (40 pmol each) were digested completely with trypsin by following established protocols. The digested products were divided into two parts, and one part was treated with alkaline phosphatase. By comparing matrix-assisted laser desorption ionization-time of flight spectra recorded before and after alkaline phosphatase treatment, we concluded that a serine residue present in the tryptic peptide spanning from Thr-165 to Arg-183 (Fig. 3A) was phosphorylated. This tryptic peptide contains three serine residues at positions 166, 174, and 175. In this tryptic peptide, the possible serine phosphorylation sites for CK I are Ser-174 and Ser-175, as predicted from the known phosphorylation motif (D/E)nX-X-S/T for CK I (where X is any amino acid) (9). The greater the number of acidic amino acids in the vicinity of the serine residue, the stronger is the site of serine phosphorylation since CK I is one of the small family of acidotropic protein kinases (6).
FIG. 3.
Characterization of the phosphorylation sites of mammalian eIF6. (A) Amino acid sequence of mammalian eIF6. The tryptic phosphopeptide that was phosphorylated by purified eIF6 kinase as determined by mass spectrometry is boxed, and Ser-174 and Ser-175 are shown in bold. (B) His-tagged recombinant wild-type (WT) and mutant (S174A; S175A; and S174A, S175A) mammalian eIF6 proteins were purified from IPTG-induced bacterial cell lysates as described in Materials and Methods and then analyzed by SDS-PAGE followed by either Coomassie blue staining or immunoblotting with anti-mammalian eIF6 antibodies. (C) Phosphorylation of recombinant wild-type and mutant (S174A; S175A; and S174A, S175A) mammalian eIF6 proteins by [γ-32P]ATP and purified rabbit reticulocyte eIF6 kinase (RRL kinase; Mono S fraction). (D) Phosphorylation of recombinant mammalian wild-type eIF6, mutant eIF6 (S174A, S175A), and mutant eIF6 (S166A) by recombinant CK Iδ.
To verify that Ser-174 and/or Ser-175 is indeed the site for in vitro eIF6 phosphorylation, we carried out site-directed mutagenesis in the eIF6-encoding sequence of mammalian cDNA such that each of these two serine residues, either alone or in combination, was converted to alanine in the bacterially expressed eIF6 protein. The purified mutant eIF6 proteins (Fig. 3B) were then used as substrates for in vitro phosphorylation by purified eIF6 kinase and by the recombinant CK Iδ isoform. As shown in Fig. 3C and D, mutation of Ser-174 and Ser-175 to alanine completely abolished in vitro phosphorylation both by purified rabbit reticulocyte eIF6 kinase (CK Iα) and by the purified recombinant CK Iδ isoform. Among these two sites, Ser-174 appears to be the major phosphorylation site since mutation of Ser-175 alone to alanine abolished phosphorylation by only about 15% while mutation of Ser-174 to alanine reduced phosphorylation by about 90% (Fig. 3C). Although Ser-166 is not in the known phosphorylation motif for CK I, we also mutated this serine residue to alanine. The purified mutant (S166A) eIF6 protein was efficiently phosphorylated both by purified reticulocyte kinase (data not shown) and by purified CK Iδ (Fig. 3D), as expected. Taken together, these results show that Ser-174 is the primary site of phosphorylation in vitro while Ser-175 is the minor site of phosphorylation.
Phosphorylation of eIF6 in vivo in yeast cells.
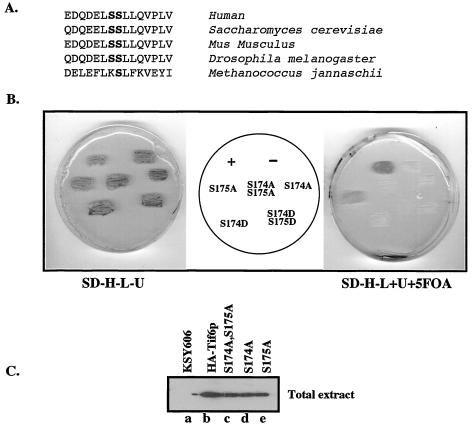
Sequence analysis of eIF6 proteins from human, mouse, Drosophila, S. cerevisiae, and Archaebacteria showed that the sequence of this tryptic peptide is highly conserved from yeast to mammals but absent in Archaebacteria (Fig. 4A). Based on the assumption that the same serine residues (Ser-174 and Ser-175) in yeast Tif6p are also phosphorylated in vivo in yeast cells, we mutated the codons for serine into codons for either alanine or aspartate at these sites in Tif6p present in a LEU2-based CEN plasmid pRS315-TIF6(-HA) by using site-directed mutagenesis. Five different mutant plasmids were constructed in which the tif6 allele contained mutation of Ser-174 and/or Ser-175 to either alanine or aspartate. We then used the plasmid-shuffling technique to first determine whether the mutant Tif6p proteins expressed from these plasmids can functionally substitute for the corresponding wild-type yeast proteins in vivo. Strain KSY606 (2) with an inactivated chromosomal TIF6, covered by TIF6 on a centromeric URA3 plasmid, was transformed individually either with each of the five mutant Tif6p expression plasmids or with the wild-type expression plasmid pRS315-TIF6(-HA). His+ Leu+ Ura+ transformants were selected on appropriate plates and then replica-plated onto plates with SD medium lacking His, Leu, and Ura (SD−His−Leu−Ura plates) (Fig. 4B, left panel) and another similar plate which, in addition, contained uracil and 5-fluoroorotic acid (5-FOA) (Fig. 4B, right panel) to select for the loss of the original plasmid pRS315-TIF6 expressing wild-type Tif6p.
FIG. 4.
Effect of mutation of Tif6p at Ser-174 and Ser-175 on yeast cell growth and viability. (A) Conservation of the amino acid residues surrounding the serine CK I phosphorylation sites of eIF6 (Tif6p). Ser-174 and Ser-175 are in bold. (B) Haploid yeast strain KSY606, carrying an inactive chromosomal TIF6 gene and harboring the wild-type URA3-based TIF6 expression plasmid pRS316-TIF6 that expresses untagged Tif6p, was transformed with different LEU2-based TIF6 expression plasmids. These LEU2 plasmids expressed either HA-tagged wild-type Tif6p (+), HA-tagged mutant Tif6p (Ser-to-Ala or Ser-to-Asp mutation as shown), or no Tif6p ORF (−). Transformants were initially selected on SD−His−Leu−Ura plates (SD−H−L−U) and then replica plated onto (i) SD−His−Leu−Ura and (ii) SD−His−Leu plates with the addition of Ura and 5-FOA (SD−H−L+U+5FOA). The plates were then incubated at 30°C for 3 and 5 days, respectively. (C) Haploid yeast cells harboring both the URA3 plasmid pRS316-TIF6 and the different recombinant LEU2 expression plasmids expressing either the HA-tagged wild-type or HA-tagged mutant (S174A; S175A; and S174A, S175A) Tif6p proteins were recovered from the SD−His−Leu−Ura plates and grown to mid-logarithmic phase in SD medium. Cell lysates were then analyzed by Western blotting by using anti-HA antibodies. Lysates prepared from KSY606 cells that expressed untagged wild-type Tif6p were also analyzed as a control.
Figure 4B shows that cells transformed with control LEU2 plasmid pRS315-TIF6(-HA) (which expresses wild-type Tif6p from its own endogenous promoter) grew on 5-FOA plates as expected. Cells transformed with the vector pRS315 failed to grow. Cells transformed with the mutant Tif6p expression plasmids (mutant S174A and mutant S174A, S175A) also failed to grow on 5-FOA plates, while cells transformed with the mutant S175A gave rise to microcolonies, indicating a slow-growth phenotype. The functional defect of the mutant Tif6p proteins was not due to lack of expression of mutant Tif6p in yeast cells. Analysis of cell extracts prepared from His+ Ura+ Leu+ transformants carrying wild-type as well as mutant genes showed that both the HA-tagged wild-type and the mutant Tif6p proteins were expressed quite well in these cells (Fig. 4C). Furthermore, mutation of Ser-174 and Ser-175 of Tif6p to aspartate, to mimic negatively charged phosphate residues, also failed to support yeast cell growth (Fig. 4B). Taken together, these results show that the serine residue at position 174 of Tif6p is essential for cell growth and viability.
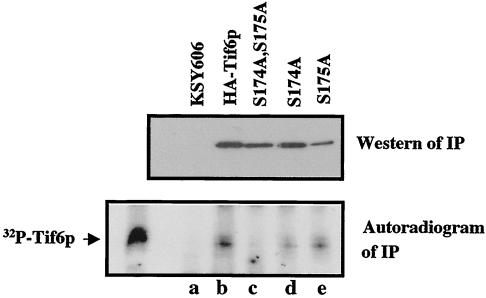
To investigate the phosphorylation status of Tif6p expressed from the mutant Tif6p expression plasmids, we grew the yeast strains (the His+ Ura+ Leu+ transformants shown in Fig. 4B, left panel), harboring both the untagged wild-type TIF6 gene in a URA3 plasmid and the HA-tagged mutant or wild-type TIF6 in a LEU2 plasmid, in low-phosphate medium. These cells were labeled with [32P]orthophosphate, and Tif6p present in the cell lysates was immunoprecipitated with anti-HA antibodies (Fig. 5). Whereas the wild-type HA-Tif6p was phosphorylated with 32P as expected, there was virtually no incorporation of 32P into the mutant HA-Tif6p (S174A, S175A) (Fig. 5, lower panel, compare lanes b and c). In contrast, the amount of 32P in the mutant Tif6p protein (S175A) was only somewhat reduced (approximately 15% reduction) (Fig. 5, lane e) while in the Tif6p (S174A) mutant, the 32P level was drastically reduced (about 75% reduction) compared to that in the wild-type protein (Fig. 5, lane d). Taken together, these results suggest that Tif6p, like its mammalian homologue eIF6, is also phosphorylated at Ser-174 and Ser-175 residues. Furthermore, phosphorylatable serine residues of Tif6p at these sites are essential for cell growth and viability (see Fig. 4).
FIG. 5.
Effect of mutation of Tif6p at Ser-174 and Ser-175 on its in vivo phosphorylation. The haploid yeast strains described in the legend to Fig. 4C were each grown separately in a 25-ml low-phosphate medium. At A600 of 0.5, the cells were treated with 5 mCi of [32P]orthophosphate (28). Following incubation for 3 h at 30°C, Tif6p was immunoprecipitated (IP) from each cell lysate by using agarose-conjugated anti-HA antibodies. The immunocomplexes formed in each case were washed; subjected to SDS-PAGE, followed by transfer onto a polyvinylidene difluoride membrane; and probed with anti-HA antibodies to detect HA-tagged Tif6p (upper panel). The blot was also analyzed by using autoradiography (lower panel).
Lack of phosphorylation of Tif6p affects its subcellular localization.
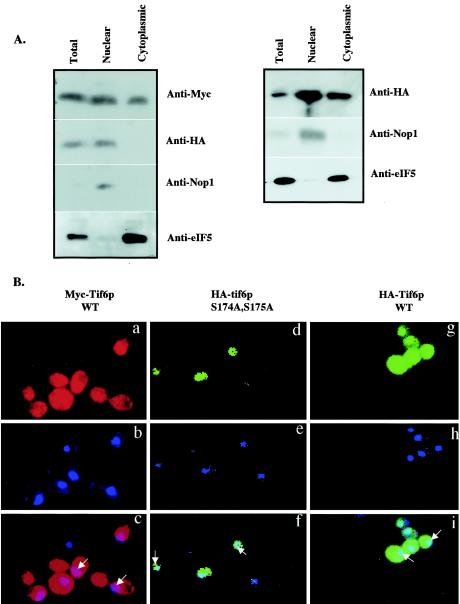
Cell fractionation as well as indirect immunofluorescence studies have shown previously that Tif6p is distributed throughout both cytoplasm and nuclei of yeast cells (2). To investigate whether mutation of Tif6p at Ser-174 and Ser-175, which results in the lack of phosphorylation of Tif6p, affects its subcellular localization, we constructed a strain in which the wild-type Tif6p is expressed as a Myc-tagged Tif6p fusion protein and the mutant Tif6p (S174A, S175A) is expressed as a HA-tagged protein. Cell extracts of this strain, UBY608, as well as a control strain UBY610 in which wild-type Tif6p is expressed as both a HA-tagged and a Myc-tagged protein from two CEN plasmids, were fractionated into nuclear and cytoplasmic fractions, and the proteins in these fractions were analyzed by Western blotting by using anti-c-Myc antibodies and anti-HA antibodies. Figure 6A shows that wild-type Tif6p tagged either with Myc or with HA (in UBY610) was present in both nuclear and cytoplasmic extracts, in agreement with previous results (2). In contrast, HA-tagged mutant Tif6p was present only in the nuclear fraction and was not detected in the cytoplasmic fraction. The validity of the fractionation was verified by using antibodies for eIF5 (a marker for cytoplasmic proteins) and Nop1p (a marker for nucleolar proteins) (Fig. 6A).
FIG. 6.
Subcellular distribution of wild-type Tif6p and the serine phosphorylation mutant of Tif6p. (A, left panel) Lysates of haploid yeast strain UBY608 carrying an inactive chromosomal TIF6 gene and harboring both the TRP1 plasmid pRS314-mycTIF6 (which expresses Myc-tagged Tif6p from its natural promoter) and a recombinant LEU2 plasmid pRS315-TIF6 (S174A, S175A), which expresses mutant HA-Tif6p protein (S174A, S175A) from its natural promoter, were fractionated into postnuclear supernatant (cytosolic fraction) and nuclear pellet as described by Aris and Blobel (1). (Right panel) Similar analysis was carried out with lysates of yeast strain UBY610 expressing both the Myc-tagged Tif6p and theHA-tagged wild-type Tif6p from its natural promoter. Approximately 30 μg of protein from each fraction was subjected to Western blot analysis by using anti-Myc, anti-HA, anti-Nop1, and anti-eIF5 antibodies as probes. (B) Localization of wild-type Myc-Tif6p and wild-type and mutant HA-Tif6p fusion proteins. Indirect immunofluorescence staining was performed with UBY608 cells expressing both the wild-type (WT) Myc-Tif6p (panels a to c) and the mutant HA-Tif6p fusion proteins (panels d to f). Similar indirect immunofluorescence staining was also carried out with UBY610 cells expressing both wild-type Myc-Tif6p and wild-type HA-Tif6p from two different CEN plasmids. Chromatin DNA of yeast cells was stained with DAPI. HA-Tif6p (d and g) was detected by immunofluorescence staining with monoclonal mouse anti-HA antibody and anti-mouse antibodies conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.). For Myc-Tif6p (a), the primary antibody used was anti-Myc antibodies and the secondary antibody used was anti-rabbit antibodies conjugated with Alexa Fluor 568 (Molecular Probes). In the third panels (c, f, and i), the two top panels (a and b, d and e, and g and h, respectively) were merged. Arrows indicate the positions of the nuclei.
To visualize directly the presence of Tif6p in subcellular fractions, exponentially growing cells of UBY608, expressing both wild-type Myc-Tif6p and mutant HA-Tif6p, were examined by indirect immunofluorescence by using anti-Myc and anti-HA antibodies followed by decoration with anti-rabbit red Alexa Fluor 568 (for wild-type Myc-Tif6p) and with anti-mouse green Alexa Fluor 488 (for mutant HA-Tif6p). Control cells, UBY610 expressing wild-type HA-Tif6p as well as wild-type Myc-Tif6p, were examined similarly. The nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). The fluorescence photograph showed that wild-type Tif6p tagged either with Myc (in UBY608) or with HA (in UBY610) was distributed evenly in the nuclei and the cytoplasm (Fig. 6B, panels a to c and g to i). In contrast, mutant HA-tagged Tif6p in UBY608 cells was localized only in the nuclei (Fig. 6B, panels d to f). No signal was obtained when cells in which untagged Tif6p was expressed were treated with a combination of the primary and secondary antibodies (data not shown).
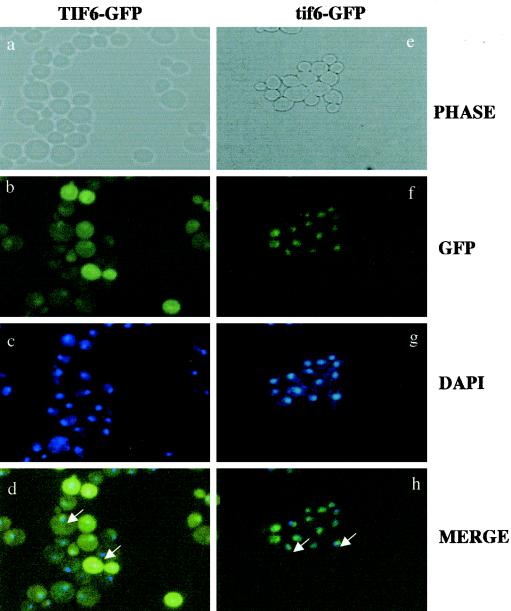
Similar localization of wild-type and Ala mutant Tif6p proteins was also observed when the wild-type or mutant protein was expressed as a C-terminally GFP-tagged Tif6p fusion protein in yeast cells (Fig. 7). Taken together, these results show that mutations of serine residues at positions 174 and 175 of Tif6p that abolish phosphorylation of Tif6p prevent its passage from the nucleus to the cytoplasm.
FIG. 7.
Localization of GFP-tagged wild-type and mutant Tif6p proteins. GFP and DAPI fluorescence of UBY612 cells (expressing GFP-tagged wild-type Tif6p) (a to d) and that of UBY613 cells (expressing GFP-tagged mutant Tif6p) (e to h) are shown. The arrow indicates the position of the cytoplasm.
Effect of phosphorylation of Tif6p on pre-rRNA processing.
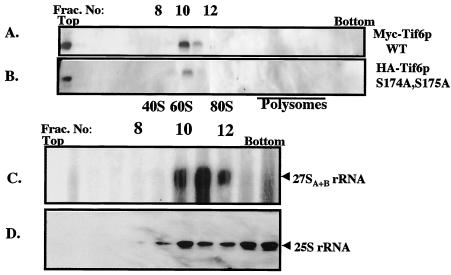
To date, the most well-characterized function of Tif6p in yeast cells is its requirement in the processing of pre-rRNA into mature 25S and 5.8S rRNAs that are the constituents of the 60S ribosomal subunit (2). Specifically, depletion of Tif6p from yeast cells blocks processing of 27SB pre-rRNA into 25S rRNA and 7S pre-rRNA, the latter being the precursor of 5.8S rRNA (2). In agreement with this essential function of Tif6p in pre-rRNA processing, it has been demonstrated that Tif6p is associated with the 60S preribosomal particles (also called 66S preribosomes) in the nucleolus (4, 10). To determine whether mutation of Ser-174 and Ser-175 of Tif6p to alanine that results in the formation of unphosphorylated Tif6p affects its ability to associate with the 60S preribosomal particles, cell extracts of an exponentially growing yeast strain (UBY608) expressing both the wild-type Myc-Tif6p and mutant HA-tagged Tif6p (S174A, S175A) were subjected to sucrose gradient centrifugation. Polysome-ribosome fractions were then analyzed for the association of wild-type and mutant Tif6p proteins with the 60S and/or 66S ribosomal particles. As shown in Fig. 8, both the wild-type (Fig. 8A) and mutant (Fig. 8B) Tif6p proteins were associated with the 60S preribosomal particles. Further confirmation that this fraction contains 60S preribosomal particles was obtained by showing the presence of 27S pre-rRNA in this fraction (Fig. 8C). The presence of 25S rRNA was used as a marker to identify the positions of 60S ribosomes in the gradient fractions (Fig. 8D). These results show that lack of phosphorylatable serine residues at positions 174 and 175 of Tif6p does not abolish the ability of Tif6p to associate with the 60S preribosomal particles.
FIG. 8.
Effect of lack of phosphorylation of Tif6p on its ability to bind to pre-60S ribosomal particles. (A) Cell extract prepared from UBY608 cells expressing both the wild-type Myc-Tif6p and Ala mutant HA-Tif6p was subjected to centrifugation on a 7 to 47% sucrose gradient as described previously (26). Wild-type (WT) Myc-Tif6p (A) and Ala mutant HA-Tif6p (B) in the gradient fractions were detected by immunoblot analysis. (C and D) RNA isolated from each gradient fraction was subjected to Northern blot analysis by using appropriate DNA probes that specifically detect 27SA+B and 25S rRNAs. The positions of 40S and 60S ribosomal subunits as well as those of 80S ribosomes and polysomes are indicated. Frac. No, fraction number.
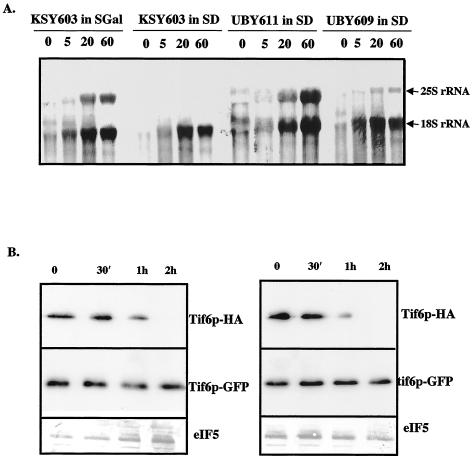
Experiments were also carried out to determine whether the presence of phosphorylatable serine residues at positions 174 and 175 of Tif6p is required for proper processing of pre-rRNA into mature 25S and 18S rRNAs. For this purpose, we constructed a yeast strain UBY609 (tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6] p[LEU2 tif6-GFP]) in which both the wild-type Tif6p and the Ala mutant Tif6p (S174A, S175A) were expressed from two different CEN plasmids. In this strain, while the wild-type Tif6p was expressed from the GAL10 promoter as an N-terminally ubiquitinylated HA-tagged fusion protein, the mutant Tif6p was expressed from its own promoter as a C-terminally GFP-tagged fusion protein. In addition, we constructed another control yeast strain, UBY611 (tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6] p[LEU2 TIF6-GFP]) which is similar to UBY609 except that it expresses the wild-type GFP-tagged Tif6p from its own promoter. Exponentially growing cultures of these strains and the parental strain KSY603 (tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6]) in SGal medium were transferred to SD-Ura (glucose) medium, and cells were allowed to grow for 120 min to deplete the wild-type HA-Tif6p from KSY603, UBY609, and UBY611 cells. As shown in Fig. 9B, there was a rapid decline of wild-type HA-Tif6p in both UBY609 and UBY611 cells. Within 2 h, the wild-type Tif6p was barely detectable. This was also true for KSY603 cells, as previously observed (2) (data not shown). In contrast, the expression of the GFP-tagged Ala mutant Tif6p in UBY609 cells as well as that of GFP-tagged wild-type Tif6p in UBY611 cells remained uninhibited under these conditions (Fig. 9B). Cells were then pulse-labeled with [5,6-3H]uracil for 3 min and chased for 5, 20, and 60 min with an excess of unlabeled uracil. As a control, the strain KSY603 growing in SGal medium was also subjected to pulse-chase labeling under similar experimental conditions. Analysis of total [3H]RNA samples by formaldehyde-agarose gel electrophoresis showed that in control KSY603 cells growing in SGal medium as well as in UBY611 cells growing in SD medium, the pre-rRNA was chased into mature 25S and 18S rRNAs as expected (Fig. 9A), while in Tif6p-depleted KSY603 cells growing in SD medium, virtually no 25S rRNA was formed (Fig. 9A). These observations are in agreement with previous results (2). More importantly, in UBY609 cells, growing in glucose medium and expressing only the Ala mutant Tif6p (Fig. 9B), the formation of both 25S and 18S rRNAs was readily observed, although there was clearly a significant inhibition in the formation of mature 25S rRNA without affecting the level of 18S rRNA (Fig. 9A). Additionally, from three such independent sets of experiments, we have calculated the ratios of 25S rRNA to 18S rRNA formed in strains UBY609 and UBY611. The results show there was about 50 to 70% inhibition of the formation of 25S rRNA in strain UBY609 expressing Ala mutant Tif6p compared to that in strain UBY611 expressing wild-type Tif6p.
FIG. 9.
Effect of mutation of Ser-174 and Ser-175 of Tif6p on its ability to function in pre-rRNA processing. (A) Exponentially growing cultures (100 ml each) of UBY609 {[GAL10::Ub-HA-TIF6] p[LEU2 tif6-GFP(S174A, S175A)]} and strains UBY611 ([GAL10::Ub-HA-TIF6] p[LEU2 TIF6-GFP]) and KSY603 [GAL10::Ub-TIF6] in SGal-Ura medium (A600 = 0.5) were transferred to equal volumes of SD-Ura medium and allowed to grow for 120 min. Each culture was centrifuged, suspended in 1 ml of SD-Ura medium, pulsed with 200 μCi of [5,6-3H]uracil for 3 min, and then chased with an excess of unlabeled uracil (1 mg/ml) for the indicated times. A culture of KSY603 grown in SGal-Ura medium was also pulsed and chased in SGal-Ura medium as a control. Total cellular RNA was isolated from each batch of cells. For each time point, an RNA sample containing about 10,000 cpm of 3H radioactivity was analyzed in a 1.2% formaldehyde-agarose gel followed by fluorography as described previously (2). The positions of mature 18S and 25S rRNAs are indicated. (B, left panel) Immunoblot analysis of wild-type GFP-Tif6p and wild-type HA-Tif6p fusion proteins isolated at the indicated times from UBY611 cells following transfer from SGal-Ura medium to SD-Ura medium. (Right panel) Immunoblot analysis of mutant GFP-tagged Tif6p and wild-type HA-tagged Tif6p. Western blot analysis of eIF5 represents loading controls in gels.
DISCUSSION
The synthesis of 60S ribosomes is a highly complex process requiring the participation of a large number (>100) of transacting factors that are required for pre-rRNA processing, pre-rRNA modification, and ribosome assembly (5, 13, 29). Tif6p, the yeast homologue of mammalian eIF6, is one of these essential 60S ribosomal assembly proteins that associates with the 60S preribosomal particles and is required for the subsequent maturation of the 60S ribosomal particle (2, 4, 10).
In this study, using in vitro biochemical techniques, we initially show that mammalian eIF6 is phosphorylated at Ser-174 and Ser-175 by mammalian CK I. Among these residues, Ser-174 is the major site of phosphorylation, accounting for nearly 90% of the total in vitro phosphorylation, while Ser-175 is phosphorylated quite inefficiently. The high conservation of the amino acid sequence surrounding these serine residues in nucleated cells from yeast to mammals suggested the possibility that these residues of eIF6 (Tif6p) are also phosphorylated in yeast cells. Indeed, we observed that while wild-type yeast eIF6 (Tif6p) isolated from 32P-labeled yeast cells was phosphorylated, mutation of Tif6p at the same residues, present in a highly conserved consensus CK I sequence motif, abolished phosphorylation of Tif6p. In yeast cells also, mutation of Ser-174 to alanine in Tif6p led to a marked reduction (about 75%) in phosphorylation of Tif6p, while mutation of Ser-175 inhibited phosphorylation only marginally. These observations, which showed a strong correlation between in vitro phosphorylation of mammalian eIF6 and in vivo phosphorylation of Tif6p in yeast, have been taken as evidence that these two serine residues, Ser-174 and Ser-175, are the sites of in vivo phosphorylation in yeast cells. It should be noted that the mammalian and the yeast eIF6 proteins are 72% identical and 85% similar to each other (25), suggesting a strong conservation in structure and function of the proteins.
The question naturally arises whether replacement of highly conserved serine residues with alanine in Tif6p causes alteration in the overall structure of Tif6p so as to prevent phosphorylation at other sites in the protein. The following evidence (data not shown) strongly suggests that this is not the case. First, derivation of the Stokes' radius by Sephadex G-75 gel filtration showed that both the wild-type eIF6 and the Ala mutant recombinant eIF6 have the same values as that obtained for native eIF6 isolated from rabbit reticulocyte lysates. Second, an important biochemical property of eIF6 isolated from both mammalian and wheat germ cell extracts is its ability to bind specifically to mature 60S ribosomal subunits in vitro (21, 22, 25-27). Both the wild-type and the Ala mutant mammalian eIF6 proteins can also bind to mature 60S ribosomal subunits quite efficiently. Additionally, as shown in Fig. 8, in yeast cells, both the wild-type and the Ala mutant Tif6p proteins were found to be associated with the pre-60S ribosomal particles. Furthermore, the crystal structures of yeast and archaebacteria eIF6 proteins (8) show a very compact domain without any unstructured regions, indicating that any binding surface requires intact, properly folded protein. In view of our finding that the binding property of eIF6 with regard to 60S particles in vivo and in vitro remains intact, these observations support strongly the notion that these mutations do not affect the overall tertiary structure of the protein.
Systematic investigation of the known properties of Tif6p in yeast showed that nuclear export of Tif6p requires the presence of the phosphorylatable serine residues, Ser-174 and Ser-175. In yeast cells expressing the wild-type as well as the mutant (S174A, S175A) Tif6p proteins, while the wild-type protein is distributed in both the nuclei and the cytoplasm, the Ala mutant Tif6p is localized exclusively in the nuclei (Fig. 6 and 7). In fact, consistent with our observation that mutation of Ser-174 alone to alanine in Tif6p led to a drastic reduction in phosphorylation of Tif6p in yeast cells (Fig. 5) as well as to a loss of cell growth and viability (Fig. 4), we have observed that in yeast cells expressing mutant Tif6p in which only Ser-174 was mutated to alanine, the mutant protein was localized exclusively in the nuclei (data not shown). In contrast, Tif6p in which only the minor phosphorylatable site, Ser-175, was mutated to alanine was distributed both in the nuclei and the cytoplasm (data not shown). Taken together, these results show a strong correlation between phosphorylation of Tif6p and its transport from the nucleus to the cytoplasm as well as its ability to support yeast cell growth.
An important function of Tif6p in yeast cells is its association with 60S preribosomal particles (4, 10, 17) and participation in the processing of the 27S pre-rRNA to form mature 25S and 5.8S rRNAs (2). In yeast cells depleted of Tif6p, the processing of 27SB pre-rRNA is blocked and virtually no 25S rRNA (and 5.8S rRNA) is formed (2). We observed that phosphorylatable serine residues Ser-174 and Ser-175 are not essential for the ability of Tif6p to bind to the 60S preribosomal particles (Fig. 8A and B). Likewise, when the ability of the mutant Tif6p to participate in the pre-rRNA processing reaction was measured, we observed that in contrast to Tif6p-depleted yeast cells, where virtually no 25S rRNA was formed, yeast cells expressing only the Ala mutant Tif6p showed formation of 25S rRNA, albeit at a somewhat lower level than cells expressing the wild-type Tif6p (Fig. 9). It is not immediately apparent why the lack of phosphorylation of Tif6p leads to a reduction in the formation of mature 25S rRNA. It has been postulated that the nuclear export of 60S ribosomes to the cytoplasm, which requires association of the protein NMD3 containing a nuclear export signal to the 60S preribosomal particles (7, 11), also requires Tif6p (12, 24). The possibility exists that lack of phosphorylation of Tif6p affects nuclear export of pre-60S ribosomal particles containing bound Tif6p. In the absence of such an export of pre-60S ribosomal particles, the 25S rRNA formed may be degraded. This may explain our observation that in the absence of nuclear export of Tif6p, the formation of mature 25S rRNA was eventually affected. Additionally, our observation that Tif6p in which Ser-174 and Ser-175 were mutated to aspartate residues also failed to substitute for the wild-type Tif6p in sustaining yeast cell growth and viability suggests that dephosphorylation of Tif6p may also be essential for its function. It should be noted that nucleocytoplasmic transport of many proteins is regulated by signals that result in the change in their phosphorylation states (for review, see reference 15). We postulate that phosphorylation and dephosphorylation of Tif6p may also be involved in nucleocytoplasmic shuttling of the protein, which is essential for its function.
Finally, because in vitro biochemical work has shown that mammalian eIF6 is phosphorylated in vitro at Ser-174 and Ser-175 by CK I and that mutation to alanine of the same residues of yeast eIF6, present in a highly conserved consensus CK I sequence, abolishes phosphorylation of eIF6 in yeast cells, it is presumed that yeast CK I is responsible for phosphorylation of Tif6p at the same residues. Both mammalian and yeast cells contain multiple isoforms of CK I that are associated with different compartments of the cell (9). These isoforms have highly conserved catalytic domains but differ from one another in having divergent amino- and carboxy-terminal domains. These noncatalytic domains are responsible for discrete cellular localization of each isoform, which is essential for the respective function of each isoform (9). In yeast there are five isoforms of CK I, only one (HRR25) of which is present in the nucleus (9). This isoform is the yeast homologue of mammalian CK Iδ, which is also predominantly nuclear and is shown in this work to phosphorylate mammalian eIF6. It remains to be determined whether the protein kinase responsible for phosphorylation of yeast eIF6 is indeed CK I and, if so, which isoform(s) of CK I is responsible for phosphorylation of Tif6p in yeast and at which subcellular location phosphorylation of Tif6p occurs. Additionally, it will be important to investigate the effect of mutational inactivation of the kinase on Tif6p function as well as on its nucleocytoplasmic distribution.
Acknowledgments
We are especially indebted to Steven Almo of our institution for many helpful discussions regarding the effect of mutations on the tertiary structure of eIF6. We thank Jonathan Warner and Romit Majumdar for critically reading the manuscript.
This work was supported by grant GM15399 from the National Institutes of Health and by Cancer Core Support Grant P30CA13330 from the National Cancer Institute.
Footnotes
This paper is dedicated to the loving memory of Amar Bhadhuri. He was an outstanding scientist and an extraordinary human being who inspired many students and scientists in India, by advice and example, to do research for the advancement of knowledge.
REFERENCES
- 1.Aris, J. P., and G. Blobel. 1991. Isolation of yeast nuclei. Methods Enzymol. 194:735-749. [DOI] [PubMed] [Google Scholar]
- 2.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevesich, J., J. Chaudhuri, and U. Maitra. 1993. Characterization of mammalian translation initiation factor 5 (eIF-5). Demonstration that eIF-5 is a phosphoprotein and is present in cells as a single molecular form of apparent M(r) 58,000. J. Biol. Chem. 268:20659-20667. [PubMed] [Google Scholar]
- 4.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 5.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 6.Flotow, H., and P. J. Roach. 1991. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 266:3724-3727. [PubMed] [Google Scholar]
- 7.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groft, C. M., R. Beckmann, A. Sali, and S. K. Burley. 2000. Crystal structures of ribosome anti-association factor IF6. Nat. Struct. Biol. 7:1156-1164. [DOI] [PubMed] [Google Scholar]
- 9.Gross, S. D., and R. A. Anderson. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 10.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 11.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27:580-585. [DOI] [PubMed] [Google Scholar]
- 13.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiti, T., A. Bandyopadhyay, and U. Maitra. 2003. Casein kinase II phosphorylates translation initiation factor 5 (eIF5) in Saccharomyces cerevisiae. Yeast 20:97-108. [DOI] [PubMed] [Google Scholar]
- 15.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 16.Merrick, W. C., and J. W. B. Hershey. 1996. The pathway and mechanism of eukaryotic protein synthesis, p. 31-69. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raychaudhuri, P., E. A. Stringer, D. M. Valenzuela, and U. Maitra. 1984. Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr = 25,000). J. Biol. Chem. 259:11930-11935. [PubMed] [Google Scholar]
- 19.Rose, M. D., F. Winston, and P. Heiter. 1989. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Rowles, J., C. Slaughter, C. Moomaw, J. Hsu, and M. H. Cobb. 1991. Purification of casein kinase I and isolation of cDNAs encoding multiple casein kinase I-like enzymes. Proc. Natl. Acad. Sci. USA 88:9548-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell, D. W., and L. L. Spremulli. 1980. Mechanism of action of the wheat germ ribosome dissociation factor: interaction with the 60 S subunit. Arch. Biochem. Biophys. 201:518-526. [DOI] [PubMed] [Google Scholar]
- 22.Russell, D. W., and L. L. Spremulli. 1979. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J. Biol. Chem. 254:8796-8800. [PubMed] [Google Scholar]
- 23.Sanvito, F., S. Piatti, A. Villa, M. Bossi, G. Lucchini, P. C. Marchisio, and S. Biffo. 1999. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 144:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 25.Si, K., J. Chaudhuri, J. Chevesich, and U. Maitra. 1997. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc. Natl. Acad. Sci. USA 94:14285-14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si, K., and U. Maitra. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valenzuela, D. M., A. Chaudhuri, and U. Maitra. 1982. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6). J. Biol. Chem. 257:7712-7719. [PubMed] [Google Scholar]
- 28.Warner, J. R. 1991. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 194:423-428. [DOI] [PubMed] [Google Scholar]
- 29.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]
- 30.Wood, L. C., M. N. Ashby, C. Grunfeld, and K. R. Feingold. 1999. Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J. Biol. Chem. 274:11653-11659. [DOI] [PubMed] [Google Scholar]