Abstract
Lymphocyte granule serine proteases (granzymes) play a critical role in protecting higher organisms against intracellular infections and cellular transformation. The proteases have also been implicated in the generation of tissue damage in a variety of chronic human conditions, including autoimmunity and transplant rejection. Granzyme B (GrB), one cytotoxic member of this family, achieves its effect through cleavage and activation of caspases as well as through caspase-independent proteolysis of cellular substrates. The 100,000-molecular-weight (100K) assembly protein of human adenovirus type 5 (Ad5-100K) was previously defined as a potent and specific inhibitor of human GrB. We now show that although human, mouse, and rat GrB proteases are well conserved in terms of structure, substrate specificity, and function, Ad5-100K inhibitory activity is directed exclusively against the human protease. Biochemical analysis demonstrates that the specificity of the 100K protein for human GrB resides in two distinct interactions with the protease: (i) a unique sequence within the reactive site loop (P1)Asp48-(P1′)Pro49 in Ad5-100K which interacts with the active site and (ii) the presence of an additional inhibitor-enzyme interaction likely outside the enzyme catalytic site (i.e., an exosite). We have located this extended macromolecular interaction site in Ad5-100K within amino acids 688 to 781, and we have demonstrated that this region is essential for stable inhibitor-enzyme complex formation as well as efficient inhibition of human GrB. This novel component of the inhibitory mechanism of the 100K protein identifies a distinct target for selective inhibitor design, a finding which may be of benefit for diseases in which GrB plays a pathogenic role.
Cytotoxic lymphocytes, which include cytotoxic T cells and natural killer cells, represent the primary defense against viral and intracellular bacterial infections and are involved in organ transplant rejection and antitumor responses (29, 49). Upon recognition by the cytotoxic effector cell, apoptosis of the target cell is induced through pathways initiated by the Fas receptor or through granule exocytosis (6, 26, 37, 41, 45). Targeted proteolysis of cellular substrates is a central mechanism underlying both of these pathways (4, 5, 8, 9, 12, 20-22, 65, 66). Granule-mediated killing utilizes several granule components with unique activities. The most abundant components present in cytotoxic granules are perforin (a pore-forming protein) and a family of serine proteases termed granzymes (26, 30, 41). Five granzymes (A, B, H, K, and M) in humans, eight granzymes (A to G and M) in mice, and seven granzymes in rats (A, B, C, I, J, K, and M) have been described to date (30, 40).
While there is significant evidence to implicate granzymes in target cell cytolysis, the mechanisms involved remain incompletely understood. Granzyme B (GrB), one of the most abundant proteases found in cytotoxic granules (42), catalyzes the cleavage and activation of several downstream caspases, inducing apoptotic changes in target cells (46). In addition, GrB can initiate caspase-independent pathways of cell death (31, 47, 54) through direct proteolysis and activation of molecules such as DFF45/ICAD (56) as well as cleavage of several substrates involved in cellular homeostasis (4, 13, 23). GrB is unique among mammalian serine proteases for its strict requirement for aspartic acid in the substrate P1 position. By use of combinatorial substrate libraries, the amino acid preferences at additional subsites have been defined for human GrB (58) and rat GrB (24) and are very similar for the two proteases. Rat and human GrB proteases are also quite similar at the structural level, with the major difference residing in the Asp-specific S1 subsite, which is significantly larger in the human enzyme (44, 61).
Adenoviruses are DNA viruses that have adopted several strategies to prevent the apoptosis of infected target cells (27, 62). These include (i) forced degradation of Fas and TRAIL receptor 1 by RID (previously named E3-10.4K/14.5K) (59, 60), (ii) direct interaction with and inhibition of members of the Fas signaling complex (E1B19K and E314.7) (15, 28, 39), (iii) binding with p53 and inhibition of p53-induced apoptosis (E1B55K and E4orf6) (19, 35, 48, 55, 63, 64), and (iv) expression of the antiapoptotic bcl-2 homologue E1B19K (11, 16, 38). Andrade et al. recently defined the 100,000-molecular-weight (100K) assembly protein of human adenovirus type 5 (Ad5-100K) as a potent and specific inhibitor of human GrB. This inhibitory effect is absolutely dependent on Asp48 in the AD5-100K protein, which is found within the GrB consensus motif Ile-Glu-Gln-Asp48 (2). The AD5-100K protein is a nonstructural protein which has essential functions in the adenovirus life cycle, including virus assembly and activation of late viral protein synthesis (14, 25, 43), and is present and well conserved in all members of the Adenoviridae family (18).
In these studies, we show that although human, mouse, and rat GrB proteases are well conserved in terms of structure, substrate specificity, and function, Ad5-100K has inhibitory activity exclusively against the human protease. Biochemical analysis demonstrates that the specificity for human GrB inhibition by Ad5-100K is the result of a unique cleavage preference for the site at Asp48 in Ad5-100K by the human but not the mouse or rat protease, the presence of Pro49 at the P1′ position, and the requirement for an additional inhibitor-enzyme interaction outside the enzyme catalytic site (i.e., an extended macromolecular recognition domain, or exosite). This additional component of the inhibitory mechanism may provide a critical target for the development of highly potent and specific inhibitors of GrB, which may be useful therapeutic tools for chronic human diseases in which GrB likely plays a role (e.g., graft rejection and autoimmunity) (3, 13, 36).
MATERIALS AND METHODS
Materials.
Human purified GrB from YT cells and recombinant rat GrB from Pichia pastoris were kindly provided by Nancy A. Thornberry (Merck & Co., Inc., Rahway, N.J.). Recombinant mouse GrB from P. pastoris was obtained from Sigma. Patient sera were used to immunoblot nuclear mitotic apparatus protein (NuMA), poly(ADP-ribose) polymerase (PARP), and Mi-2 (13). Rabbit polyclonal antibodies to the 100K protein were raised against a full-length recombinant His fusion protein (2). Monoclonal antibodies against epitope tag T7 and GrB were purchased from Novagen (Madison, Wis.) and Kamiya Biomedical Company (Seattle, Wash.), respectively. Immunoblotted proteins were detected by using a Supersignal substrate system (Pierce, Rockford, Ill.).
Cell culture, viral infection, and cell transfection.
NIH 3T3 and 293T cells were cultured by standard procedures. Confluent NIH 3T3 cell monolayers were infected with murine adenovirus type 1 (MAV-1) at 40 PFU/cell, followed by incubation at 37°C in a 5% CO2 humidified incubator for 48 h. Biochemical analysis was performed after these procedures. 293T cells were transfected by using Lipofectamine 2000 (Gibco BRL) as recommended by the manufacturer.
GrB cleavage of endogenous substrates in cell lysates.
Cell lysates were prepared as described previously (4). Lysates were incubated with 5 mM iodoacetamide for 10 min at 4°C to inactivate endogenous caspases and then incubated at 37°C for 60 min in the presence or absence of purified GrB. Equimolar amounts of human, mouse, and rat GrB proteases were confirmed based upon equal proteolytic activities against macromolecular substrates (i.e., NuMA, PARP, and Mi-2). The reactions were stopped by boiling in sodium dodecyl sulfate (SDS) sample buffer, and samples were electrophoresed on SDS-10% polyacrylamide gels containing 0.058% bisacrylamide. Intact autoantigens and their specific GrB cleavage fragments were visualized by immunoblotting as described previously (13).
Expression vectors, 100K protein cloning, and purification.
Ad5-100K was cloned and purified as described previously (2). For cloning of the 100K protein of MAV-1 (MAV1-100K), total RNA was purified from MAV-1-infected NIH 3T3 cells 48 h postinfection with Trizol (Gibco BRL) as described by the manufacturer. First-strand cDNA synthesis was performed with SuperScript II (Gibco BRL) according to the manufacturer's directions. Two microliters of this cDNA was used as a template in a PCR with Pfu and Taq DNA polymerases (1:1) and with oligonucleotides containing restriction enzyme adapters. The PCR-generated cDNA fragment containing the entire MAV1-100K open reading frame was cloned into pcDNA3.1(+) (Invitrogen). 100K protein hybrids and deletion mutants were generated by either site-directed mutagenesis (Stratagene) or PCR. All of the constructs, as well as green fluorescent protein (GFP) cDNA from pEGFP-N3, were cloned into prokaryotic expression vector pET28(+) (Novagen), generating N-terminal His6-T7-tagged fusion proteins that were further purified as previously described (2). The purity of the preparations was ∼70 to 90%, as assessed by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining (data not shown).
GrB cleavage of substrates generated by coupled IVTT.
[35S]methionine-labeled substrates were generated by in vitro transcription-translation (IVTT; Promega), and cleavage reactions were performed with ICE buffer (10 mM HEPES-KOH, 2 mM EDTA, 1% NP-40 [pH 7.4]). After incubation at 37°C for 30 min, reactions were terminated and samples were electrophoresed on SDS-10% polyacrylamide gels. Radiolabeled proteins and their fragments were visualized by fluorography. To better resolve the difference between full-length Ad5-100K and its 95-kDa human GrB cleavage fragment, N-terminal His6-T7-tagged Ad5-100K (which retards the migration of Ad5-100K to an apparent molecular mass of 110 kDa) was used as a substrate for the human, mouse, and rat GrB cleavage reactions.
Analysis of GrB inhibition by purified 100K protein and its mutants.
A fixed amount of purified GrB (25 nM) was preincubated with increasing amounts of purified recombinant Ad5-100K (rAd5-100K) or its mutants in ICE buffer at 37°C for 10 min. The residual proteolytic activity of GrB was determined by adding [35S]methionine-labeled human Mi-2 and incubating the mixture for an additional 15 min at 37°C. As a control, the radiolabeled substrate was incubated with buffer alone. The reactions were terminated by boiling in SDS sample buffer. Samples were analyzed by electrophoresis on SDS-8% polyacrylamide gels, and radiolabeled proteins and their fragments were visualized by fluorography.
Calculation of catalytic constant values.
Catalytic constant (kcat/Km) values were determined by using subsaturating substrate concentrations. Substrate and product bands on autoradiograms were scanned by densitometry to calculate the percentages of substrate cleavage. To calculate kcat/Km, these values were fitted to the following first-order rate equation: 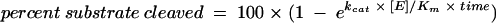 . In this equation, [E] is the concentration of enzyme.
. In this equation, [E] is the concentration of enzyme.
RESULTS
Ad5-100K inhibits human GrB but not mouse or rat GrB.
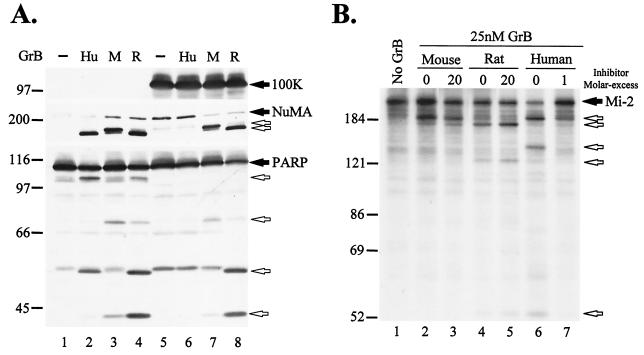
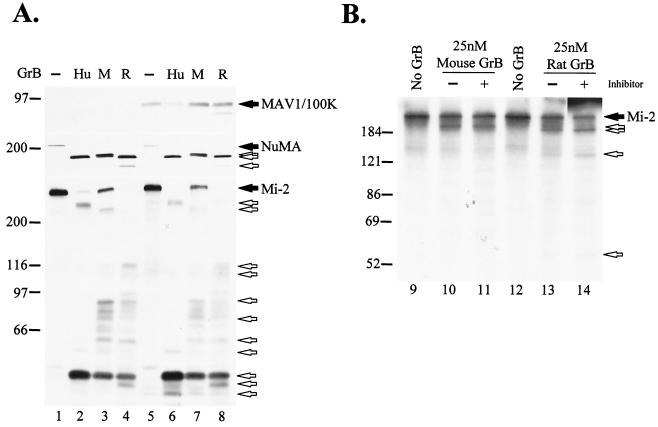
Proteolysis of signature death substrates is a characteristic feature of cytotoxic lymphocyte granule-induced death (4). It was previously shown that GrB directly and efficiently cleaves several downstream caspase substrates, generating unique fragments (4, 13). To compare the effects of Ad5-100K on the activities of human GrB, mouse GrB, and rat GrB, we studied the proteolytic activities of these proteases against macromolecular substrates in lysates from cells expressing Ad5-100K and in which caspases were inactivated by incubation with iodoacetamide (13). Interestingly, even though all three granzymes directly cleaved NuMA and PARP in lysates from mock-transfected cells, the cleavage patterns for fragments generated by the three enzymes were different (Fig. 1A, compare lanes 2, 3, and 4). When GrB activity was tested in lysates from cells expressing the 100K protein, the cleavage of NuMA and PARP by human GrB was abolished (Fig. 1A, lane 6), as previously described (2). In contrast, Ad5-100K did not significantly inhibit the cleavage of NuMA or PARP by mouse GrB and rat GrB (Fig. 1A, lanes 7 and 8, respectively).
FIG. 1.
Ad5-100K inhibits human but not mouse or rat GrB. (A) Cell lysates from mock-infected (lanes 1 to 4) or Ad5-100K plasmid-transfected (lanes 5 to 8) 293T cells were incubated in the absence or presence of 100 nM human (Hu), 160 nM mouse (M), or 130 nM rat (R) GrB. Ad5-100K, NuMA, and PARP were detected by immunoblotting. Filled arrows denote intact proteins, and unfilled arrows denote GrB-specific fragments. (B) GrB (25 nM) was preincubated in the presence or absence of rAd5-100K (at the indicated enzyme/inhibitor molar ratios) at 37°C for 10 min, and residual GrB proteolytic activity was determined by cleavage of IVTT-generated radiolabeled Mi-2. As a control, the radiolabeled substrate was incubated with buffer alone (lane 1). Filled and unfilled arrows denote intact Mi-2 and its fragments, respectively. The experiments were performed on at least two separate occasions, with similar results. Numbers at left of panels indicate molecular weights.
The inhibitory activity of the 100K protein against GrB from different species was further examined by using purified components. Increasing amounts of purified rAd5-100K were titrated against a fixed amount of purified GrB, and the residual proteolytic activity of GrB against [35S]methionine-labeled human Mi-2 was determined. Mi-2 was cleaved by all three granzymes with a similar second-order rate constant (kcat/Km, ∼104 M−1 s−1). Interestingly, as previously observed for NuMA and PARP, the patterns of Mi-2 cleavage by the three granzymes were different. In the presence of equimolar amounts of rAd5-100K and human GrB, the cleavage of Mi-2 was decreased by 90% (Fig. 1B, compare lanes 6 and 7). In contrast, rAd5-100K did not inhibit the mouse (Fig. 1B, compare lanes 2 and 3) and rat (Fig. 1B, compare lanes 4 and 5) proteases, even when a 20-fold molar excess of the inhibitor was used. These data therefore demonstrate that even though Ad5-100K is a potent and specific inhibitor of human GrB, this molecule has no effect on the activities of the mouse GrB and rat GrB orthologues. In addition, these data highlight the striking observation that even though GrB proteases from different species have similar structures, their cleavage specificities for macromolecular substrates appear quite distinct. While structural differences in GrB proteases (e.g., different glycosylation patterns or free versus complexed with serglycin) that relate to the different sources of the enzymes used (i.e., native versus recombinant from baculovirus or P. pastoris) may influence protease substrate-inhibitor interactions, the cleavage specificities of human GrB (native or recombinant) and mouse GrB (native or recombinant) were determined exclusively by the species of origin and were independent of the source of purification (data not shown).
Ad5-100K is differentially cleaved by human GrB, mouse GrB, and rat GrB.
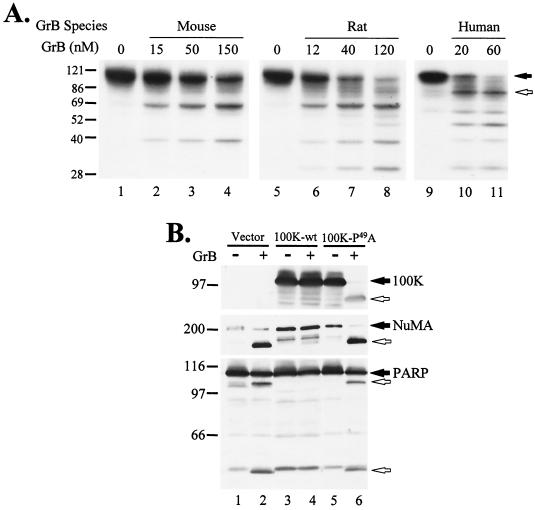
It was previously shown that 100K protein inhibition of human GrB is dependent on Asp48 in Ad5-100K, which is found within a classic GrB consensus motif (IEQD48-P) (2). Ad5-100K cleavage at Asp48 by human GrB is very efficient and generates a prominent 95-kDa fragment (Fig. 2A, lanes 10 and 11) which is abolished when Asp48 is changed to Ala (2). Since human GrB, mouse GrB, and rat GrB cleave the macromolecular substrates NuMA, PARP, and Mi-2 at different sites (see above), we addressed whether Ad5-100K was differentially cleaved by the human, mouse, and rat proteases. When [35S]methionine-labeled Ad5-100K was incubated in the presence of increasing concentrations of GrB, Ad5-100K was cleaved by the rat protease with an efficiency similar to that of the human protease (kcat/Km values, 1.5 × 104 and 3 × 104 M−1 s−1, respectively). In contrast, the cleavage of Ad5-100K by mouse GrB was approximately 1 order of magnitude less efficient than that by human GrB (kcat/Km value, 3 × 103 M−1 s−1). Interestingly, Ad5-100K cleavage at Asp48 by either mouse or rat GrB was very inefficient. The most prominent fragments generated by mouse GrB and rat GrB were ∼65 and 38kDa (Fig. 2A, lanes 2 to 4 and lanes 6 to 8, respectively); an additional 30-kDa fragment resulted from rat GrB cleavage (Fig. 2A, lanes 6 to 8). It is therefore likely that the efficient recognition of the IEQD48-P motif by different granzymes plays a role in the species selectivity of inhibition by Ad5-100K.
FIG. 2.
GrB-mediated cleavage and inhibition of 100K. (A) Human GrB, mouse GrB, and rat GrB cleave Ad5-100K at different sites. [35S]methionine-labeled His-Ad5-100K generated by IVTT was incubated in the absence (lanes 1, 5, and 9) or presence of mouse (lanes 2 to 4), rat (lanes 6 to 8), or human (lanes 10 and 11) GrB at the indicated concentrations. Filled and unfilled arrows denote intact Ad5-100K and the 95-kDa fragment generated by human GrB cleavage at Asp48, respectively. (B) P1′ Pro49 in Ad5-100K is essential for human GrB inhibition. Cell lysates from mock-infected (lanes 1 and 2) or Ad5-100K wild-type (wt) plasmid-transfected (lanes 3 and 4) or Ad5-100K-P49A plasmid-transfected (lanes 5 and 6) 293T cells were incubated in the absence or presence of 100 nM human GrB. Ad5-100K, NuMA, and PARP were detected by immunoblotting. Filled arrows denote intact proteins, and unfilled arrows denote their GrB-induced fragments. The experiments were repeated on two to four separate occasions, with similar results. Numbers at left of panels indicate molecular weights.
Importance of the P1′ residue in human GrB inhibition by Ad5-100K.
The presence of proline at the P1′ position in Ad5-100K is noteworthy, since proline at this position is disfavored by almost all proteases. We therefore addressed whether this proline plays a role in generating the inhibitory activity of Ad5-100K and whether this site may contribute to the species selectivity of cleavage or inhibition. When IEQD48↓P49 (arrow, cleavage site) was mutated to IEQD48↓A49 in Ad5-100K (i.e., Ad5-100K-P49A), neither its cleavage by mouse GrB and rat GrB nor its inhibitory activity against these proteases was changed (data not shown). However, Ad5-100K-P49A lost all inhibitory activity against the human protease and was very efficiently cleaved (Fig. 2B, compare lanes 4 and 6). Thus, these data strongly suggest that proline in the P1′ position of the 100K protein does not play a role in defining the species selectivity of Ad5-100K but is essential for the inhibition of human GrB. Interestingly, this inhibitory cleavage site (IEQD48↓P49) alone is not sufficient to inhibit human GrB, since deletion mutants of Ad5-100K containing this motif but lacking the C terminus were poor inhibitors of this protease (see below and Fig. 6); these results suggest that additional interactions (outside the cleavage site) are critical for the precise recognition and inhibition of human GrB by Ad5-100K. In addition, it is likely that these further species-specific enzyme substrate-inhibitor interactions outside the substrate binding pockets of the active site influence the cleavage specificity of each GrB for macromolecular substrates (e.g., NuMA, PARP, and Mi-2).
FIG. 6.
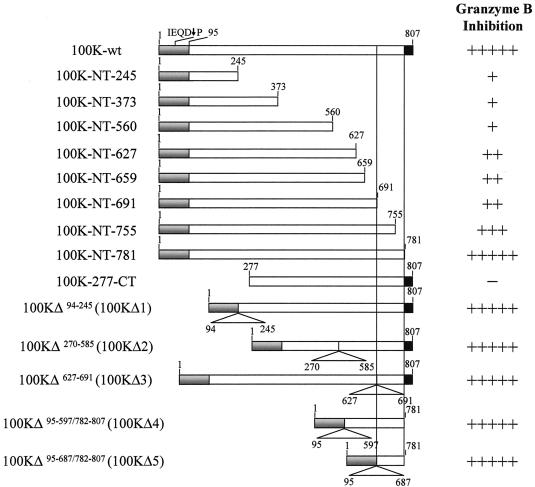
Analysis of human GrB inhibition by wild-type Ad5-100K and its deletion mutants. Schematic representation of wild-type Ad5-100K (100K-wt) and its deletion mutants. The Ad5-100K NTR is depicted in gray, the CCR is depicted in white, and the CTR is depicted in black. Human GrB inhibition was determined by preincubating increasing amounts of purified recombinant proteins with a fixed amount (25 nM) of human GrB (enzyme/inhibitor molar ratios of 1:0, 1:1, 1:3, 1:5, 1:10, and 1:20), and the residual GrB activity was determined by cleavage of IVTT-generated radiolabeled substrate Mi-2 (data not shown). Maximum GrB inhibition was defined as the residual GrB activity obtained at a 1:1 ratio of wild-type Ad5-100K and human GrB. Inhibition is represented as follows: +++++, 1:1 ratio; ++++, 1:3 ratio; +++, 1:5 ratio; ++, 1:10 ratio; +, 1:20 ratio; −, no inhibition at any ratio (from 1:1 to 1:20). Results are representative of at least two separate experiments.
The 100K assembly protein from MAV-1 is a substrate for mouse GrB without inhibitory activity against this protease.
Although the 100K assembly protein is well conserved among different adenovirus subgenera, no homology is observed at the amino- and carboxy-terminal extremes of these molecules (Fig. 3A). Therefore, the N-terminal region (NTR) (where the motif for human GrB inhibition in Ad5-100K is located) is unique for the 100K protein from different subgenera. The phylogeny of mastadenoviruses is largely similar to that of their hosts, supporting the view that most of these viruses have evolved exclusively with their hosts or have switched hosts only between closely related species (18). We therefore addressed whether MAV1-100K might have evolved specific inhibitory activity against mouse GrB similar to that for its human counterpart.
FIG. 3.
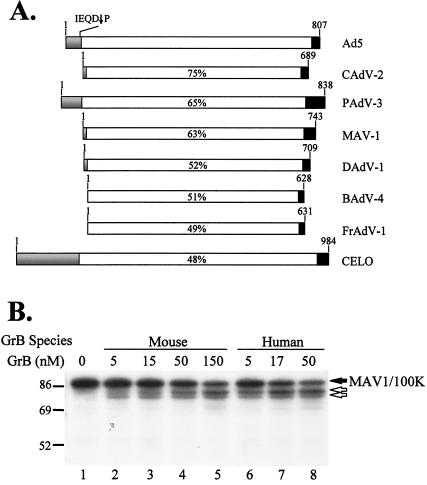
Species effects on 100K cleavage by GrB. (A) Analysis of 100K assembly protein sequences of members of the Adenoviridae family. Based on sequence homology among several adenovirus subgenera, the 100K protein can be divided into three regions, an NTR of ∼0 to 170 amino acids (shown in gray), a CCR of ∼600 to 700 amino acids (shown in white), and a CTR of ∼20 to 100 amino acids (shown in black). The percentage of homology between the various adenovirus CCRs and the Ad5-100K CCR is shown inside each CCR. The adenovirus subgenera analyzed included the following: Ad5, canine adenovirus type 2 (CAdV-2), porcine adenovirus type 3 (PAdV-3), MAV-1, duck adenovirus type 1 (DAdV-1), bovine adenovirus type 4 (BAdV-4), frog adenovirus type 1 (FrAdV-1), and fowl adenovirus type 1 (CELO). (B) Cleavage of the 100K assembly protein from MAV-1 by mouse GrB and human GrB. [35S]methionine-labeled MAV1-100K generated by IVTT was incubated in the absence or presence of increasing concentrations of mouse or human GrB. The filled arrow denotes intact MAV1-100K, and unfilled arrows denote GrB cleavage fragments. The experiments were performed twice on separate occasions, with identical results. Numbers at left of panel B indicate molecular weights.
Interestingly, [35S]methionine-labeled MAV1-100K generated in vitro was cleaved by mouse GrB (kcat/Km, 4.8 × 103 M−1 s−1) (Fig. 3B, lanes 2 to 5) and human GrB (kcat/Km, 2.3 × 104 M−1 s−1) (Fig. 3B, lanes 6 to 8), generating similar patterns of fragments, with prominent bands of ∼83 and 80 kDa. Since MAV1-100K was expressed at low levels in transfected cells (data not shown), we determined whether mouse GrB inhibitory activity was present in lysates from MAV-1-infected NIH 3T3 cells by analyzing the cleavage of endogenous mouse NuMA and Mi-2. No inhibitory activity against human, mouse, or rat GrB was observed in lysates from infected cells (Fig. 4A, lanes 5 to 8) compared to lysates from mock-infected cells (Fig. 4A, lanes 1 to 4).
FIG. 4.
Neither MAV-1 nor its assembly protein has inhibitory activity against mouse and rat GrB. (A) MAV-1 does not encode inhibitors for human, mouse, or rat GrB. Cell lysates from mock-infected (lanes 1 to 4) or MAV-1-infected (lanes 5 to 8) NIH 3T3 cells were incubated in the absence or presence of 100 nM human (Hu), 160 nM mouse (M), or 130 nM rat (R) GrB. MAV1-100K, NuMA, and Mi-2 were detected by immunoblotting. Filled arrows denote intact proteins, and unfilled arrows denote prominent GrB-specific fragments. (B) Purified rMAV1-100K does not inhibit the proteolytic activities of mouse GrB and rat GrB. Mouse or rat GrB (25 nM) was preincubated in the absence or presence of rMAV1-100K (enzyme/inhibitor molar ratio, 1:20) at 37°C for 10 min, and the residual GrB proteolytic activity was determined by cleavage of IVTT-generated radiolabeled Mi-2. As a control, the radiolabeled substrate was incubated with buffer alone (lane 9). Filled and unfilled arrows denote intact Mi-2 and its fragments, respectively. The experiments were performed on two separate occasions, with identical results. Numbers at left of panels indicate molecular weights.
Since MAV1-100K levels achieved in infected cells were still low compared to the levels of 100K protein expressed in human Ad5-infected cells (data not shown), we further examined the effect of MAV1-100K on the proteolytic activity of the different GrB proteases by using purified components. Increasing amounts of purified recombinant MAV1-100K (rMAV1-100K) were titrated against a fixed amount of purified GrB, and the residual activity was determined by quantifying the cleavage of [35S]methionine-labeled Mi-2. No inhibitory activity of rMAV1-100K was observed against mouse or rat GrB, even at a 20-fold molar excess of the inhibitor (Fig. 4B, lanes 11 and 14, respectively). Interestingly, at a 20-fold molar excess, rMAV1-100K inhibited human GrB (see below). MAV1-100K is thus a substrate for mouse GrB but has no inhibitory activity against this protease.
Evolution of Ad5-100K as an inhibitor of human GrB.
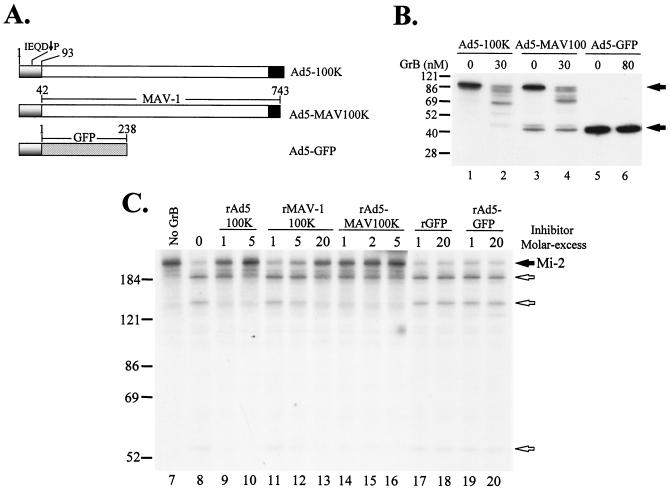
The 100K protein is a multifunctional protein with essential functions in the adenovirus life cycle. Sequence analysis of several adenovirus subgenera distinguished a major conserved region of ∼600 to 700 amino acids located centrally in the protein (central conserved region [CCR]) and a nonconserved region located at each terminus of the molecule (Fig. 3A). Interestingly, the NTR (which is unique to each adenovirus subgenus) contains the human GrB inhibitory motif in Ad5-100K. To better understand the importance of the Ad5-100K NTR and CCR in human GrB inhibition, we constructed a chimeric molecule (Ad5-MAV100K) containing the NTR of Ad5-100K (amino acids 1 to 93) and both the CCR and the C-terminal region (CTR) of MAV1-100K (amino acids 42 to 743) (Fig. 5A). Similarly, we also addressed whether the NTR of Ad5-100K alone can inhibit human GrB by fusing this region to GFP (Ad5-GFP) (Fig. 5A).
FIG. 5.
The chimera Ad5-MAV100K is a potent inhibitor of human GrB. (A) Schematic representation of Ad5-100K and the chimeras containing the NTR from Ad5-100K (Ad5-MAV100K and Ad5-GFP). The Ad5-100K NTR (amino acids 1 to 93) is depicted in gray, the CCR is depicted in white, the CTR is depicted in black, and full-length GFP is depicted by stippling. (B) Cleavage of Ad5-100K, Ad5-MAV100K, and Ad5-GFP by human GrB. [35S]methionine-labeled substrates generated by IVTT were incubated in the presence of 0, 30, or 80 nM human GrB. Filled arrows denote intact molecules. (C) The CCR and the Ad5-100K NTR are important in human GrB inhibition. Increasing amounts of rAd5-100K, rMAV1-100K, recombinant GFP (rGFP), or the chimeras rAd5-MAV100K and rAd5-GFP were preincubated with 25 nM human GrB (at the indicated enzyme/inhibitor molar ratios) as described in the legend to Fig. 4, and the residual GrB proteolytic activity was determined by cleavage of IVTT-generated radiolabeled Mi-2. As a control, the radiolabeled substrate was incubated with buffer alone (lane 7). Filled and unfilled arrows denote intact Mi-2 and its fragments, respectively. All experiments were performed on at least two separate occasions, with similar results. Numbers at left of panels B and C indicate molecular weights.
To address whether the chimeric molecules (which contained the human GrB consensus site) were substrates for human GrB, we initially determined their cleavage patterns in vitro. Both Ad5-100K and Ad5-MAV100K were cleaved by human GrB with similar efficiencies (Fig. 5B, lanes 2 and 4, respectively). In contrast, Ad5-GFP was not cleaved by human GrB, even at higher enzyme concentrations (Fig. 5B, lane 6). The presence of the NTR (containing the human GrB cleavage site) alone is therefore insufficient to render the molecule efficiently cleaved by GrB.
The human GrB inhibitory activities of the different molecules then were determined by using purified components in vitro and assaying GrB activity by quantifying Mi-2 cleavage. Equimolar amounts of rAd5-100K inhibited GrB significantly; a fivefold molar excess of the inhibitor completely abolished GrB activity (Fig. 5C, lanes 9 and 10). Interestingly, recombinant Ad5-MAV100K (rAd5-MAV100K) demonstrated a similarly potent inhibitory activity against human GrB (Fig. 5C, lanes 14 to 16). When similar studies were performed with mouse GrB and rat GrB, no inhibitory activity was observed, even with a 20-fold molar excess of the inhibitor (data not shown). Of note, although rMAV1-100K did not show inhibitory activity against the mouse and rat proteases, even when present at a molar excess (Fig. 4B), inhibitory activity against human GrB was observed at a 20-fold molar excess of the inhibitor (Fig. 5C, lane 13). In contrast, recombinant GFP (Fig. 5C, lanes 17 and 18) or recombinant Ad5-GFP (rAd5-GFP) (Fig. 5C, lanes 19 and 20) had no inhibitory effects on human GrB. Taken together, these data demonstrate that elements within both the NTR of Ad5-100K and the CCR and CTR are required for efficient and potent inhibition of human GrB.
Identification of a C-terminal domain in the 100K protein that is required for efficient inhibition and stable complex formation between Ad5-100K and human GrB.
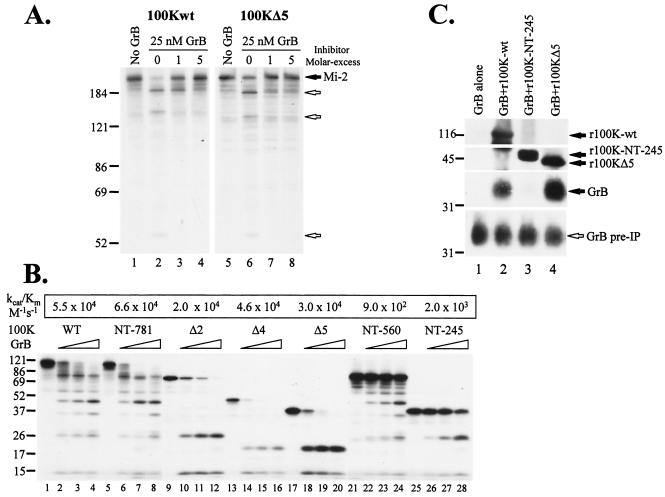
To determine whether an extended macromolecular recognition domain, in addition to the reactive-site loop (containing IEQD48-P), in Ad5-100K is required for human GrB inhibition, we generated a series of Ad5-100K deletion mutants (Fig. 6). Increasing amounts of wild-type rAd5-100K or deletion mutants were titrated against a fixed amount of purified human GrB, and the residual protease activity was determined by cleavage of [35S]methionine-labeled Mi-2. All deletion mutants that included the NTR of Ad5-100K showed some inhibitory activity against human GrB, although their potencies varied greatly (Fig. 6). Interestingly, the most potent inhibitors were those in which amino acids 688 to 781 at the C terminus (CT688-781) were included (except for 100K-277-CT, in which the NTR is absent). Indeed, deletion mutant r100KΔ5, which contains only the NTR plus CT688-781, was as efficient as wild-type rAd5-100K in inhibiting GrB. Thus, equimolar amounts of rAd5-100K or r100KΔ5 inhibited GrB significantly, and a fivefold molar excess of the inhibitor completely abolished GrB activity (Fig. 7A, lanes 3 and 4 and lanes 7 and 8, respectively).
FIG. 7.
Amino acids 688 to 781 (CT688-781) in Ad5-100K are required for efficient cleavage and/or recognition, inhibition, and stable complex formation between Ad5-100K and human GrB. (A) Wild-type Ad5-100K (100Kwt) and deletion mutant 100KΔ5 inhibit human GrB with similar efficiencies. Increasing amounts of purified recombinant proteins were preincubated with 25 nM human GrB, and the residual GrB activity was determined by cleavage of IVTT-generated radiolabeled Mi-2. Filled and unfilled arrows denote intact Mi-2 and its fragments, respectively. (B) The CT688-781 domain is required for efficient cleavage of wild-type 100K protein and its deletion mutants. [35S]methionine-labeled substrates generated by IVTT were incubated in the presence of 0, 20, 60, or 200 nM human GrB for 30 min at 37°C. After the reactions were terminated, samples were electrophoresed. The intact polypeptide and the cleavage products were detected by fluorography. WT, wild type. (C) The CT688-781 domain is required for stable Ad5-100K/human GrB complex formation. Equimolar amounts (300 nM) of human GrB and recombinant wild-type protein (r100K-wt) (lane 2), recombinant 100K-NT-245 (r100K-NT-245) (lane 3), or recombinant 100KΔ5 (r100KΔ5) (lane 4) were coincubated for 1 h at 4°C. As a control, GrB was incubated with buffer alone (lane 1). After incubation, an aliquot of each sample was boiled in SDS sample buffer and analyzed by immunoblotting with a monoclonal antibody against human GrB (bottom panel). The rest of the samples were immunoprecipitated with a polyclonal antibody against the 100K protein. After electrophoresis, r100K-wt, r100K-NT-245, and r100KΔ5 were visualized by immunoblotting with a monoclonal antibody against epitope tag T7 (which is present at the N terminus of all recombinant 100K proteins), and coimmunoprecipitated GrB was detected with the monoclonal antibody against GrB. The unfilled arrow denotes human GrB before immunoprecipitation (pre-IP). Filled arrows denote r100K-wt, r100K-NT-245, r100Δ5, and GrB after immunoprecipitation. Experiments were performed on two separate occasions, with similar results. Numbers at left of panels indicate molecular weights.
We next addressed the mechanism by which the CT688-781 domain contributes to GrB inhibition. In initial studies, the efficiency of cleavage of [35S]methionine-labeled wild-type Ad5-100K and deletion mutants by human GrB was defined (Fig. 7B). While the full-length protein and deletion mutants containing the CT688-781 domain were efficiently cleaved by GrB (average kcat/Km, 4.4 × 104 M−1 s−1) (Fig. 7B, lanes 1 to 20), mutants lacking the CT688-781 domain (100K-NT-245 and 100K-NT-560) were cleaved approximately 30-fold less efficiently (average kcat/Km, 1.4 × 103 M− s−1) (Fig. 7B, lanes 21 to 28). Interestingly, the deletion mutant containing only the NTR fused to CT688-781 (i.e., 100KΔ5) was cleaved as efficiently as the full-length protein (Fig. 7B, lanes 17 to 20 and lanes 1 to 4, respectively) and was equally potent as an inhibitor of GrB (Fig. 7A). These data were somewhat surprising, in light of previous observations demonstrating that IEQD48 within the NTR is the major cleavage site within the inhibitor and likely interacts with the active site of the protease (2). These results therefore support the idea that the efficiency of cleavage at IEQD48 is improved by an interaction with the CT688-781 domain.
Since extended macromolecular interactions outside the active site (exosite) in serine proteases frequently contribute significant binding energy and specificity to the interaction, we used coimmunoprecipitation analysis to address whether the CT688-781 domain in the 100K protein might be required for the establishment of a stable inhibitory complex with GrB. GrB and Ad5-100K or several deletion mutants were coincubated and immunoprecipitated with a polyclonal antibody that is directed against the 100K protein and that recognizes wild-type and mutant proteins. The 100K proteins were visualized by immunoblotting of N-terminal epitope tag T7, which is present on all of the 100K proteins, and GrB was visualized by immunoblotting with a monoclonal antibody against GrB. Full-length Ad5-100K formed a stable complex with human GrB (Fig. 7C, lane 2) but not mouse GrB (data not shown), as demonstrated by coprecipitation of both the inhibitor and the protease. In contrast, deletion mutant 100K-NT-245 did not coprecipitate human GrB (Fig. 7C, lane 3). Mutant 100KΔ5 did coprecipitate GrB (Fig. 7C, lane 4), demonstrating that the stable interaction of the 100K protein with the protease is species specific and requires two distinct elements: (i) interaction of the reactive-site loop with the active site of the protease and (ii) an additional interaction involving the CT688-781 domain with an as-yet-unidentified region on GrB. Both interactions are critical for stable binding of the protease and the inhibitor, as well as for efficient cleavage and potent inhibition.
DISCUSSION
The 100K assembly protein of pathogenic human Ad5 is a potent and specific inhibitor of human GrB. In these studies, we have demonstrated that this inhibitory activity is uniquely directed against the human protease and does not affect the mouse and rat orthologues. This result was initially unexpected, since these enzymes share a high degree of sequence homology (70% identity and 80% similarity), are structurally very similar (44, 61), share similar tetrapeptide substrate specificities (at least the human and rat proteases) (24, 58), and likely have the same functional roles in different species (1, 17, 23, 24, 30, 32, 40, 53, 58). However, in spite of such similarities, each GrB has a unique specificity for macromolecular substrates. This unique specificity of these enzymes for different cleavage sites is exemplified by the cleavage of Ad5-100K, in which only the human protease and not the murine and rat proteases can efficiently cleave at Asp48. When the residues within the GrB recognition motif (IEQD48) in Ad5-100K were mutated to a more optimal GrB cleavage site preferred by the rat protease (i.e., IETD48 and IEPD48), the efficiency of cleavage at Asp48 by the mouse or rat protease was unchanged; this result suggests that additional interactions between human GrB and Ad5-100K which are absent from (or are different in) the mouse and rat proteases are critical for the precise recognition and inhibition of human GrB (see below).
This species-specific interaction of GrB with its substrate or inhibitor has also been observed for other serpin inhibitors of GrB. Thus, the murine serpin SPI6 has potent inhibitory activity against murine GrB (33, 34) but is a relatively inefficient inhibitor of human GrB (50). In contrast, the human homologue of SPI6 (PI-9) has potent inhibitory activity against the human protease (52). Interestingly, GrB cleavage of PARP, as well as the activation of caspase 3 and Bid, does not appear to be species specific, since interspecies enzymes and substrates have been broadly used (i.e., rat, human, and mouse enzymes all cleave human, mouse, bovine, and monkey substrates) (1, 17, 23, 32, 53), although their cleavage efficiencies have not been addressed.
Although the 100K protein from mouse adenovirus does not inhibit mouse or rat GrB (even at a 20-fold molar inhibitor excess), it has some inhibitory activity against human GrB. Interestingly, fusion of the NTR of Ad5-100K with MAV1-100K (but not GFP) increased the inhibitory activity of MAV1-100K against human GrB by approximately 20-fold, demonstrating that there are at least two critical regions which define efficient 100K protein inhibition against human GrB: (i) a unique sequence within the reactive-site loop [(P1)Asp48-(P1′)Pro49] in Ad5-100K which interacts with the active site and (ii) the presence of an additional inhibitor-enzyme interaction likely outside the catalytic site. Indeed, by analyzing the inhibitory effects of different Ad5-100K deletion mutants, we defined a small region (amino acids 688 to 781) located toward the end of Ad5-100K which is crucial for the efficient inhibition of human GrB, likely through stabilizing the protease-inhibitor interaction (see below). This requirement for an extended interaction outside the GrB catalytic site is not exclusive for the Ad5-100K inhibitor but is likely required for all efficient GrB substrate interactions. This notion is supported by prior studies showing that the hydrolysis of synthetic substrates catalyzed by GrB is highly dependent on the length and sequence of the substrates; in addition, even a peptide comprising an optimized P4-P2′ sequence resulted in a molecule that was not cleaved efficiently by GrB, indicating that substrate recognition involves features beyond this recognition motif (24, 30).
All protein proteinase inhibitors prevent access of substrates to the proteinase catalytic site through steric hindrance. Members of one class of inhibitors (e.g., most serpins) achieve this goal through binding of a peptide segment directly to the catalytic site in a substrate- or product-like manner; the selectivity of inhibition is achieved through the utilization of the substrate recognition sites in the proteinase. Although these inhibitors make additional contacts with their cognate proteinases, these secondary contacts are in most cases not very specific and apparently not important for the tightness of binding (10). In contrast, other inhibitors (e.g., hirudin and related inhibitors) bind mainly through extended macromolecular interaction sites (exosites) adjacent to the catalytic residues of their cognate proteinases, utilizing substrate binding and other sites (10). Since Asp48 within the NTR of Ad5-100K is directly cleaved by GrB and the motif Asp48↓Pro49 is absolutely required for protease inhibition, it is likely that the consensus sequence containing IEQD48↓P49 is bound to the GrB active site. Interestingly, deletion mutants containing the N-terminal reactive-site loop but lacking the CT688-781 region are inefficiently cleaved by human GrB and are poor inhibitors of the protease. We therefore propose that the CT688-781 region in Ad5-100K interacts with human GrB through an exosite (likely specific for human GrB but not mouse or rat GrB). The binding site on GrB for CT688-781 is not yet known, nor is the mechanism whereby this additional interaction participates in the establishment of rapid and potent inhibition of the protease. Since CT688-781 alone neither has inhibitory activity against human GrB nor stably binds this protease (data not shown) but is required for potent inhibition by the NTR and since both domains are required for stable complex formation with GrB, these data support the proposal that the function of CT688-871 is to stabilize the interaction between the protease and the inhibitor, allowing stable complex formation and protease inhibition at the active site. Although the proline in the P1′ position likely disfavors GrB cleavage at IEQD through steric hindrance of the scissile bond by the proline secondary amide, this proline is essential for human GrB inhibition. Interestingly, the domain containing IEQD-P alone is insufficient for inhibition and requires the presence of CT688-871. It is therefore possible that this docking site (CT688-871) serves to enhance the recognition or cleavage of this suboptimal site. Thus, the optimal tetrapeptide sequence upstream of the cleavage site allows occlusion of the active site, while the P1′ proline disfavors a conformation which allows for efficient cleavage, dramatically slowing down the catalytic activity of the protease (kcat, 5.2 × 10−5/s) (2).
Since exosite interactions define the affinity and binding specificity within enzyme-substrate complexes (7), different exosite specificities among human GrB, mouse GrB, and rat GrB could explain the differences in cleavage site recognition for the same substrate. These differences in specificity of the proteases in different species raise significant obstacles for the use of GrB-deficient mice as a model from which to infer the role of GrB in human inflammatory and autoimmune pathology.
Despite their distinct catalytic mechanisms, GrB and the caspase family members share a novel and stringent specificity for aspartic acid in the P1 position (24, 57, 58). The GrB optimal tetrapeptide recognition motif (P4-P1) is also similar to that of caspases involved in the initiation of apoptosis (caspases 6, 8, 9, and 10), presumably reflecting their common role as initiators of the caspase-driven proteolytic cascade (24, 58). Despite these similarities, there are also striking differences in the substrate recognition properties of the two families of apoptotic proteases. For example, caspases cleave tetrapeptides and macromolecules with similar, high efficiencies (58). In contrast, GrB cleaves tetrapeptides and synthetic substrates comprising an optimized P4-P2′ sequence poorly relative to macromolecules, supporting the concept that substrate recognition involves features beyond the limited cleavage motif (13, 24, 30, 51). The data presented here define the substrate- and/or inhibitor-specific component of such extended macromolecular recognition. The definition of the corresponding exosite in GrB will reveal novel methods for selective inhibitor design, which may be useful for therapy of the inflammatory and autoimmune processes in which GrB has been implicated.
Acknowledgments
MAV-1 was a kind gift from Katherine R. Spindler, University of Georgia. Human GrB and rat GrB were generously provided by Nancy A. Thornberry.
F.A. holds the Mariana De Garay endowed chair for research on rheumatoid arthritis and related conditions and is supported by the Lupus Foundation of America. L.A.C.-R. is supported by NIH grant AR-44684. A.R. is supported by NIH grant DE-12354 and a Burroughs Wellcome Foundation translational research award.
REFERENCES
- 1.Alimonti, J. B., L. Shi, P. K. Baijal, and A. H. Greenberg. 2001. Granzyme B induces BID-mediated cytochrome c release and mitochondrial permeability transition. J. Biol. Chem. 276:6974-6982. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, F., H. G. Bull, N. A. Thornberry, G. W. Ketner, L. A. Casciola-Rosen, and A. Rosen. 2001. Adenovirus L4-100K assembly protein is a granzyme B substrate that potently inhibits granzyme B-mediated cell death. Immunity 14:751-761. [DOI] [PubMed] [Google Scholar]
- 3.Andrade, F., L. Casciola-Rosen, and A. Rosen. 2000. Apoptosis in systemic lupus erythematosus. Clinical implications. Rheum. Dis. Clin. North Am. 26:215-227. [DOI] [PubMed] [Google Scholar]
- 4.Andrade, F., S. Roy, D. Nicholson, N. Thornberry, A. Rosen, and L. Casciola-Rosen. 1998. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity 8:451-460. [DOI] [PubMed] [Google Scholar]
- 5.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305-1308. [DOI] [PubMed] [Google Scholar]
- 6.Barry, M., and R. C. Bleackley. 2002. Cytotoxic T lymphocytes: all roads lead to death. Nat. Rev. Immunol. 2:401-409. [DOI] [PubMed] [Google Scholar]
- 7.Baugh, R. J., C. D. Dickinson, W. Ruf, and S. Krishnaswamy. 2000. Exosite interactions determine the affinity of factor X for the extrinsic Xase complex. J. Biol. Chem. 275:28826-28833. [DOI] [PubMed] [Google Scholar]
- 8.Beresford, P. J., Z. Xia, A. H. Greenberg, and J. Lieberman. 1999. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity 10:585-594. [DOI] [PubMed] [Google Scholar]
- 9.Beresford, P. J., D. Zhang, D. Y. Oh, Z. Fan, E. L. Greer, M. L. Russo, M. Jaju, and J. Lieberman. 2001. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J. Biol. Chem. 276:43285-43293. [DOI] [PubMed] [Google Scholar]
- 10.Bode, W., and R. Huber. 1992. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 204:433-451. [DOI] [PubMed] [Google Scholar]
- 11.Boyd, J. M., S. Malstrom, T. Subramanian, L. K. Venkatesh, U. Schaeper, B. Elangovan, C. D'Sa-Eipper, and G. Chinnadurai. 1994. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 79:341-351. [DOI] [PubMed] [Google Scholar]
- 12.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 13.Casciola-Rosen, L., F. Andrade, D. Ulanet, W. B. Wong, and A. Rosen. 1999. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J. Exp. Med. 190:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cepko, C. L., and P. A. Sharp. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407-415. [DOI] [PubMed] [Google Scholar]
- 15.Chen, P., J. Tian, I. Kovesdi, and J. T. Bruder. 1998. Interaction of the adenovirus 14.7-kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J. Biol. Chem. 273:5815-5820. [DOI] [PubMed] [Google Scholar]
- 16.Chinnadurai, G. 1983. Adenovirus 2 Ip+ locus codes for a 19 kd tumor antigen that plays an essential role in cell transformation. Cell 33:759-766. [DOI] [PubMed] [Google Scholar]
- 17.Darmon, A. J., D. W. Nicholson, and R. C. Bleackley. 1995. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature 377:446-448. [DOI] [PubMed] [Google Scholar]
- 18.Davison, A. J., K. M. Wright, and B. Harrach. 2000. DNA sequence of frog adenovirus. J. Gen. Virol. 81:2431-2439. [DOI] [PubMed] [Google Scholar]
- 19.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 20.Fan, Z., P. J. Beresford, D. Y. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659-672. [DOI] [PubMed] [Google Scholar]
- 21.Fan, Z., P. J. Beresford, D. Zhang, and J. Lieberman. 2002. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol. 22:2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan, Z., P. J. Beresford, D. Zhang, Z. Xu, C. D. Novina, A. Yoshida, Y. Pommier, and J. Lieberman. 2003. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 4:145-153. [DOI] [PubMed] [Google Scholar]
- 23.Froelich, C. J., W. L. Hanna, G. G. Poirier, P. J. Duriez, D. D'Amours, G. S. Salvesen, E. S. Alnemri, W. C. Earnshaw, and G. M. Shah. 1996. Granzyme B perforin-mediated apoptosis of jurkat cells results in cleavage of poly(ADP-ribose) polymerase to the 89-kDa apoptotic fragment and less abundant 64-kDa fragment. Biochem. Biophys. Res. Commun. 227:658-665. [DOI] [PubMed] [Google Scholar]
- 24.Harris, J. L., E. P. Peterson, D. Hudig, N. A. Thornberry, and C. S. Craik. 1998. Definition and redesign of the extended substrate specificity of granzyme B. J. Biol. Chem. 273:27364-27373. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henkart, P. A. 1994. Lymphocyte-mediated cytotoxicity: two pathways and multiple effector molecules. Immunity 1:343-346. [DOI] [PubMed] [Google Scholar]
- 27.Horwitz, M. S. 2001. Adenovirus immunoregulatory genes and their cellular targets. Virology 279:1-8. [DOI] [PubMed] [Google Scholar]
- 28.Imai, Y., T. Kimura, A. Murakami, N. Yajima, K. Sakamaki, and S. Yonehara. 1999. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature 398:777-785. [DOI] [PubMed] [Google Scholar]
- 29.Kagi, D., B. Ledermann, K. Burki, R. M. Zinkernagel, and H. Hengartner. 1995. Lymphocyte-mediated cytotoxicity in vitro and in vivo: mechanisms and significance. Immunol. Rev. 146:95-115. [DOI] [PubMed] [Google Scholar]
- 30.Kam, C. M., D. Hudig, and J. C. Powers. 2000. Granzymes (lymphocyte serine proteases): characterization with natural and synthetic substrates and inhibitors. Biochim. Biophys. Acta 1477:307-323. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald, G., L. Shi, C. Vande Velde, J. Lieberman, and A. H. Greenberg. 1999. Mitochondria-dependent and -independent regulation of granzyme B-induced apoptosis. J. Exp. Med. 189:131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, S. J., G. P. Amarante-Mendes, L. F. Shi, T. H. Chuang, C. A. Casiano, G. A. O'Brien, P. Fitzgerald, E. M. Tan, G. M. Bokoch, A. H. Greenberg, and D. R. Green. 1996. The cytotoxic cell protease granzyme B initiates apoptosis in a cell-free system by proteolytic processing and activation of the ICE/CED-3 family protease, CPP32, via a novel two-step mechanism. EMBO J. 15:2407-2416. [PMC free article] [PubMed] [Google Scholar]
- 33.Medema, J. P., J. de Jong, L. T. Peltenburg, E. M. Verdegaal, A. Gorter, S. A. Bres, K. L. Franken, M. Hahne, J. P. Albar, C. J. Melief, and R. Offringa. 2001. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. USA 98:11515-11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medema, J. P., D. H. Schuurhuis, D. Rea, J. van Tongeren, J. de Jong, S. A. Bres, S. Laban, R. E. Toes, M. Toebes, T. N. Schumacher, B. A. Bladergroen, F. Ossendorp, J. A. Kummer, C. J. Melief, and R. Offringa. 2001. Expression of the serpin serine protease inhibitor 6 protects dendritic cells from cytotoxic T lymphocyte-induced apoptosis: differential modulation by T helper type 1 and type 2 cells. J. Exp. Med. 194:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motyka, B., G. Korbutt, M. J. Pinkoski, J. A. Heibein, A. Caputo, M. Hobman, M. Barry, I. Shostak, T. Sawchuk, C. F. Holmes, J. Gauldie, and R. C. Bleackley. 2000. Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell 103:491-500. [DOI] [PubMed] [Google Scholar]
- 37.Nagata, S. 1999. Fas ligand-induced apoptosis. Annu. Rev. Genet. 33:29-55. [DOI] [PubMed] [Google Scholar]
- 38.Oltvai, Z. N., C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609-619. [DOI] [PubMed] [Google Scholar]
- 39.Perez, D., and E. White. 1998. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J. Cell Biol. 141:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham, C. T., and T. J. Ley. 1997. The role of granzyme B cluster proteases in cell-mediated cytotoxicity. Semin. Immunol. 9:127-133. [DOI] [PubMed] [Google Scholar]
- 41.Podack, E. R., H. Hengartner, and M. G. Lichtenheld. 1991. A central role of perforin in cytolysis? Annu. Rev. Immunol. 9:129-157. [DOI] [PubMed] [Google Scholar]
- 42.Poe, M., J. T. Blake, D. A. Boulton, M. Gammon, N. H. Sigal, J. K. Wu, and H. J. Zweerink. 1991. Human cytotoxic lymphocyte granzyme B: its purification from granules and the characterization of substrate and inhibitor specificity. J. Biol. Chem. 266:98-103. [PubMed] [Google Scholar]
- 43.Riley, D., and S. J. Flint. 1993. RNA-binding properties of a translational activator, the adenovirus L4 100-kilodalton protein. J. Virol. 67:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotonda, J., M. Garcia-Calvo, H. G. Bull, W. M. Geissler, B. M. McKeever, C. A. Willoughby, N. A. Thornberry, and J. W. Becker. 2001. The three-dimensional structure of human granzyme B compared to caspase-3, key mediators of cell death with cleavage specificity for aspartic acid in P1. Chem. Biol. 8:357-368. [DOI] [PubMed] [Google Scholar]
- 45.Russell, J. H., and T. J. Ley. 2002. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20:323-370. [DOI] [PubMed] [Google Scholar]
- 46.Salvesen, G., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 47.Sarin, A., M. S. Williams, M. A. Alexander-Miller, J. A. Berzofsky, C. M. Zacharchuk, and P. A. Henkart. 1997. Target cell lysis by CTL granule exocytosis is independent of ICE/Ced-3 family proteases. Immunity 6:209-215. [DOI] [PubMed] [Google Scholar]
- 48.Sarnow, P., Y. S. Ho, J. Williams, and A. J. Levine. 1982. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell 28:387-394. [DOI] [PubMed] [Google Scholar]
- 49.Smyth, M. J., J. M. Kelly, V. R. Sutton, J. E. Davis, K. A. Browne, T. J. Sayers, and J. A. Trapani. 2001. Unlocking the secrets of cytotoxic granule proteins. J. Leukoc. Biol. 70:18-29. [PubMed] [Google Scholar]
- 50.Sun, J., L. Ooms, C. H. Bird, V. R. Sutton, J. A. Trapani, and P. I. Bird. 1997. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9). J. Biol. Chem. 272:15434-15441. [DOI] [PubMed] [Google Scholar]
- 51.Sun, J., J. C. Whisstock, P. Harriott, B. Walker, A. Novak, P. E. Thompson, A. I. Smith, and P. I. Bird. 2001. Importance of the P4′ residue in human granzyme B inhibitors and substrates revealed by scanning mutagenesis of the proteinase inhibitor 9 reactive center loop. J. Biol. Chem. 276:15177-15184. [DOI] [PubMed] [Google Scholar]
- 52.Sun, J. R., C. H. Bird, V. Sutton, L. McDonald, P. B. Coughlin, T. A. De Jong, J. A. Trapani, and P. I. Bird. 1996. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier a is present in cytotoxic lymphocytes. J. Biol. Chem. 271:27802-27809. [DOI] [PubMed] [Google Scholar]
- 53.Sutton, V. R., J. E. Davis, M. Cancilla, R. W. Johnstone, A. A. Ruefli, K. Sedelies, K. A. Browne, and J. A. Trapani. 2000. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 192:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talanian, R. V., X. H. Yang, J. Turbov, P. Seth, T. Ghayur, C. A. Casiano, K. Orth, and C. J. Froelich. 1997. Granule-mediated killing: pathways for granzyme B-initiated apoptosis. J. Exp. Med. 186:1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, D. A., C. Du, M. Xu, X. Wang, and T. J. Ley. 2000. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity 12:621-632. [DOI] [PubMed] [Google Scholar]
- 57.Thornberry, N. A., and S. M. Molineaux. 1995. Interleukin-1 beta converting enzyme: a novel cysteine protease required for IL-1 beta production and implicated in programmed cell death. Protein Sci. 4:3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thornberry, N. A., T. A. Rano, E. P. Peterson, D. M. Rasper, T. Timkey, M. Garcia-Calvo, V. M. Houtzager, P. A. Nordstrom, S. Roy, J. P. Vaillancourt, K. T. Chapman, and D. W. Nicholson. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272:17907-17911. [DOI] [PubMed] [Google Scholar]
- 59.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 60.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waugh, S. M., J. L. Harris, R. Fletterick, and C. S. Craik. 2000. The structure of the pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity. Nat. Struct. Biol. 7:762-765. [DOI] [PubMed] [Google Scholar]
- 62.Wold, W. S., K. Doronin, K. Toth, M. Kuppuswamy, D. L. Lichtenstein, and A. E. Tollefson. 1999. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 11:380-386. [DOI] [PubMed] [Google Scholar]
- 63.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 64.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, D., P. J. Beresford, A. H. Greenberg, and J. Lieberman. 2001. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc. Natl. Acad. Sci. USA 98:5746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, D., M. S. Pasternack, P. J. Beresford, L. Wagner, A. H. Greenberg, and J. Lieberman. 2001. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease granzyme A. J. Biol. Chem. 276:3683-3690. [DOI] [PubMed] [Google Scholar]