Abstract
Interindividual differences of endothelial cells in response to endotoxins might contribute to the diversity in clinical outcome among septic patients. The present study was conducted to test the hypothesis that endothelial cells (EC) with high and low proinflammatory potential exist and to dissect the molecular basis underlying this phenomenon. Thirty human umbilical vein endothelial cell (HUVEC) lines were stimulated for 24 h with lipopolysaccharide (LPS) and screened for interleukin (IL)-8 production. Based on IL-8 production five low and five high producers, tentatively called types I and II responders, respectively, were selected for genome-wide gene expression profiling. From the 74 genes that were modulated by LPS in all type II responders, 33 genes were not influenced in type I responders. Among the 41 genes that were increased in both responders, 17 were expressed significantly stronger in type II responders. Apart from IL-8, significant differences in the expression of proinflammatory related genes between types I and II responders were found for adhesion molecules [intercellular adhesion molecule (ICAM-1), E-selectin)], chemokines [monocyte chemoattractant protein (MCP-1), granulocyte chemotactic protein (GCP-2)], cytokines (IL-6) and the transcription factor CCAAT/enhancer binding protein-delta (C/EBP-δ). Type I responders also displayed a low response towards tumour necrosis factor (TNF)-α. In general, maximal activation of nuclear factor (NF)-κB was achieved in type I responders at higher concentrations of LPS compared to type II responders. In the present study we demonstrate that LPS-mediated gene expression differs quantitatively and qualitatively in types I and II responders. Our results suggest a pivotal role for common transcription factors as a low inflammatory response was also observed after TNF-α stimulation. Further studies are required to elucidate the relevance of these findings in terms of clinical outcome in septic patients.
Keywords: endothelial cells, gene expression profiling, heterogeneity, LPS
Introduction
Infections with gram-negative bacteria are considered to be the most frequent cause in the onset of organ dysfunction, although other mechanisms might also be involved [1]. Despite considerable effort that has been given over the past decade to understand the systemic inflammatory response and characteristics of severe sepsis, mortality in septic patients remains high. It is clear that mortality is highly dependent on individual patient factors, e.g. pre-existing disease, hospitalization and age [2,3]. However, the reason why the course of sepsis develops more vigorously in some patients than in others has not yet been solved. Apart from health status, age and hospitalization it is believed that genetic variations within promoter, intron or exon sequences of inflammatory genes may, in part, determine the clinical course in septic patients [4].
Genetic polymorphisms have been identified in genes encoding inflammatory molecules, e.g. interleukin (IL)-1, IL-1 receptor antagonist (IL-RA), tumour necrosis factor (TNF)-α or Toll-like receptors (TLR) [5,6]. In septic patients mortality seems to be associated only with the TNFB2 allele [7–10], which is also associated with high TNF-α production by mononuclear cells. This suggests that TNF-α might play a pivotal role in mortality caused by sepsis. In clinical studies anti-TNF-α antibodies have nevertheless failed to demonstrate any significance on mortality in septic patients [11,12]. These findings therefore point towards the involvement of other factors that might, in concert with TNF-α, influence mortality in these patients.
There is compelling evidence that in addition to monocytes [13,14], endothelial cells also play an important role in the clinical outcome of sepsis. In septic patients a dysfunction in macro- and microcirculation is observed frequently. Moreover, endothelial cells have the propensity to produce high amounts of a variety of proinflammatory mediators, e.g. cytokines, chemokines and eicosanoids. As a consequence of increased expression of adhesion molecules on these cells, leukocytes of septic patients adhere much more strongly and migrate subsequently along a chemotactic gradient into the subendothelial interstitial tissue [15].
As the amount of inflammatory mediators, produced by endothelial cells, is highly heterogeneous [16,17], severity of inflammation and thus mortality in sepsis could be dependent on interindividual variations of endothelial cells to respond to bacterial toxins.
In the present study, we therefore tested the hypothesis that the response of endothelial cells to lipopolysaccharide (LPS) can be classified into general phenotypes of cells with a low and a high proinflammatory potential. Based on previous findings that serum IL-8 concentration is associated with severity of sepsis [16,18,19], and the fact that IL-8 is produced strongly by endothelial cells [20,21], endothelial cells were grouped according to their IL-8 production. The following questions were then raised: (1) is low IL-8 production associated with low production of other mediators, (2) if so, what are these mediators and (3) what is the molecular basis for this phenomenon?
Materials and methods
Cell isolation and culture
Human umbilical vein endothelial cells (HUVEC) were isolated from fresh umbilical cords, as has been described previously [22]. The cells were cultured in essential growth medium for endothelial cells (Promocell, Heidelberg, Germany) in gelatine (1%) coated culture flasks (Greiner, Frickenhausen, Germany). Confluent monolayers were subcultured by trypsin 0025 vol%/ethylenediamine tetraacetic acid (EDTA) 0·01 vol% (Promocell, Heidelberg, Germany). Characterization of endothelial cells was performed on the basis of uptake of acetylated low-density lipoprotein (LDL), a positive staining for factor VIII-related antigen and platelet-endothelial cell adhesion molecule (PECAM), and a negative staining for alpha smooth muscle actin.
Chemokine production
HUVEC (1 × 105 cells/ml) were seeded in 24-well plates and grown until confluence. The cells were stimulated for 24 h with 1 µg/ml of LPS (Sigma, Deisenhofen, Germany). In each experiment control HUVEC were included to determine basal expression of chemokines. Supernatants were collected and assessed for IL-8 production by enzyme-linked immunosorbent assay (ELISA) (R&D Systems GmbH, Wiesbaden, Germany). ELISA was performed according to the manufacturer's instructions. Each experimental condition was performed in triplicate and each experiment was confirmed at least three times.
DNA-isolation and IL-8 genotyping
DNA was isolated from HUVEC using the Wizard genomic DNA purification Kit (Promega Corporation, Madison, WI, USA). IL-8 genotyping was performed by polymerase chain reaction (PCR), as described previously [23]. In brief, 40 ng of genomic DNA was added to a 25 µl reaction mixture containing 0·2 mMdeoxyribonucleoside triphosphate (dNTPs) (Gibco brl, Eggenstein, Germany), 12·5 pmol of primer (Perkin Elmer Applied Biosystems, Weiterstadt, Germany), 0·5 units of Taq-DNA-polymerase (Invitrogen GmbH, Karlsruhe, Germany) and 1·5 mM MgCl2. After 5 min of denaturation at 94°C, amplification was performed in 30 cycles, each consisting of 1 min at 94°C, 1 min at 53°C and 1 min at 72°C. After the last cycle primer extension was performed for 5 min at 72°C. T/A alleles were assessed by Mfe I restriction enzyme digestion of the PCR products (Roche, Basel, Switzerland) and visualized by ethidium bromide in 2% agarose gels.
RNA isolation, cRNA and array hybridization
Sample preparation and processing was performed according to the Affymetrix GeneChip Expression Analysis Manual (http://www.Affymetrix.com). Briefly, endothelial cell monolayers were stimulated with LPS (1 µg/ml) or left in normal medium for 24 h. Total RNA was isolated from these cultures using Trizol®-Reagent (Gibco brl). Hereafter DNase treatment was carried out, using RNase free DNase I (Ambion, Woodward, Austin, TX, USA). RNA concentration and quality were assessed by RNA 6000 nano assays on a Bioanalyser 2100 system (Agilent, Waldbronn, Germany). Five µg of RNA was converted into cDNA using T7-(dT)24 primers and the SuperScript Choice system for cDNA synthesis (Life Technologies, Inc., Rockville, MD, USA). Biotin-labelled cRNA was prepared by in vitro transcription using the BioArray high yield RNA transcript labelling kit (Enzo Diagnostics, Farmingdale, NY, USA). The resulting cRNA was purified, fragmented and hybridized to U133A gene chips (Affymetrix, Santa Clara, CA, USA). After hybridization the chips were stained with streptavidin–phycoerythrin (MoBiTec, Goettingen, Germany) and analysed on a GeneArray scanner (Hewlett Packard Corporation, Palo Alto, CA, USA).
Reverse transcription–polymerase chain reaction (RT-PCR)
One µg of total RNA was reverse transcribed into cDNA using the SuperScript TM II Preamplification System (Life Technologies, Karlsruhe Germany) according to the manufacturer's instructions.
The sequences of the primers used for amplification are listed in Table 1. Primers were purchased from Perkin Elmer. PCR was performed in a volume of 25 µl containing 10 mM Tris HCl, 75 mM KCl, 1·5 mM MgCl2, 200 µM dNTPs, 20 pmol of each primer, 0·5 µl cDNA and 2·5 U of Taq DNA polymerase (Gibco brl). A control containing no cDNA was always included. The PCR reactions were initiated at 94°C for 3 min and amplification was performed in 28–32 cycles, each consisting of 1 min at 94°C, 1 min at the annealing temperature and 2 min at 72°C, followed by final extension for 10 min at 72°C. PCR products were subjected to electrophoresis in 1% agarose (Serva, Boehringer Ingelheim, Heidelberg, Germany).
Table 1.
List of oligonucleotides used for cDNA amplification.
| Primer | Sequence | Size of PCR product (bp) |
|---|---|---|
| GAPDH | 5′-GTCTTCACCACCATGGAGAA-3′ | 268 |
| 5′-ATCCACAGTCTTCTGGGTGG-3 | ||
| IL-8 | 5′-CGATGTCAGTGCATAAAGACA-3′ | 200 |
| 5′-TGAATTCTCAGCCCTCTTCAAAAA-3′ | ||
| IL-6 | 5′-TCAATGAGGAGACTTGCCTGGT-3′ | 114 |
| 5′-ACAGCTCTGGCTTGTTCCTCAC-3′ | ||
| MCP-1 | 5′-TGTGCCTGCTGCTCATAG-3′ | 326 |
| 5′-GAATCCTGAACCCACTCCTG-3′ | ||
| ICAM-1 | 5′-GCAATGTGCAAGAAGATAGCCA-3′ | 105 |
| 5′-ACCCGTTCTGGAGTCCAGTACA-3′ | ||
| E-selectin | 5′-TGTGAAGCTCCCACTGAGT-3′ | 307 |
| 5′-TCTGGCATAGTAGGCAAGAA-3′ |
bp: Base pairs; GAPDH: glyceraldehyde-3-phosphate-dehydrogenase; ICAM: intercellular adhesion molecule; IL: interleukin; MCP: monocyte chemoattractant protein; PCR: polymerase chain reaction.
Flow cytometry
Confluent endothelial cell monolayers were harvested by T/E and washed three times in PBS. Hereafter, monoclonal antibodies directed against intercellular adhesion molecule (ICAM-1) and E-selectin, both from Dako, Glostrup, Denmark, were added to the cells in dilutions of 1 : 20. Isotype-matched anti-idiotypic antibodies were used as negative control. After 30 min at 4°C, the cells were washed and incubated with an anti-mouse F(ab′)2, fluorescein isothiocyanate (FITC)-conjugated secondary antibody for 30 min at 4°C. Finally the cells were washed three times with PBS before analysis was performed on a FACScalibur (Becton Dickinson, Heidelberg, Germany).
Electrophorese mobility shift assay (EMSA)
Confluent monolayers of HUVEC were incubated overnight in 0·01% bovine serum albumin (BSA) containing culture medium without growth factors or fetal calf serum (FCS). The cells were stimulated for 4 h with different concentrations of LPS. Nuclear extracts and EMSA were performed essentially as described previously [24]. Briefly, nuclear factor-kappa B (NF-κB) consensus oligonucleotides (Promega, Mannheim, Germany) were labelled to a specific activity > 5 × 107 counts per min (cpm)/µg DNA. The labelled oligonucleotide was added to 10 µg of nuclear extracts in a total volume of 20 µl containing 10 mM HEPES (pH = 7·5), 0·5 mM EDTA, 70 mM KCl, 2 mM dithiothreitol (DTT), 2% glycerol, 0·025% NP-40, 4% Ficoll, 0·1 Mphenylmethylsulphonyl fluoride (PMSF), 1 mg/ml BSA and 0·1 µg/µl poly di/dc. In each experiment specificity of binding was demonstrated by adding cold consensus or mutated NF-κB oligonucluotides to the nuclear extracts. In addition supershifts were performed by adding anti-p65 and -p50 antibodies (Santa Cruz Biotechnology, Heidelberg, Germany) to the samples. DNA–protein complexes were separated on 5% non-denaturating polyacrylamide gels in low ionic strength buffer and visualized by autoradiography.
Statistical analysis
Normalization and data analysis of the chips were performed according to instructions provided by Affymetrix. Pairwise comparisons were made by using unstimulated probe arrays as baseline and LPS-stimulated probe arrays of the same cell line as the experiment. The baseline corrected data were imported into the Affymetrix Data Mining tool (dmt version 4·0) using the publishing tool (mdb version 3·0). Subsequently, the genes were filtered using Affymetrix statistical data analysis software (Affymetrix Microarray Suite version 5·0). Probe sets were excluded when the detection call was absent, when the change call gave no change (NC) in comparison analysis, or when the signal log ratio (SLR) between unstimulated and stimulated cells was between −1 and 1. SLR was used to describe the change between a target and a reference array. The change was expressed as log2 ratio. Thus, a signal log ratio of 1 equals a fold change (FC) of 2. Only genes that fulfilled the filtering criteria were used for further analysis. Functional categorization of genes was based on ontological designations in the NetAffx Analysis Center (http://www.affymetrix.com), the AmiGO gene ontology database (http://www.godatabase.org) and gene descriptions in Online Mendelian Inheritance in Man (OMIM). Statistical analysis of SLRs was performed using Stata statistical software (Mann–Whitney test). A P-value < 0·05 was considered significant.
Results
Heterogeneity in IL-8 production
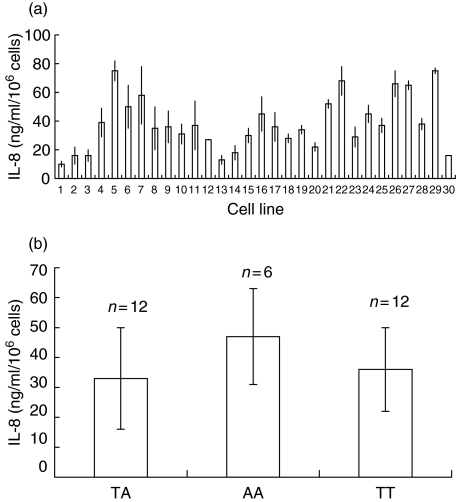
A total of 30 primary HUVEC cultures was analysed for IL-8 production. Among these, strong differences in basal and LPS-mediated IL-8 production were detected. In general, basal IL-8 production was low and varied from 0·225 to 4·13 ng/ml/106 cells (data not shown). Upon stimulation with LPS for 24 h IL-8 production was up-regulated, varying from 10 to 85 ng/ml/106 cells (Fig. 1a). Low basal IL-8 production was not associated with low IL-8 production after LPS stimulation. IL-8 production was not associated under basal nor under stimulatory conditions with the −251 T→A polymorphism in the IL-8 promoter (Fig. 1b).
Fig. 1.
Heterogeneity in lipopolysaccharide (LPS)-mediated interleukin (IL)-8 production among 30 human umbilical vein endothelial cell (HUVEC) lines. (a) Endothelial cells were stimulated for 24 h with 1 µg/ml of LPS. Supernatants were collected and assessed for IL-8 production. The results are expressed as mean IL-8 production ±s.d. of triplicate cultures. (b) Mean IL-8 production was calculated from the data set obtained in (a) for each of the different genotypes. The results are expressed as mean IL-8 production ± s.d.
Gene expression profiling
Based on LPS-mediated IL-8 production, 10 HUVEC cultures were selected for further analysis, i.e. low (n = 5) and high (n = 5) IL-8 producing HUVEC. These were tentatively called types I and II responders, respectively. All genotypes with respect to the −251 A→T polymorphism in the IL-8 promoter were present in our selection (Table 2).
Table 2.
Characteristics of the selected cell lines.
| HUVEC line | Responder type | Basal IL-8 production (ng/ml/106 cells) | LPS-mediated IL-8 production (ng/ml/106 cells) | IL-8 genotype |
|---|---|---|---|---|
| 1 | I | 0·61 ± 0·12 | 10·87 ± 2·74 | TA |
| 2 | I | 0·92 ± 0·23 | 16·44 ± 2·67 | TA |
| 3 | I | 0·95 ± 0·19 | 16·38 ± 4·63 | TA |
| 13 | I | 0·54 ± 0·34 | 16·58 ± 2·09 | TT |
| 14 | I | 0·46 ± 0·33 | 18·70 ± 1·57 | AA |
| 5 | II | 0·38 ± 0·12 | 75·45 ± 7·98 | AA |
| 22 | II | 1·23 ± 0·33 | 58·45 ± 4·50 | TT |
| 26 | II | 1·12 ± 0·11 | 66·50 ± 9·76 | TA |
| 27 | II | 1·45 ± 0·33 | 64·87 ± 11·2 | TA |
| 29 | II | 1·88 ± 0·56 | 70·34 ± 12·0 | AA |
HUVEC: human umbilical vein endothelial cell; IL: interleukin; LPS: lipopolysaccharide.
In the next step we analysed if low IL-8 production was associated with low production of other mediators. Genome-wide gene expression profiling was performed and gene expression patterns were compared between types I and II responders using two algorithms, i.e. SLR and change P-value. From all genes present on the chip 74 genes were found to be modulated significantly by LPS in all type II responders (SLR > 1 or SLR < −1, change P-value increase ≤ 0·0045 < marginal increase ≤ 0·006; 0·006 < no change < 0·994; 0·94 ≤ marginal decrease ≤ 0·9955 ≤ decrease). Whereas the majority of these genes were up-regulated, only three were down-regulated. In contrast, only 41 genes were significantly up-regulated in all type I responders and these were also found up-regulated in type II responders. Thus 33 genes were modulated specifically only in all type II responders. Among these, genes encoding for cytoskeleton proteins, immune response genes and genes encoding for transcription factors were found (Table 3). The genes that were significantly down-regulated by LPS type II responders were thrombomodulin, the intercellular adhesion molecule connexin and bone morphogenetic protein (BMP)-4.
Table 3.
Comparison of lipopolysaccharide (LPS)-mediated gene expression between type I and type II responders. Genes are grouped based on known or putative function as determined by GeneOntology classifications.
| Gene ID | GOMF | Type II SLR | Type I SLR | P-value |
|---|---|---|---|---|
| 1 | Transcription factor binding/// activity | |||
| 203973_s_at | CCAAT/enhancer binding protein (C/EBP), delta | 3·1 | 1·94 | 0·0068 |
| 201502_s_at | Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha | 2·06 | 1·6 | 0·24 |
| 209636_at | Nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (p49/p100) | 1·93 | 1·65 | 0·68 |
| 208436_s_at | Interferon regulatory factor 7 | 1·12 | ||
| 2 | Cytoskeletal protein binding | |||
| 202071_at | Syndecan 4 (amphiglycan, ryudocan) | 1·1 | ||
| 3 | GTP binding/// GTPase activity | |||
| 202086_at | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | 2·84 | ||
| 4 | ATP binding/// ATPase activity | |||
| 202307_s_at | Transporter 1, ATP-binding cassette B (MDR/TAP) | 1·22 | ||
| 204567_s_at | ATP-binding cassette G (WHITE) | 1·2 | 1·14 | 0·83 |
| 205552_s_at | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | 2·1 | ||
| 209545_s_at | Receptor-interacting serine–threonine kinase 2 | 1·27 | ||
| 218943_s_at | DEAD (Asp–Glu–Ala–Asp) box polypeptide 58 | 1·31 | ||
| 219209_at | Interferon induced with helicase C domain 1 | 1·89 | 1·2 | 0·21 |
| 5 | Complement activation | |||
| 202357_s_at | B-factor, properdin | 2·25 | 1·,93 | 0·5 |
| 208747_s_at | Complement component 1, s | 3·9 | 2·24 | 0·24 |
| 6 | Immune response | |||
| 202411_at | Interferon, alpha-inducible protein 27 | 1·55 | ||
| 203153_at | Interferon-induced protein with tetratricopeptide repeats | 3·85 | ||
| 203828_s_at | Natural killer cell transcript 4 | 2·02 | 2 | 0·93 |
| 204279_at | Proteasome subunit, beta type, 9 | 1·28 | ||
| 204415_at | Interferon, alpha-inducible protein | 1·5 | ||
| 207339_s_at | Lymphotoxin beta | 1·4 | 1·01 | 0·14 |
| 208729_x_at | Major histocompatibility complex, class I, B | 1·4 | ||
| 209795_at | CD69 antigen | 2·04 | 2·71 | 0·3 |
| 211911_x_at | Major histocompatibility complex, class IC/IB | 1·85 | ||
| 7 | Cathepsin K activity | |||
| 202450_s_at | Cathepsin K | 1·41 | 1·28 | 0·77 |
| 8 | Angiogenesis | |||
| 202510_s_at | Tumour necrosis factor, alpha-induced protein 2 | 1·12 | ||
| 9 | Protein binding/// transmembrane receptor activity | |||
| 202637_s_at | Intercellular adhesion molecule 1 (ICAM) | 3·3 | 2·15 | 0·0005 |
| 203868_s_at | Vascular cell adhesion molecule 1 (VCAM) | 3·05 | 2·1 | 0·06 |
| 205483_s_at | Interferon, alpha-inducible protein | 1·4 | ||
| 206211_at | Selectin E (endothelial adhesion molecule 1) | 5·66 | 4·28 | 0·041 |
| 207196_s_at | TNFAIP3 interacting protein 1 | 1·28 | ||
| 209417_s_at | Interferon-induced protein 35 | 1·13 | ||
| 214587_at | Collagen, type VIII, alpha 1 | 1·52 | ||
| 204105_s_at | Neuronal cell adhesion molecule | 1·16 | ||
| 214022_s_at | Interferon induced transmembrane protein 1 | 1·36 | ||
| 10 | DNA binding/// protein binding | |||
| 202643_s_at | Tumour necrosis factor,alpha-induced protein 3 | 2 | 1 | 0·0021 |
| 206321_at | Regulatory factor X, 1 (influences HLA class II) | 0·3 | ||
| 214290_s_at | Histone 2, H2aa | 0·92 | ||
| 11 | Chemokine activity | |||
| 202859_x_at | Interleukin 8 | 4·1 | 2·4 | 0·0062 |
| 203687_at | MCP-1 | 2·6 | 1·12 | 0·006 |
| 204470-at | Melanoma growth stimulating activity-alpha | 3·8 | 3 | 0·177 |
| 205476_at | Chemokine (C–C motif) ligand 20 | 4·46 | 3·56 | 0·42 |
| 207850_at | Chemokine (C–X–C motif) ligand 3 | 5 | 3·48 | 0·011 |
| 209774_x_at | Chemokine (C–X–C motif) ligand 2 | 3·95 | 2·78 | 0·017 |
| 211122_s_at | Fractalkine | 2 | 1·3 | 0·12 |
| 214974_x_at | Chemokine (C–X–C motif) ligand 5 | 4·45 | 4·1 | 0·43 |
| 215101_s_at | ENA-78 | 5·28 | 3·84 | 0·06 |
| 823_at | Chemokine (C–X3–C motif) ligand 1 | 1·9 | 1·05 | 0·08 |
| 204533_at | IP-10 | 1·7 | ||
| 206336_at | Granulocyte chemotactic protein 2 | 4·6 | 2·77 | 0·031 |
| 210163_at | Chemokine (C–X–C motif) ligand 11 | 2·39 | 1·54 | 0·01 |
| 12 | Cathepsin S activity | |||
| 202902_s_at | Cathepsin S | 1·87 | 1·2 | 0·077 |
| 13 | Ion transporter activity/// ion binding | |||
| 212067_s_at | Complement component 1, r | 1·7 | ||
| 203124_s_at | Solute carrier family 11, member 2 | 1 | ||
| 204404_at | Solute carrier family 12 member 2 | 1·5 | 2·8 | 0·04 |
| 205680_at | Stromelysin 2 | 1·72 | 1·34 | 0·29 |
| 216841_s_at | Superoxide dismutase 2, mitochondrial | 3·9 | 2·6 | 0·033 |
| 217873_at | Calcium binding protein 39 | 1·12 | ||
| 220091_at | Solute carrier family 2 | 1·33 | ||
| 14 | Blood coagulation | |||
| 203887_s_at | Thrombomodulin | −1·90 | ||
| 213506_at | Thrombin receptor-like 1 | 1·64 | 0·98 | 0·01 |
| 15 | Cell proliferation | |||
| 33304_at | Interferon stimulated gene 20 kDa | 1·54 | 1 | 0·07 |
| 16 | Phospholipase–cyclooxygenase–lipase pathway | |||
| 204748_at | Prostaglandin G/H synthase and cyclooxygenase | 2·56 | 1·22 | 0·003 |
| 209785_s_at | Phospholipase A2, group IVC | 2·08 | 2·15 | 0·75 |
| 219181_at | Lipase, endothelial | 2·2 | 1·08 | 0·01 |
| 17 | Transferase activity | |||
| 221765_at | UDP-glucose ceramide glucosyltransferase | 1·4 | ||
| 18 | Intercellular junction assembly | |||
| 204904_at | Connexin 37 | −2·15 | ||
| 19 | Cytokine activity | |||
| 205207_at | Interleukin 6 | 1·84 | 1·2 | 0·046 |
| 207442_at | Colony stimulating factor 3 (granulocyte) | 5·5 | 2·6 | 0·07 |
| 210229_s_at | Colony stimulating factor 2 (granulocyte-macrophage) | 4·68 | 3·4 | 0·26 |
| 211518_s_at | Bone morphogenetic protein 4 | −1·63 | ||
| 219424_at | Epstein–Barr virus induced gene 3 | 4·11 | 3·45 | 0·09 |
| 20 | Proteolysis and peptidolysis | |||
| 205890_s_at | Ubiquitin D | 6·12 | 4·3 | 0·05 |
| 21 | G-protein coupled receptor protein signalling pathway | |||
| 212977_at | Chemokine orphan receptor 1 | 1·8 | ||
| 22 | Transferase activity | |||
| 213988_s_at | Spermidine/spermine N1-acetyltransferase | 1·27 |
Bold type: enlisted are all the genes that were significantly up-regulated by lipolysaccharide (LPS) in all type I and all type II responders. Italic bold type: enlisted are all the genes that were expressed in both types I and II responders, but were expressed significantly more strongly in type II responders. Note that the P-value refers to the comparison between types I and II responders. Plain text: enlisted are genes that were significantly modulated by LPS in all type II responders only. The results are expressed as mean signal log ratio (SLR).
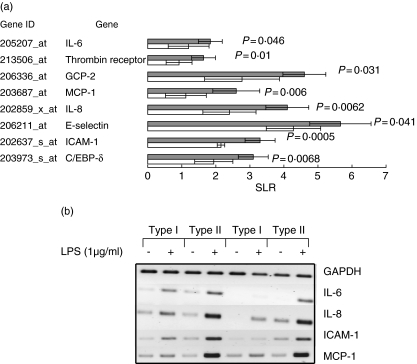
In order to compare whether the modulation of gene expression was quantitatively different for the 41 genes that were up-regulated in type I and type II responders the mean SLR for these genes was calculated in all five type I and type II responders. Comparisons between type I and type II responders revealed that 17 of 41 genes were more strongly up-regulated significantly in type II responders (Table 3). Significant differences between type I and type II responders for genes that might be implicated in inflammatory processes are depicted in Fig. 2a and include adhesion molecules (ICAM-1 and E-selectin), chemokines (MCP-1, IL-8, GCP-2), cytokines (IL-6), co-agglutination-related genes (thrombin receptor) and transcription factor (C/EBP-δ). Validation of the gene profiling data for IL-8, IL-6, MCP-1, E-selectin and ICAM-1 was performed by RT-PCR in all types I and II responders. Figure 2b shows the results of a representative experiment using two different types I and II responders.
Fig. 2.
Differences in gene expression between types I and II responders. (a) Significant differences in lipopolysaccharide (LPS)-mediated gene expression between type I and type II responders for genes implicated in inflammatory processes. Filled bars: type II responders, open bars: type I responders. The results are expressed as mean signal log ratio (SLR) (n = 5 for each responder type) ± s.d. (b) Validation of gene expression profiling was performed by reverse transcription–reverse polymerase chain reaction (RT-PCR) for interleukin (IL)-8, IL-6, monocyte chemoattractant protein (MCP)-1, E-selectin and intercellular adhesion molecule (ICAM)-1. Total RNA was isolated from unstimulated (−) or LPS (1 µg/ml, 24 h)-stimulated (+) cells. A representative experiment using two different types I and II responders is depicted. Similar findings were found in all types I and II responders.
Differences between types I and II responders are not restricted to LPS stimulation
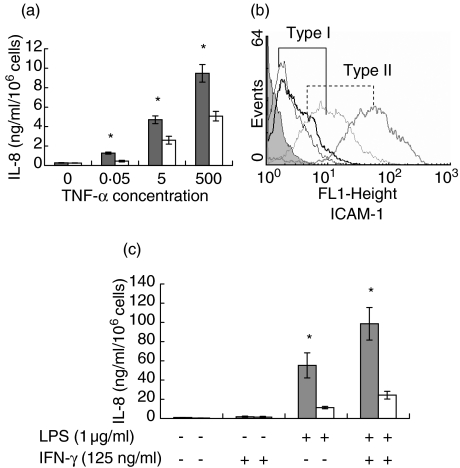
In order to study if the differences between types I and II responders were specific for LPS stimulation, the production of IL-8 and the expression of ICAM was also investigated after TNF-α stimulation. As described for LPS, we found that TNF-α-mediated IL-8 production was significantly higher in type II than in type I responders. In fact, stimulation with 500 U/ml of TNF-α in type I compared to 5 U/ml in type II responders was required for the production of an equal amount of IL-8 (Fig. 3a). Similarly, TNF-α-mediated ICAM-1 expression was clearly higher in all type II responders (Fig. 3b).
Fig. 3.
Susceptibility to tumour necrosis factor (TNF)-α stimulation in type I and type II responders. (a) Type I and type II responder cell lines were stimulated for 24 h with different concentrations of TNF-α. The supernatants were collected and assessed for interleukin (IL)-8 production by enzyme-linked immunosorbent assay (ELISA). The results are expressed as mean IL-8 ± s.d. production for each responder type (n = 5 for both types I and II). Hatched bars: type II responders, open bars: type I responders, *P < 0·05 compared to type I. (b) Constitutive (thin lines) and TNF-α mediated (bold lines) intercellular adhesion molecule (ICAM)-1 expression in type I (histograms in black) and type II (histograms in grey) responders. The negative control is depicted as filled histogram. The results of a representative experiment of types I and II responders is depicted. Similar findings were found in all types I and II responders. (c) Endothelial cells of types I and II responders were pretreated with 125 ng/ml of interferon (IFN)-γ (+) or left untreated (−) for 24 h. Hereafter the cells were washed and stimulated with lipopolysaccharide (LPS) (1 µg/ml) (+) or not (−). Twenty-four h hereafter supernatants were collected and assessed for IL-8 production. The results are expressed as mean IL-8 ± s.d. production for each responder type (n = 5 for both types I and II). Hatched bars: type II responders, open bars: type I responders, *P < 0·05 compared to type I.
To study if type I responders could be converted into type II responders, HUVEC were stimulated simultaneously with LPS and IFN-γ. This influenced neither the amount of IL-8 production (LPS versus LPS + IFN-γ) nor the type of responder (data not shown). Interestingly, however, pretreatment of HUVEC with IFN-γ 24 h before stimulation with LPS induced a nearly twofold up-regulation in IL-8 production in both responder types (Fig. 3c).
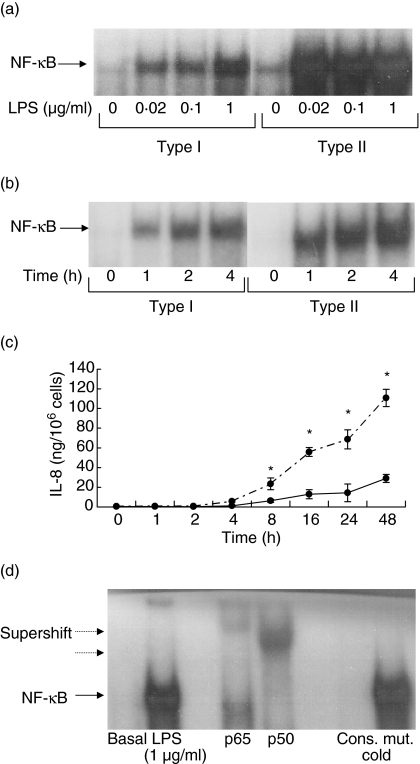
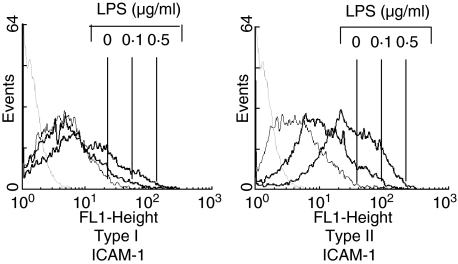
To gain more insight into the mechanism underlying the differences between types I and II responders, activation of NF-κB was investigated. In all types I and II responders NF-κB was activated by LPS in a dose- and time-dependent fashion. In all type II responders maximal activation was achieved with lower concentrations of LPS (0·02 µg/ml), while maximal activation of NF-κB in type I responders was observed at higher LPS concentrations (1 µg/ml) (Fig. 4a). No difference in the kinetic of NF-κB activation between types I and II responders was observed (Fig. 4b). In line with this, no difference in the kinetic of IL-8 production was found between both responders, although type II responders produced significantly more IL-8 (Fig. 4c). Activated NF-κB consisted of p65 and p50 subunits as demonstrated by supershift analysis (Fig. 4d). No differences between types I and II responders were found in this regard (data not shown). To demonstrate the functional relevance of NF-κB activation by low concentrations of LPS, the expression of ICAM-1 was determined in types I and II responders using suboptimal concentrations of LPS. While ICAM-1 was clearly up-regulated with 0·1 µg of LPS in all type II responders, this was not found in type I responders (Fig. 5).
Fig. 4.
Electrophoresis mobility shift assay (EMSA) for NF-κB. (a) Susceptibility towards lipopolysaccharide (LPS) stimulation as measured by NF-κB activation in types I and II responders. The data of representative experiment are depicted. Similar findings were observed in all types I and II responders. Endothelial cells were stimulated for 4 h using different concentrations of LPS. (b) Time response of NF-κB activation in types I and II responders. Endothelial cells were stimulated with 1 µg/ml of LPS. At different time-points the cells were harvested for preparation of nuclear extracts. The data of representative experiment are depicted. Similar findings were observed in all types I and II responders. Endothelial cells were stimulated for 4 h using different concentrations of LPS. (c) Kinetics of LPS-mediated interleukin (IL)-8 production in types I and II responders. Human umbilical vein endothelial cell (HUVEC) were stimulated for different time-periods with 1 µg/ml of LPS. Hereafter the supernatants were collected and assessed for IL-8 production. The results are expressed as mean IL-8 ± s.d. production for each responder type (n = 5 for both types I and II). Dotted line: type II responders, bold line: type I responders, *P < 0·05 compared to type I. (d) The specificity of the shifted bands was demonstrated by incubating a positive sample either with an excess of cold consensus (cons.) or mutated (mut.) NF-κB oligonucleotides before adding labelled consensus NF-κB oligonucleotides. Note that the NF-κB shifted band (arrow) consisted of both p50 and p65 as demonstrated by super-shift (dotted arrow).
Fig. 5.
Lipopolysaccharide (LPS) mediated intercellular adhesion molecule (ICAM)-1 expression in types I and II responders. Endothelial cells were stimulated for 24 h with different concentrations of LPS. ICAM-1 expression was measured by fluorescence activated cell sorter (FACS) analysis. The data of a representative experiment are depicted. Similar results were obtained for all types I and II responders. The negative control is depicted as grey dotted histogram.
Discussion
Sepsis and septic shock are major causes of morbidity and mortality in patients admitted to intensive care units. The systemic response to infection encompasses both pro- and anti-inflammatory phases that are marked by sequential generation of pro- and anti-inflammatory cytokines [25]. Among the proinflammatory cytokines TNF-α and IL-1β play a pivotal role [26]. These cytokines can, either alone or in conjunction with bacterial toxins, activate the vascular endothelium finally resulting in inflammation [27,28]. Interindividual differences in the endothelial inflammatory response to bacterial toxins might be considered as an additional factor influencing the clinical course of septic patients. In the present study we investigated if and to what extent primary cultures of endothelial cells display a heterogeneous response to LPS stimulation, resulting in cells with low and high proinflammatory characteristics.
To test this hypothesis we have grouped endothelial cell cultures according to the amount of IL-8 produced upon LPS stimulation and questioned whether low IL-8 production was associated with low expression of other inflammatory genes. The main findings of this study are first, that endothelial cells with low and high proinflammatory characteristics as such do not exist, although some proinflammatory genes are expressed significantly lower in so-called type I responders; secondly, that this was not specific for LPS and also observed after TNF-α stimulation; thirdly, that gene expression after LPS stimulation in type I and type II responders differ qualitatively and quantitatively from each other. Fourthly, endothelial cells of type I responders required higher LPS concentrations for maximal NF-κB activation.
The search for predictors of mortality in septic patients is of crucial clinical relevance in order to treat these patients interindividually. Although a variety of factors has been suggested to be associated with mortality in sepsis [29–32], improvements in clinical outcome have been sporadic and, with few exceptions, are related to improvements in supportive care rather than to specific therapies. While in some studies soluble adhesion molecules, i.e. sICAM-1 and sVCAM-1 in serum of septic patients, were associated with mortality [29,30], this was not confirmed by others [33,34]. The relevance of circulating pro- and anti-inflammatory cytokines seems to be more consistent in this regard [31,32,35].
Apart from monocytes, the vascular endothelium is a large source from which circulating cytokines can be derived. This is particularly true for IL-8 and IL-6 [36,37]. Although it is difficult to estimate to what extent endothelial cells contribute to the amount of these cytokines in septic patients, the severity of sepsis is clearly associated with both [38,39]. It also remains to be elucidated whether low IL-8 and IL-6 production in Gram-negative sepsis is an intrinsic patient factor or whether this merely reflects bacterial load. Our data are in line with the former. LPS stimulation of some endothelial cells, i.e. type I responders, resulted in low IL-8 production while type II responders produced high amounts of IL-8. In addition low IL-8 producers also expressed low IL-6 mRNA. Interestingly, our data also revealed that in addition to LPS the up-regulation of IL-8 and ICAM-1 was less pronounced in type I responders when stimulated with TNF-α. Hence differences between types I and II responders were unlikely, due to functional differences in TLR or their level of expression. Moreover, no differences, either at mRNA or protein levels, for TLR4 and TLR2 between types I and II responders were identified (data not shown).
A more likely explanation could be the expression of transcription factors. Among the transcription factors that were influenced by LPS only the up-regulation of C/EBP-δ was reduced significantly in type I compared to type II responders. C/EBP-δ has been implicated in the regulation of proinflammatory cytokines such as IL-6, IL-8 and MCP-1 [40–42] and is activated by both TNF-α and LPS [43]. Because gene analysis was performed 24 h after LPS stimulation, differences in mRNA expression for other transcription factors may also exist between type I and type II responders, e.g. NF-κB [44] and activator protein (AP)-1 [45]. These transcription factors are known to be transcribed early after stimulation. In a study by Bohrer et al. [46] investigating 25 septic patients, non-survivors could be distinguished from survivors by an increased activation of NF-κB in mononuclear cells. This might be related to our findings using endothelial cells. Although no differences in NF-κB p65 or p50 mRNA expression were detected among the cell lines, activation of NF-κB was different between types I and II responders. Maximal activation of NF-κB was already obtained with low concentrations of LPS in type II responders. In contrast, type I responders required 500-fold higher concentrations of LPS for maximal NF-κB activation.
The clinical relevance of our findings with respect to types I and II responders remains to be elucidated and further studies are planned. Nevertheless, our study demonstrates that in addition to peripheral blood monocytes [47] heterogeneity in LPS responsiveness also occurs at the level of endothelial cells and thus might influence severity of the inflammatory response in septic patients.
Acknowledgments
This study was supported by a grant of the Forschungsfonds of the University of Mannheim und the GRK 880/2004 of the Deutsche Forschungsgesellschaft.
References
- 1.Venet C, Zeni F, Viallon A, et al. Endotoxaemia in patients with severe sepsis or septic shock. Intens Care Med. 2000;26:538–44. doi: 10.1007/s001340051201. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–10. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Mirzanejad Y, Roman S, Talbot J. Pneumococcal bacteremia in two tertiary care hospitals in Winnipeg. Can Chest. 1996;109:173–8. doi: 10.1378/chest.109.1.173. [DOI] [PubMed] [Google Scholar]
- 4.Zehnbauer B. Population genetics in critical illness. Crit Care Med. 2005;33:242–3. doi: 10.1097/01.ccm.0000150763.21694.8a. [DOI] [PubMed] [Google Scholar]
- 5.Holmes CL, Russell JA, Walley KR. Genetic polymorphisms in sepsis and septic shock: role in prognosis and potential for therapy. Chest. 2003;124:1103–34. doi: 10.1378/chest.124.3.1103. [DOI] [PubMed] [Google Scholar]
- 6.Kellum JA, Angus DC. Genetic variation and risk of sepsis. Minerva Anesthesiol. 2003;69:245–53. [PubMed] [Google Scholar]
- 7.Weitkamp JH, Stüber F, Bartmann P. Pilot study assessing TNF gene polymorphism as a prognostic marker for disease progression in neonates with sepsis. Infection. 2000;28:92–6. doi: 10.1007/s150100050053. [DOI] [PubMed] [Google Scholar]
- 8.Reid CL, Perrey C, Pravica V, et al. Genetic variation in proinflammatory and anti-inflammatory cytokine production in multiple organ dysfunction syndrome. Crit Care Med. 2002;30:2216–21. doi: 10.1097/00003246-200210000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Calvano JE, Um JY, Agnese DM, et al. Influence of the TNF-alpha and TNF-beta polymorphisms upon infectious risk and outcome in surgical intensive care patients. Surg Infect. 2003;4:163–9. doi: 10.1089/109629603766956951. [DOI] [PubMed] [Google Scholar]
- 10.Stuber F, Petersen M, Bokelmann F, et al. A genomic polymorphism within the tumor necrosis factor locus influences plasma tumor necrosis factor-alpha concentrations and outcome of patients with severe sepsis. Crit Care Med. 1996;24:381–4. doi: 10.1097/00003246-199603000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart K, Karzai W. Anti-tumor necrosis factor therapy in sepsis: update on clinical trials and lessons learned. Crit Care Med. 2001;29:S121–5. doi: 10.1097/00003246-200107001-00037. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J, Carlet J. Intersept: an international, multicenter, placebo-controlled trial of monoclonal antibody to human tumor necrosis factor-alpha in patients with sepsis. Crit Care Med. 1996;23:1431–40. doi: 10.1097/00003246-199609000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Haugen TS, Nakstad B, Lyberg T. Heterogeneity of procoagulant activity and cytokine release in subpopulations of alveolar macrophages and monocytes. Inflammation. 1999;22:15–23. doi: 10.1023/a:1020283316002. [DOI] [PubMed] [Google Scholar]
- 14.Taylor PR, Gordon S. Monocyte heterogeneity and innate immunity. Immunity. 2003;19:2–4. doi: 10.1016/s1074-7613(03)00178-x. [DOI] [PubMed] [Google Scholar]
- 15.Hack CE, Zeerleder S. The endothelium in sepsis: source of and target for inflammation. Crit Care Med. 2001;29:S21–7. doi: 10.1097/00003246-200107001-00011. [DOI] [PubMed] [Google Scholar]
- 16.Sablotzki A, Dehne MG, Friedrich I, et al. Different expression of cytokines in survivors and non-survivors from MODS following cardiovascular surgery. Eur J Med Res. 2003;8:71–6. [PubMed] [Google Scholar]
- 17.Gebb S, Stevens T. On lung endothelial cell heterogeneity. Microvasc Res. 2004;68:1–12. doi: 10.1016/j.mvr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Simmons EM, Himmelfarb J, Sezer MT, et al. Plasma cytokine levels predict mortality in patients with acute renal failure. Kidney Int. 2004;65:1357–65. doi: 10.1111/j.1523-1755.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 19.Igonin AA, Armstrong VW, Shipkova M, et al. Circulating cytokines as markers of systemic inflammatory response in severe community-acquired pneumonia. Clin Biochem. 2004;37:204–9. doi: 10.1016/j.clinbiochem.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Oude Nijhuis CS, Vellenga E, Daenen SM, et al. Endothelial cells are main producers of interleukin 8 through Toll-like receptor 2 and 4 signaling during bacterial infection in leukopenic cancer patients. Clin Diagn Lab Immunol. 2003;10:558–63. doi: 10.1128/CDLI.10.4.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Stavchansky SA, Bowden RA, et al. Effect of interleukin-1-beta and tumor necrosis factor-alpha on gene expression in human endothelial cells. Am J Physiol Cell Physiol. 2003;284:C1577–83. doi: 10.1152/ajpcell.00243.2002. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe EA, Nachman RL, Becker CG. Culture of human endothelial cells derived from umbilical veins: identification by morphologic and immunologic criteria. J Clin Invest. 1973;5:2745–56. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull J, Thomson A, Kwiatkowski D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax. 2000;55:1023–7. doi: 10.1136/thorax.55.12.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa H, Rafiee P, Heidemann J, et al. Mechanisms of endotoxin tolerance in human intestinal microvascular endothelial cells. J Immunol. 2003;170:5956–64. doi: 10.4049/jimmunol.170.12.5956. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Bojorquez LN, Dehesa AZ, Reyes-Teran G, et al. Molecular mechanisms involved in the pathogenesis of septic shock. Arch Med Res. 2004;35:465–79. doi: 10.1016/j.arcmed.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Remick DG. Cytokine therapeutics for the treatment of sepsis: why has nothing worked? Curr Pharm Des. 2003;9:75–82. doi: 10.2174/1381612033392567. [DOI] [PubMed] [Google Scholar]
- 27.Martinez MA, Pena JM, Fernandez A, et al. Time course and prognostic significance of hemostatic changes in sepsis: relation to tumor necrosis factor-alpha. Crit Care Med. 1999;27:1303–8. doi: 10.1097/00003246-199907000-00017. [DOI] [PubMed] [Google Scholar]
- 28.Zanotti S, Kumar A, Kumar A. Cytokine modulation in sepsis and septic shock. Expert Opin Invest Drugs. 2002;11:1061–75. doi: 10.1517/13543784.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 29.Kayal S, Jais JP, Aguini N, et al. Elevated circulating E-selectin, intercellular adhesion molecule 1, and von Willebrand factor in patients with severe infection. Am J Respir Crit Care Med. 1998;5:7776–84. doi: 10.1164/ajrccm.157.3.9705034. [DOI] [PubMed] [Google Scholar]
- 30.Whalen MJ, Doughty LA, Carlos TM, et al. Intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 are increased in the plasma of children with sepsis-induced multiple organ failure. Crit Care Med. 2000;28:2600–7. doi: 10.1097/00003246-200007000-00070. [DOI] [PubMed] [Google Scholar]
- 31.Lekkou A, Karakantza M, Mouzaki A, et al. Cytokine production and monocyte HLA-DR expression as predictors of outcome for patients with community-acquired severe infections. Clin Diagn Lab Immunol. 2004;11:161–7. doi: 10.1128/CDLI.11.1.161-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gogos CA, Drosou E, Bassaris HP, et al. Pro- versus anti-inflammatory cytokine profile in patients with severe sepsis: a marker for prognosis and future therapeutic options. J Infect Dis. 2000;181:176–80. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- 33.Weigand MA, Schmidt H, Pourmahmoud M, et al. Circulating intercellular adhesion molecule-1 as an early predictor of hepatic failure in patients with septic shock. Crit Care Med. 1999;27:2656–61. doi: 10.1097/00003246-199912000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Knapp S, Thalhammer F, Locker GJ, et al. Prognostic value of MIP-1 alpha, TGF-beta 2, sELAM-1 and sVCAM-1 in patients with Gram-positive sepsis. Clin Immunol Immunopathol. 1998;87:139–44. doi: 10.1006/clin.1998.4523. [DOI] [PubMed] [Google Scholar]
- 35.van Dissel JT, van Langevelde P, Westendorp RG, et al. Anti-inflammatory cytokine profile and mortality in febrile patients. Lancet. 1998;351:950–3. doi: 10.1016/S0140-6736(05)60606-X. [DOI] [PubMed] [Google Scholar]
- 36.Franscini N, Bachli EB, Blau N, et al. Gene expression profiling of inflamed human endothelial cells and influence of activated protein C. Circulation. 2004;110:2903–9. doi: 10.1161/01.CIR.0000146344.49689.BB. [DOI] [PubMed] [Google Scholar]
- 37.Krishnaswamy G, Kelley J, Yerra L, et al. Human endothelium as a source of multifunctional cytokines: molecular regulation and possible role in human disease. J Interferon Cytokine Res. 1999;19:91–104. doi: 10.1089/107999099314234. [DOI] [PubMed] [Google Scholar]
- 38.Hack CE, Hart M, Schijndel RJ, et al. Interleukin-8 in sepsis: relation to shock and inflammatory mediators. Infect Immun. 1992;60:2835–42. doi: 10.1128/iai.60.7.2835-2842.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oda S, Hirasawa H, Shiga H, et al. Sequential measurement of IL-6 blood levels in patients with systemic inflammatory response syndrome (SIRS) /sepsis. Cytokine. 2005;29:169–75. doi: 10.1016/j.cyto.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Gao H, Parkin S, Johnson PF, et al. C/EBP has a stimulatory role on the IL-6 and IL-8 promotors. J Biol Chem. 2002;227:38827–37. doi: 10.1074/jbc.M206224200. [DOI] [PubMed] [Google Scholar]
- 41.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–82. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 42.Weber M, Sydlik C, Quirling M, et al. Transcriptional inhibition of interleukin-8 expression in tumor necrosis factor-tolerant cells: evidence for involvement of C/EBP beta. J Biol Chem. 2003;278:23586–93. doi: 10.1074/jbc.M211646200. [DOI] [PubMed] [Google Scholar]
- 43.Reddy KV, Serio KJ, Hodulik CR, et al. 5-lipoxygenase-activating protein gene expression. Key role of CCAAT/enhancer-binding proteins (C/EBP) in constitutive and tumor necrosis factor (TNF) alpha-induced expression in THP-1 cells. J Biol Chem. 2003;278:13810–8. doi: 10.1074/jbc.M211102200. [DOI] [PubMed] [Google Scholar]
- 44.Chan EL, Murphy JT. Reactive oxygen species mediate endotoxin-induced human dermal endothelial NF-kappa B activation. J Surg Res. 2003;111:120–6. doi: 10.1016/s0022-4804(03)00050-7. [DOI] [PubMed] [Google Scholar]
- 45.Pendurthi UR, Williams JT, Rao LV. Inhibition of tissue factor gene activation in cultured endothelial cells by curcumin. Suppression of activation of transcription factors Egr-1, AP-1, and NF-kappa B. Arterioscler Thromb Vasc Biol. 1997;17:3406–13. doi: 10.1161/01.atv.17.12.3406. [DOI] [PubMed] [Google Scholar]
- 46.Bohrer H, Qiu F, Zimmermann T, et al. Role of NF-kappaB in the mortality of sepsis. J Clin Invest. 1997;100:972–85. doi: 10.1172/JCI119648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lush CW, Cepinskas G, Kvietys PR. LPS tolerance in human endothelial cells: reduced PMN adhesion, E-selectin expression, and NF-kappaB mobilization. Am J Physiol Heart Circ Physiol. 2000;278:H853–61. doi: 10.1152/ajpheart.2000.278.3.H853. [DOI] [PubMed] [Google Scholar]