Abstract
Chromosome 22q11.2 deletion syndrome is a common disorder characterized by thymic hypoplasia, conotruncal cardiac defect and hypoparathyroidism. Patients have a risk of infections and autoimmunity associated with T lymphocytopenia. To assess the immunological constitution of patients, the numerical changes and cytokine profile of circulating T cells were analysed by flow cytometry and real-time polymerase chain reaction (PCR). CD3+, CD4+, T cell receptor (TCR)αβ+ or CD8αα+ cell counts were lower, and CD56+ cell counts were higher in patients than in controls during the period from birth to adulthood. The ageing decline of CD3+ or CD4+ cell counts was slower in patients than in controls. The proportion of CD8αα+ cells increased in controls, and the slope index was larger than in patients. On the other hand, both the number and proportion of Vα24+ cells increased in patients, and the slope indexes tended to be larger than in controls. The positive correlation of the number of T cells with CD8αα+ cells was observed only in patients, and that with Vα24+ cells was seen only in controls. No gene expression levels of interferon (IFN)-γ, interleukin (IL)-10, transforming growth factor (TGF)-β, cytotoxic T lymphocyte antigen 4 (CTLA4) or forkhead box p3 (Foxp3) in T cells differed between patients and controls. There was no significant association between the lymphocyte subsets or gene expression levels and clinical phenotype including the types of cardiac disease, hypocalcaemia and frequency of infection. These results indicated that T-lymphocytopenia in 22q11.2 deletion patients became less severe with age under the altered composition of minor subsets. The balanced cytokine profile in the limited T cell pool may represent a T cell homeostasis in thymic deficiency syndrome.
Keywords: 22q11.2 deletion, cytokine, DiGeorge syndrome, T cell subsets, T cell homeostasis
Introduction
22q11.2 deletion syndrome is one of the most common genetic disorders occurring in one of 4000–6000 live births [1]. DiGeorge syndrome/velocardiofacial syndrome is a prototype of the deletion presenting conotruncal cardiac anomalies, thymic hypoplasia, hypoparathyroidism, velopharyngeal incompetence (VPI) and mental retardation [2]. This deletion is also associated with CHARGE [coloboma, heart disease, atresia choanae, retarded growth and retarded development or central nervous system (CNS) anomalies, genital hypoplasia and ear anomalies and/or deafness] association, Opitz/GBBB syndrome and conotruncal anomaly face syndrome. Most patients have a deletion of the same 3 Mb region on 22q11.2 [3]. Critical T cell deficiency occurs rarely in athymic patients with the deletion (complete DiGeorge syndrome) [4], while varying degrees of lymphopenia arising from the thymic hypoplasia is seen in more than 80% of patients (partial DiGeorge syndrome) [5]. Diminished T cell count with biased T cell receptor (TCR) repertoire is a typical finding of the deletion syndrome [6,7], which may predict susceptibility to infections [8]. On the other hand, the decline of peripheral T cells in patients might be blunted with age [9,10]. Extrathymic T cells can proliferate in complete DiGeorge syndrome [11]. The ageing alteration of thymus-dependent and -independent T cells has not been clarified in this deletion syndrome.
Effective control of cardiac anomalies and infections has led to the improved survival of patients. However, another caveat is the complication of allergic diseases and autoimmune disorders such as rheumatoid arthritis, haemolytic anaemia and thrombocytopenia [9,12–17]. There have been a few reports on the immunodysregulation in this syndrome, including high lymphoproliferative activity [18], increased apoptosis of T cells [19] and deficient regulatory T (Tr) cells [20]. The expression of cytokines and T helper (Th) 1/Th 2 status remains unknown. The numerical and functional analyses of patients’ T cells may delineate the precise thymic function in humans.
In this study, we followed age-related changes of T cell subsets in 22q11.2 deletion patients focusing on TCRγδ+, CD8αα+ or Vα24+ cells arising partly from the extrathymic pathway, and analysed quantitatively the gene expression of interferon (IFN)-γ, interleukin (IL)-10, transforming growth factor (TGF)-β, cytotoxic T-lymphocyte antigen (CTLA) 4 and forkhead box p3 (Foxp3) in circulating T cells. Unique quantitative and qualitative maturations of the limited T cell pool in thymic deficiency are discussed.
Patients, materials and methods
Patients
A total of 15 patients carrying the 22q11.2 deletion were eligible for the study, based on the results of fluorescence in situ hybridization (FISH) using the N25 (D22S75) or Tuple1 DNA probes [3]. There was no other abnormal karyotype assessed by G-banding. Clinical characteristics of patients are shown in Table 1. The male : female ratio was 6 : 9. All patients showed complex cardiac and arch anomalies, psychomotor retardation and lymphopenia. There was no complication of thrombocytopenia. VPI was defined in seven of 15 patients. At surgical intervention of cardiac disease, thymic hypoplasia was confirmed in 12 of 15 patients. Patients 11 and 12 had severe brain damage due to head trauma and hypoxic encephalopathy, respectively; their neurological evaluations were excluded for the study. All patients were free from serious infections, and received live-attenuated vaccines safely. Three patients had immunological abnormalities but had not developed autoimmune diseases; positive anti-nuclear antibody in patient 3, a low immunoglobulin (Ig) G3 level in patient 7 and an isolated low IgG level in patient 8. Patient 12 died of haemophagocytic lymphohistiocytosis (HLH) at 26 months of age. Age-matched controls included 32 subjects who had no underlying disease or active infection. Peripheral blood was collected from patients without infection after informed consent was obtained.
Table 1.
Clinical characteristics of patients with chromosome 22q11.2 deletion syndrome
| No. | Sex | Age*(years, months) | Thymus | Cardiac defect/arch anomaly | VPI | PMR | Hypocalcaemia | Prone to infection | Immunological abnormality |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 4 | Trace | VSD/IAA | no | Slight | Yes | No | |
| 2 | F | 8 | Small | VSD/IAA | yes | Moderate | Yes | Yes | |
| 3 | F | 14 | Small | VSD/RAA | no | Slight | Yes | No | Positive ANA |
| 4 | F | 8 | Trace | VSD/LAA | yes | Severe | Yes | Yes | |
| 5 | F | 21 | Trace | TOF/RAA | yes | Slight | Yes | No | |
| 6 | M | 10 | Small | TOF/RAA | no | Slight | No | Yes | |
| 7 | F | 13 | Small | VSD/IAA | no | Slight | Yes | Yes | Low IgG3 levels |
| 8 | M | 1,6 | Small | TOF/LAA | no | Slight | No | Yes | Low IgG levels |
| 9 | M | 2,0 | NR | VSD/LAA | yes | Slight | No | No | |
| 10 | M | 13 | Small | TOF/LAA | no | Slight | No | Yes | |
| 11 | F | 2,2 | NR | TOF/RAA | no | Severe** | Yes | No | |
| 12 | F | 2,0 | Small | TOF/RAA | yes | Severe** | Yes | No | HLH |
| 13 | F | 7,6 | Small | VSD/LAA | yes | Moderate | No | Yes | |
| 14 | M | 1,0 | NR | TOF/LAA | no | Severe | Yes | No | |
| 15 | M | 1,3 | Trace | TOF/RAA | yes | Slight | No | Yes |
IAA: interruption of aortic arch, LAA: left aortic arch, RAA: right aortic arch, VSD: ventricular septal defect, TOF: tetralogy of Fallot, VPI: velopharyngeal incompetence; PMR: psychomotor retardation, ANA: anti-nuclear antibodies, Ig: immunoglobulin, HLH: haemophagocytic lymphohistiocytosis, NR: not recorded.
Age represents the time of real-time polymerase chain reaction (PC) study for the quantification of cytokine genes in fractionated T cells.
Neurological functions of patients 11 and 12 were severely impaired because of intracranial haemorrhage and hypoxic encephalopathy, respectively.
Flow cytometry and cell sorting
Flow cytometry was carried out using epics-xl (Immunotech Coulter, Miami, FL, USA) [21]. The forward light-scatter gate was set to analyse viable cells and exclude background artefacts. Multicolour staining was carried out using fluorescein isothiocyanate-, phycoerythrin-, phycoerythrin-cyanin 5·1-conjugated monoclonal antibodies against CD3, CD4, CD8, CD8α, CD19, CD56, TCRαβ, TCRγδ and TCRVα24 (Immunotech Coulter). Mononuclear cells (MNCs) were separated from peripheral blood using Ficoll-Paque (ICN Biomedicals, Inc., Aurora, OH, USA), and were then fractionated into CD3+ cells using the magnetic cell sorting system (Miltenyl Biotec, Auburn, CA, USA) [22]. T cells with > 97% of purity were stored at −80°C to extract RNA.
RNA extractions and cDNA synthesis
Total RNA was extracted from the sorted T cells by RNA extraction kit, Isogen (Nippon Gene, Osaka, Japan). cDNA was synthesized with a first-strand cDNA synthesis kit (Amersham Pharmacia Biotech, Tokyo, Japan) at 37°C for 60 min with random hexamers. The cDNA were used for the following polymerase chain reactions (PCR).
Quantitative PCR by TaqMan method
Real-time PCR was used to quantify the gene dosages expressed in T cell fractions. The quantified genes were mRNA of cytokines of IFN-γ, IL-10 and TGF-β, a co-stimulatory molecule of CTLA4 and a key regulatory T cell molecule of Foxp3. The PCR primers and TaqMan probes were designed with the assistance of the computer program primer express (PE Biosystems, Foster City, CA, USA), based on information of the sequences for all genes from the GenBank database. Nucleotide sequences of PCR primers and the TaqMan probe were as follows; IFN-γ forward primer (F): 5′-ACGAGATGACTTCGAAAAGCTG-3′, IFN-γ reverse primer (R): 5′-TTTAGCTGCTGGCGACAGTTC-3′, IFN-γTaqMan probe: 5′-CGGTAACTGACTTGAATGTC CAACGCAA-3′, IL-10 F: 5′-ACCTGCCTAACATGCTTC GAG-3′, IL-10 R: 5′-CCAGCTGATCCTTCATTTGAAAG-3′, IL-10 TaqMan probe: 5′-TCTCCGAGATGCCTTCAG CAGAGTGA-3′, TGF-β F: 5′-ATTGCTTCAGCTCCACG GA−3′, TGF-β R: 5′-CCCGGGTTATGCTGGTTGTAC-3′, TGF-βTaqMan probe: 5′-CAGCTGTACATTGACTTCCG CAAGGACCT-3′, CTLA4 F: 5′-CAGTGGAAATCAAGT GAACCTCAC-3′; CTLA4 R: 5′-GCACGGTTCTGGAT CAAT TACA-3′; CTLA4 TaqMan probe: 5′-TGGAGCTCAT GTACCCACCGCCATACTA-3′, and Foxp3 F: 5′-GAGAAGCTGAGTGCCATGCA-3′, Foxp3 R: 5′-AGGAGC CCTTGTCGGATGAT-3′, Foxp3 TaqMan probe: 5′-CACA GATGAAGCCTTGGTCAGTGCCA-3′ (anti-sense strand). Each TaqMan probe was labelled at the 5′ end with the reporter dye molecule FAM (6-carboxyfluoresenscein; emission I, 538 nm). Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) TaqMan control reagents kit (PE Biosystems) was used as an internal control. TaqMan probes were labelled with the quencher flour TAMRA (6-carboxy-tetramethyl rhodamine; emission I, 582 nm) at the 3′ end via a linker arm nucleotide.
Cellular mRNA was quantified by abi prism 7700 Sequence Detector (PE Biosystems) [21,22]. Briefly, the PCR primer set and TaqMan probe were mixed with TaqMan universal PCR master mix (PE Biosystems). PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, 50 cycles of amplification at 94°C for 15 s and 60°C for 1 min. During each cycle of the PCR, the 5′→3′ exonuclease activity of Ampli-Taq Gold DNA polymerase cleaves the TaqMan probe, thereby increasing the fluorescence of the reporter dye at the appropriate wavelength. The increase in fluorescence was proportional to the concentration of template in the PCR mixture. To calculate the relative amount of gene in cells, each value was divided by that of the internal control. It then was defined as a relative ratio to that (1·00) of phytohaemagglutinin (PHA)-stimulated MNCs (relative units). All analyses were performed in duplicate samples and repeated for confirmation.
Statistical analysis
Group means were compared between two groups by Mann–Whitney U-test. Pearson's correlation coefficients and Fisher's Z-transformation were used for the association study. Analysis of covariance was used to compare the slope index of regression equations in the relation of age and the proportion or absolute number of lymphocyte subsets. Calculations were performed using bmdp statistical software (BMDP Statistical Software, Inc., Los Angeles, CA, USA) on sparc station 20 (OS: solaris 2·5.1, Sun Microsystems, Mountain View, CA, USA) [21,22].
Results
Comparison of lymphocyte subsets
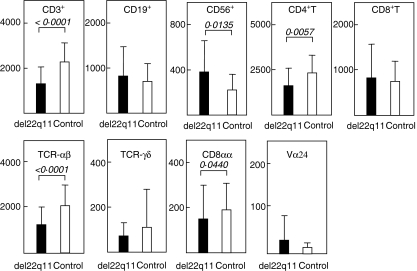
Surface markers of peripheral lymphocytes in 15 patients were examined and compared with those in 32 age-matched controls. Blood sampling was performed more than twice during the observation period (median: 42·6 months, range: 2–247 months of age). The absolute numbers of lymphocytes are shown in Fig. 1. The proportion and number of CD3+ cells (both P < 0·0001), CD4+ T cells (P = 0·0069 and 0·0057, respectively) and αβT cells (both P < 0·0001) were each lower in patients than those in controls throughout the observation period. The CD4/CD8 ratio was lower in patients than in controls (P = 0·0388). The number but not proportion of CD8αα+ cells (P = 0·0440) was significantly lower in patients than in controls. In contrast, the proportion and number of CD56+ cells (P< 0·0001 and 0·0135, respectively) were higher in patients than in controls. The proportion but not the number of CD19+ cells (P = 0·0113) was higher in patients than in controls. There were no differences in the proportion and number of γδT cells or Vα24+ cells between patients and controls during the observation period.
Fig. 1.
Absolute number of peripheral blood lymphocytes during the observation period (median: 42·6 months, ranges: 2–247 months of age). The mean ± s.d. cell counts are shown in patients with 22q11.2 deletion (▪) and age-matched controls (□). CD3+ cell (P< 0·0001), CD4+ T cell (P= 0·0057), T cell receptor (TCR)αβ cell (P< 0·0001) and CD8αα+ cell (P= 0·0440) counts were lower in patients than in controls. On the other hand, CD56+ cell counts in patients were higher than in controls (P= 0·0135). There were no statistical differences in CD19+ cells, CD8+ T cells, TCRγδ cells and Vα24+ cells between patients and controls. The Mann–Whitney U-test was used for the statistical analyses. Sample number of patients: CD3+, CD19+, CD56+, CD4+ T, CD8+ T, αβT and γδT cells: 43, each of controls: 32.
Changes of T cell subsets during maturation
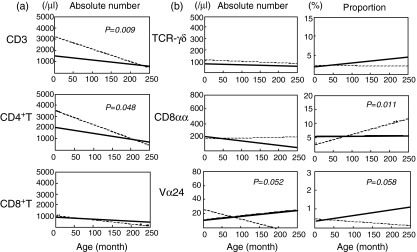
The ageing alterations of T cell subsets were then assessed by the analysis of covariance. Figure 2a shows the changes in absolute number of T cells. Figure 2b presents the changes in number and proportion of T cell subsets including the extrathymic population. The decline slope of numbers but not proportion of CD3+ cells was slower in patients than in controls (P= 0·009, patient: y = −3·8678x + 1605, control: y = −12·086x + 3431). CD4+ T cells showed the decline pattern of numbers similar to CD3+ cells (P= 0·048, patient: y = −5·5848x + 2139, control: y = −12·411x + 3619). The similar decline pattern of αβT cell counts did not reach statistical significance (data not shown). There were no differences in the slope index of numbers or proportion of CD8+ T cells, CD19+ cells or CD56+ cells between patients and controls (data not shown).
Fig. 2.
Correlation between absolute number of T cell subsets and age. The regression lines were derived from the relationship between number or proportion of each subset and age, in patients: (a) solid line n = 43, (b) n = 29–42 and in controls (dotted line, n = 32). The slope index of regression equations was compared between patients and controls assessed by the analysis of covariance. (a) The decline slope of CD3+ cell number was slower in patients than in controls (P= 0·009, patient: y = −3·8678x + 1605, control: y = −12·086x + 3431). CD4+ T cells showed the decline pattern similar to CD3+ cells (P= 0·048, patient: y = −5·5848x + 2139, control: y = −12·411x + 3619). (b) The decline slope of proportion of CD8αα+ cells was slower in patients than controls (P= 0·011, patient: y = 0·0014x + 5·5213, control: y = 0·0324x + 2·9278). The slope indexes in both of Vα24+ cells in patients tended to be larger than in controls (number: P = 0·052, patient: y = 0·0549x + 11·997, control: y = −0·1367x + 27·006; proportion: P = 0·058, patient: y = 0·0033x + 0·3478, control: y = −0·0014x + 0·5238), although these did not reach statistical significance. No slope indexes of proportion or number of T cell receptor (TCR)γδ cells differed between patients and controls.
The ageing slopes of γδT cells did not differ between patients and controls (Fig. 2b). The proportion of CD8αα+ cells increased in controls during the observation period, the slope index of which was larger than that in patients (P= 0·011, patient: y = 0·0014x + 5·5213, control: y = 0·0324x + 2·9278). The difference in the slope index of CD8αα+ cell number between patients and controls did not reach significance. On the other hand, the number and proportion of Vα24+ cells increased in patients and decreased in controls. The slope indexes in both of Vα24+ cells in patients tended to be larger than in controls (number: P = 0·052, patient: y = 0·0549x + 11·997, control: y = −0·1367x + 27·006; proportion: P = 0·058, patient: y = 0·0033x + 0·3478, control: y = −0·0014x + 0·5238) (Fig. 2b).
Interrelation of lymphocyte subsets in patients and controls
To assess the mechanism of T cell kinetics in patients, we examined the association among all subpopulations in patients and controls. Table 2 shows the results of correlation matrix analyses on each cell number in controls (A) and patients (B). The absolute number of T cells correlated positively with that of CD8αα+ cells (correlation coefficient: CC = 0·386, P = 0·0342) in patients but not in controls. On the other hand, the number of T cells showed a positive correlation with that of Vα24+ cells (CC = 0·657, P < 0·0001) only in controls. The Vα24+ cells showed a positive correlation with six of eight subsets in controls, while all the significant correlations were lost in patients. In addition, the positive correlation between CD8+ T cells or αβT cells and CD56+ cells was significant in patients but not controls. These significant correlations seen only in patients or only in controls may indicate the disease-specific or control-specific associations (in italics, Table 2). In the correlation matrix, 17 significant associations were found in both patients and controls (not italics, Table 2).
Table 2.
Correlation matrix of the absolute number of lymphocyte subpopulations
| (A) Control | (B) Patient | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3 | CD19 | CD56 | CD4 | CD8 | αβT | γδT | CD8αα | Vα24 | CD3 | CD19 | CD56 | CD4 | CD8 | αβT | γδT | CD8αα | Vα24 | |
| CD3 CC | 0·568 | 0·421 | 0·962 | 0·588 | 0·664 | n.s. | n.s. | 0·657 | 0·654 | 0·551 | 0·743 | 0·814 | 0·938 | n.s. | 0·386 | n.s. | ||
| P-value | 0·0005 | 0·0157 | < 0·0001 | 0·0007 | < 0·0001 | < 0·0001 | < 0·0001 | < 0·0001 | < 0·0001 | < 0·0001 | < 0·0001 | 0·0342 | ||||||
| CD19 CC | 0·516 | 0·535 | 0·740 | 0·679 | 0·424 | 0·349 | n.s. | 0·499 | 0·859 | 0·502 | 0·808 | n.s. | 0·514 | n.s. | ||||
| P-value | 0·0021 | 0·0028 | < 0·0001 | < 0·0001 | 0·0149 | 0·0499 | 0·0005 | < 0·0001 | 0·0005 | < 0·0001 | 0·0031 | |||||||
| CD56 CC | 0·466 | n.s. | n.s. | 0·377 | 0·632 | 0·395 | 0·622 | 0·471 | 0·566 | n.s. | 0·636 | n.s. | ||||||
| P-value | 0·0115 | 0·0329 | < 0·0001 | 0·0245 | < 0·0001 | 0·0012 | < 0·0001 | < 0·0001 | ||||||||||
| CD4 CC | 0·469 | 0·646 | n.s. | 0·401 | 0·551 | 0·442 | 0·807 | n.s. | 0·481 | n.s. | ||||||||
| P-value | 0·0109 | 0·0001 | 0·0338 | 0·0020 | 0·0027 | < 0·0001 | 0·0065 | |||||||||||
| CD8 CC | 0·421 | 0·510 | n.s. | 0·533 | 0·769 | 0·374 | n.s. | n.s. | ||||||||||
| P-value | 0·0248 | 0·0049 | 0·0030 | < 0·0001 | 0·0142 | |||||||||||||
| αβT CC | n.s. | n.s. | 0·359 | n.s. | n.s. | n.s. | ||||||||||||
| P-value | 0·0430 | |||||||||||||||||
| γδT CC | n.s. | 0·607 | n.s. | n.s. | ||||||||||||||
| P-value | 0·0002 | |||||||||||||||||
| CD8αα CC | n.s. | n.s. | ||||||||||||||||
| P-value | ||||||||||||||||||
| Vα24 CC | ||||||||||||||||||
| P-value | ||||||||||||||||||
This matrix shows Pearson's correlation coefficients (CC) analysed by Fisher's Z-transformation. The results of analysis using Pearson's CC were paralleled with those using Spearman's CC. Control-specific or patient-specific associations are shown in italic type. NS: not significant.
The mRNA expression levels in T cells
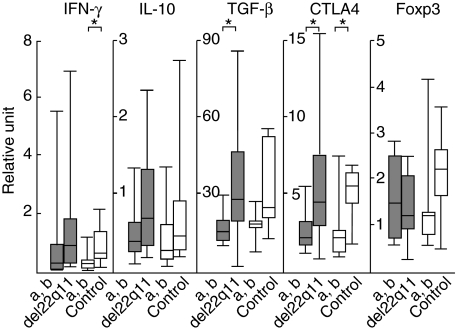
To evaluate the functional property of T cells in patients, the amounts of mRNA in highly purified T cells were compared between patients (n = 15) and age-matched controls (n = 14) by real-time PCR. The median age at the time of gene expression study was 4 years, ranging from 1 to 21 years (Table 1). IFN-γ represents as a Th1 cytokine, IL-10 and TGF-β as Th2 and/or regulatory cytokines, CTLA4 and Foxp3 as Tr cell-associated molecules. As no expression levels differed between patients and controls (data not shown), the levels were compared between the age group (a) 0–71 months and (b) 72–250 months to assess the ageing alteration (Fig. 3). The gene expression levels of IFN-γ, IL-10, TGF-β and CTLA4 over 71 months of age tended to be higher than those below the age in both patients and controls (see legend to Fig. 3). Foxp3 levels tended to show a decreasing pattern with age in patients only. However, there were no differences of the expression levels in the same age group between patients and controls. The covariance analysis showed no ageing alterations of expression levels between patients and controls. When the Th1/Th2 shift was assessed by comparison of the IFN-γ/IL-10 or IFN-γ/TGF-β ratios, there was neither difference between patient and controls nor any change between the two age groups (data not shown).
Fig. 3.
Quantification of interferon (IFN)-γ, interleukin (IL)-10, transforming growth factor (TGF)-β, cytotoxic T lymphocyte antigen-4 (CTLA4) and forkhead box p3 (Foxp3) genes in circulating T cells of patients and controls more or less than 6 years of age; (a) < 72 months, (b) ≥ 72 months. Total RNA was extracted from CD3+ cell fractions obtained from patients (closed bar) and age-matched controls (open bar). No gene expression levels of any cytokine or associated molecule differed in the group more than 71 months of age or in the group less than 72 months of age between patients and controls. Multiple comparisons with Bonferroni's correction abolished the significant differences in each single comparison (*) (IFN-γ controls: P = 0·028, transforming growth factor (TGF)-β patients: P = 0·030, CTLA4 patients: P = 0·040, controls: P = 0·049). Gene dosage was quantified by real-time polymerase chain reaction (PCR). The expression level of cytokine gene was divided by that of the internal control, glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and then defined as a ratio to that (1·00) of phytohaemagglutinin (PHA)-stimulated mononuclear cells (MNC) (relative units). The median bar in the box-and-whiskers plot represents 10%, 25%, 50%, 75% and 90% levels. The Mann–Whitney U-test was used to compare group means.
Clinical features and lymphocyte parameters
The proportion and number of lymphocytes or the gene expression levels in T cells studied above showed no significant association with various clinical features including types of conotruncal anomalies [ventricular septal defect (VSD) versus tetralogy of Fallot (TOF) and interruption of aortic arch (IAA) versus left aortic arch (LAA) versus right aortic arch (RAA); see footnotes in Table 1], presence or absence of velopharyngeal incompetance (VPI), infection proneness and hypocalcaemia. There were no remarkable values regarding lymphocyte subpopulations in patients having immunological abnormalities (patients 3, 7, 8 and 12, Table 1).
Discussion
The notable finding of the present study was that circulating T cells decreased with age at a slower rate in 22q11.2 deletion patients than in controls, and the kinetics were linked to the altered composition of minor T cell populations, arising partly from the extrathymic pathway. No impaired expression of cytokine genes in patients indicated a functional maturation of peripheral T cells. All but one (patient 12) of the patients studied here were free from serious infections after 1 year of age, and received live-attenuated vaccines with no specific adverse effects. The slower decreasing rate and balanced cytokine profile of circulating T cells in patients suggest an immune homeostasis of the limited T cell pool in thymic deficiency syndrome.
Early studies on the immune function of patients with a clinical diagnosis of DiGeorge syndrome revealed diminished T cell counts and preserved lymphoproliferative responses but an unpredictable clinical course [5,23,24]. The majority of patients were less susceptible to infection than expected from the degree of lymphopenia and hypoplastic thymus. Sullivan et al. [25] reported that the peripheral T cell counts were lower at birth in 19 patients having 22q11.2 deletion than age-matched controls, but the difference was less significant at 1 year of age. Mitogen responses were not depressed in patients, otherwise higher than in controls [18]. There was no correlation between immunological findings and phenotypic features [26]. The present study showed that the slower decline of T cells from birth to adulthood was dominant in CD4+ fractions, consistent with recent reports [9,27]. The CD4+ and CD8+ TCR Vβ repertoire of mature T cells in patients was appreciably normal but with limited diversity [6–8]. The development of thymus-dependent T cells may not be impaired severely as a result of defective thymus. The γδT cells, CD8αα+ cells and Vα24+ cells arising partly from the extrathymic pathway play a vital role in the innate immunity and immunoregulatory responses [28–30]. The paucity of information on the thymus-independent T cells in patients [9,11] allowed us to study the dynamics of the numerically minor but functionally influential populations consisting of both intra- and extrathymic lineages. The number and proportion of γδT cells in patients were not higher than those in controls in contrast to those of CD56+ natural killer (NK) cells (Fig. 1), despite sharing similar biological properties [28]. The different kinetics of γδT cells and NK cells were expected, as in previous reports [9]. The ageing decline of circulating γδT cell counts in patients was similar to that in healthy controls [31]. However, it was of note that γδT cells, CD8αα+ cells and Vα24+ cells showed distinct age-related changes in patients and controls (Fig. 2b). There was no association among these populations that were believed to include extrathymic T cells (Table 2).
A major concern is the mechanism of blunting slope of T cells in patients. Piliero et al. [10] studied the age-related changes of T cells up to elderly patients, and reported that the slower T cell decrease continued throughout their lives. They revealed that accelerated expansion of CD45RA+ to CD45RA– T cells contributed to the slower decline of CD4+ T cells in patients assessed by the balance of thymic output and apoptosis [10]. The unexpected increase of activated T cells has been distinguished in patients with this syndrome [6]. These findings are consistent with homeostatic T cell proliferation in patients with limited T cell production due to thymic hypoplasia, but not secondary to recurrent infections [10,32]. In addition to the slower shrinkage of CD4+ T cells in patients, the present study indicated as the disease-specific findings (1) the slowly increasing proportion of CD8αα+ cells and (2) the rapidly increasing proportion and number of Vα24+ cells (Fig. 2b). Konno et al. [33] revealed that peripheral CD8αα+ cells were derived from thymus-dependent pathway in healthy subjects, in contrast to the extrathymic differentiation of intraepithelial CD8αα+ cells expressing mainly TCRγδ [34,35]. Circulating CD8αα+ T cells increased up to 10% with age of CD8+ TCRαβ T cells in normal adults [33], which were consistent with the data in Fig. 2b. These cells were CD45RO–CCR7– memory effector cells positive for perforin, T cell intracellular antigen (TIA-1) and IFN-γ but negative for granzyme B and IL-2, arising from CD8αα+ TCRαβ T cells to expand oligoclonally [33]. In this context, the effector memory CD8αα+ cells in the circulation could expand poorly with age in patients. The positive correlation between CD8αα+ cells and T cells (Table 2) might suggest the intrathymic differentiation of CD8αα+ cells in patients with this syndrome.
NK T cells, which are characterized by the expression of invariant TCR Vα24/Vβ11-expressing cells in humans, interact with cells presenting glycolipids in the CD1d-restricted manner. This subset plays a regulatory role by producing IFN-γ and IL-4, and participates in the responses in infection, tumour-immunity and autoimmunity [30]. Vα24+NK T cells are classified into the major population of CD4+CD8– or CD4–CD8– double-negative (DN) phenotypes arising from intrathymic differentiation, and the minor one of CD4–CD8+ Vα24+ cells from extrathymic differentiation. DelaRosa et al. [36] reported that CD4+ or DN Vα24+ cells decrease and CD4–CD8+ Vα24+ cells increase with age. The physiological decrease of circulating Vα24+ cells might be secondary to age-associated changes in the T cell compartment with characteristic CD3+CD28+ shrinkage and CD3+CD28– expansion [36]. On the contrary, 22q11.2 deletion patients showed an increasing tendency of Vα24+ cells with age (Fig. 2b). Moreover, significant associations of Vα24+ cells with other subsets in controls were lost in patients, and there was no association between Vα24+ cells and CD8αα+ cells (Table 2). Taken together, the major subset of Vα24+ cells expand actively in patients against the decreasing T cell compartment. The ageing increase of NK T cells may exert immunoregulatory effects on the accelerated T cell activation and, if any, against the development of autoimmunity in this syndrome. In this reasoning, the majority of circulating CD8αα+ cells and Vα24+ cells in patients appear to stem from thymus-dependent lineages.
The high incidence of allergic diseases and autoimmune disorders in 22q11.2 deletion has not been clarified. Sullivan et al. [20] reported that absolute number of CD4+CD25+ T cells was lower in patients than in healthy infants. We showed no statistical difference in the gene expression levels of cytokines between patients and controls. On the other hand, Foxp3 levels only in patients tended to decrease after 6 years of age. Foxp3 gene is the master gene of CD4+CD25+ Tr cells, and the Tr cells arise from fetal thymus [37]. It may raise the possibility that patients have an insufficient production of Tr cells with age. Our study supported the appreciably preserved T cell functions in patients [9,10], and implicated the potential role of immunoregulatory CD4+CD25+ cells and NK T cells arising from thymus-dependent pathway. Peripheral T cell expansion should be taken into account for assessing the function of circulating T cell pool. Further studies including sufficient number of patients with overt autoimmune disorders and quantifying the TCR excision circles (TREC) are required to understand the mechanisms of autoimmunity in 22q11.2 deletion patients. Immunological analyses of 22q11.1 deletion syndrome are useful not only for the clinical management of patients, but also shed some light on the thymic function in immune homeostasis.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research to Y. Kanaya and S. Ohga from the Ministry of Education, Science, Sports and Culture of Japan. We are grateful to Tamami Tanaka PhD and Akihiko Nomura MD (Kyushu University) for helpful discussion, and to Naoki Fusazaki MD, Shiro Ishikawa MD and Junichiro Fukushige MD (Fukuoka Municipal Children's Hospital and Medical Center for Infectious Diseases, Fukuoka, Japan) for kindly providing blood samples.
References
- 1.Botto LD, May K, Fernhoff PM, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–7. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan KE. The clinical, immunological, and molecular spectrum of chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Curr Opin Allergy Clin Immunol. 2004;4:505–12. doi: 10.1097/00130832-200412000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Kariyazono H, Ohno T, Ihara K, et al. Rapid detection of the 22q11.2 deletion with quantitative real-time PCR. Mol Cell Probes. 2001;15:71–3. doi: 10.1006/mcpr.2000.0340. [DOI] [PubMed] [Google Scholar]
- 4.Markert ML, Hummell DS, Rosenblatt HM, et al. Complete DiGeorge syndrome: persistence of profound immunodeficiency. J Pediatr. 1998;132:15–21. doi: 10.1016/s0022-3476(98)70478-0. [DOI] [PubMed] [Google Scholar]
- 5.Martin Mateos MA, Perez Duenas BP, Iriondo M, Krauel J, Gean Molins E. Clinical and immunological spectrum of partial DiGeorge syndrome. J Invest Allergol Clin Immunol. 2000;10:352–60. [PubMed] [Google Scholar]
- 6.Pierdominici M, Marziali M, Giovannetti A, et al. T cell receptor repertoire and function in patients with DiGeorge syndrome and velocardiofacial syndrome. Clin Exp Immunol. 2000;121:127–32. doi: 10.1046/j.1365-2249.2000.01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierdominici M, Mazzetta F, Caprini E, et al. Biased T cell receptor repertoires in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Clin Exp Immunol. 2003;132:323–31. doi: 10.1046/j.1365-2249.2003.02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancrini C, Romiti ML, Finocchi A, et al. Post-natal ontogenesis of the T cell receptor CD4 and CD8 Vbeta repertoire and immune function in children with DiGeorge syndrome. J Clin Immunol. 2005;25:265–74. doi: 10.1007/s10875-005-4085-3. [DOI] [PubMed] [Google Scholar]
- 9.Jawad AF, McDonald-Mcginn DM, Zackai E, Sullivan KE. Immunologic features of chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) J Pediatr. 2001;139:715–23. doi: 10.1067/mpd.2001.118534. [DOI] [PubMed] [Google Scholar]
- 10.Piliero LM, Sanford AN, McDonald-McGinn DM, Zackai EH, Sullivan KE. T cell homeostasis in humans with thymic hypoplasia due to chromosome 22q11.2 deletion syndrome. Blood. 2004;103:1020–5. doi: 10.1182/blood-2003-08-2824. [DOI] [PubMed] [Google Scholar]
- 11.Collard HR, Boeck A, McLaughlin TM, et al. Possible extrathymic development of nonfunctional T cells in a patient with complete DiGeorge syndrome. Clin Immunol. 1999;91:156–62. doi: 10.1006/clim.1999.4691. [DOI] [PubMed] [Google Scholar]
- 12.Gennery AR, Barge D, O'Sullivan JJ, Flood TJ, Abinun M, Cant AJ. Antibody deficiency and autoimmunity in 22q11.2 deletion syndrome. Arch Dis Child. 2002;86:422–5. doi: 10.1136/adc.86.6.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staple L, Andrews T, McDonald-McGinn D, Zackai E, Sullivan KE. Allergies in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/ velocardiofacial syndrome) and patients with chronic granulomatous disease. Pediatr Allergy Immunol. 2005;16:226–30. doi: 10.1111/j.1399-3038.2005.00259.x. [DOI] [PubMed] [Google Scholar]
- 14.Bruno B, Barbier C, Lambilliotte A, Rey C, Turck D. Auto-immune pancytopenia in a child with DiGeorge syndrome. Eur J Pediatr. 2002;161:390–2. doi: 10.1007/s00431-002-0976-y. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence S, McDonald-McGinn DM, Zackai E, Sullivan KE. Thrombocytopenia in patients with chromosome 22q11.2 deletion syndrome. J Pediatr. 2003;143:277–8. doi: 10.1067/S0022-3476(03)00248-8. [DOI] [PubMed] [Google Scholar]
- 16.Okiyama N, Yamamoto T, Watanabe K, Yokozeki H, Nishioka K. Juvenile dermatomyositis in association with 22q11.2 deletion syndrome. Br J Dermatol. 2005;152:1370–2. doi: 10.1111/j.1365-2133.2005.06622.x. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan KE, McDonald-McGinn DM, Driscoll DA, et al. Juvenile rheumatoid arthritis-like polyarthritis in chromosome 22q11.2 deletion syndrome (DiGeorge anomalad/velocardiofacial syndrome/conotruncal anomaly face syndrome) Arthritis Rheum. 1997;40:430–6. doi: 10.1002/art.1780400307. [DOI] [PubMed] [Google Scholar]
- 18.Sediva A, Bartunkova J, Zachova R, et al. Early development of immunity in diGeorge syndrome. Med Sci Monit. 2005;11:CR182–7. [PubMed] [Google Scholar]
- 19.Gupta S, Aggarwal S, Nguyen T. Increased spontaneous apoptosis in T lymphocytes in DiGeorge anomaly. Clin Exp Immunol. 1998;113:65–71. doi: 10.1046/j.1365-2249.1998.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan KE, McDonald-McGinn D, Zackai EH. CD4(+) CD25(+) T cell production in healthy humans and in patients with thymic hypoplasia. Clin Diagn Lab Immunol. 2002;9:1129–31. doi: 10.1128/CDLI.9.5.1129-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohga S, Nomura A, Ihara K, et al. Cytokine imbalance in hyper-IgE syndrome: reduced expression of transforming growth factor-beta and interferon-gamma genes in circulating activated T cells. Br J Haematol. 2003;121:324–31. doi: 10.1046/j.1365-2141.2003.04267.x. [DOI] [PubMed] [Google Scholar]
- 22.Ohga S, Nomura A, Takada H, et al. Dominant expression of interleukin-10 and transforming growth factor-beta genes in activated T cells of chronic active Epstein–Barr virus infection. J Med Virol. 2004;74:449–58. doi: 10.1002/jmv.20197. [DOI] [PubMed] [Google Scholar]
- 23.Barrett DJ, Ammann AJ, Wara DW, Cowan MJ, Fisher TJ, Stiehm ER. Clinical and immunologic spectrum of the DiGeorge syndrome. J Clin Lab Immunol. 1981;6:1–6. [PubMed] [Google Scholar]
- 24.Bastian J, Law S, Vogler L, et al. Prediction of persistent immunodeficiency in the DiGeorge anomaly. J Pediatr. 1989;115:391–6. doi: 10.1016/s0022-3476(89)80837-6. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan KE, McDonald-McGinn D, Driscoll DA, Emanuel BS, Zackai EH, Jawad AF. Longitudinal analysis of lymphocyte function and numbers in the first year of life in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Clin Diagn Lab Immunol. 1999;6:906–11. doi: 10.1128/cdli.6.6.906-911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan KE, Jawad AF, Randall P, et al. Lack of correlation between impaired T cell production, immunodeficiency, and other phenotypic features in chromosome 22q11.2 deletion syndromes. Clin Immunol Immunopathol. 1998;86:141–6. doi: 10.1006/clin.1997.4463. [DOI] [PubMed] [Google Scholar]
- 27.Chinen J, Rosenblatt HM, Smith EO, Shearer WT, Noroski LM. Long-term assessment of T cell populations in DiGeorge syndrome. J Allergy Clin Immunol. 2003;111:573–9. doi: 10.1067/mai.2003.165. [DOI] [PubMed] [Google Scholar]
- 28.Colonna-Romano G, Potestio M, Aquino A, Candore G, Lio D, Caruso C. Gamma/delta T lymphocytes are affected in the elderly. Exp Gerontol. 2002;37:205–11. doi: 10.1016/s0531-5565(01)00185-1. [DOI] [PubMed] [Google Scholar]
- 29.Imhof BA, Dunon D, Courtois D, Luhtala M, Vainio O. Intestinal CD8 alpha alpha and CD8 alpha beta intraepithelial lymphocytes are thymus derived and exhibit subtle differences in TCR beta repertoires. J Immunol. 2000;165:6716–22. doi: 10.4049/jimmunol.165.12.6716. [DOI] [PubMed] [Google Scholar]
- 30.Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: facts, functions and fallacies. Immunol Today. 2000;21:573–83. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 31.Argentati K, Re F, Donnini A, et al. Numerical and functional alterations of circulating gammadelta T lymphocytes in aged people and centenarians. J Leukoc Biol. 2002;72:65–71. [PubMed] [Google Scholar]
- 32.Hakim FT, Memon SA, Cepeda R, et al. Age-dependent incidence, time course, and consequences of thymic renewal in adults. J Clin Invest. 2005;115:930–9. doi: 10.1172/JCI22492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konno A, Okada K, Mizuno K, et al. CD8alpha alpha memory effector T cells descend directly from clonally expanded CD8alpha+ betahigh TCRalpha beta T cells in vivo. Blood. 2002;100:4090–7. doi: 10.1182/blood-2002-04-1136. [DOI] [PubMed] [Google Scholar]
- 34.Lin T, Matsuzaki G, Kenai H, et al. Characteristics of fetal thymus-derived T cell receptor gamma delta intestinal intraepithelial lymphocytes. Eur J Immunol. 1994;24:1792–8. doi: 10.1002/eji.1830240811. [DOI] [PubMed] [Google Scholar]
- 35.Lin T, Matsuzaki G, Kenai H, Nomoto K. Progenies of fetal thymocytes are the major source of CD4–CD8+ alpha alpha intestinal intraepithelial lymphocytes early in ontogeny. Eur J Immunol. 1994;24:1785–91. doi: 10.1002/eji.1830240810. [DOI] [PubMed] [Google Scholar]
- 36.DelaRosa O, Tarazona R, Casado JG, et al. Valpha24+ NKT cells are decreased in elderly humans. Exp Gerontol. 2002;37:213–7. doi: 10.1016/s0531-5565(01)00186-3. [DOI] [PubMed] [Google Scholar]
- 37.Takahata Y, Nomura A, Takada H, et al. CD25+CD4+ T cells in human cord blood: an immunoregulatory subset with naive phenotype and specific expression of forkhead box p3 (Foxp3) gene. Exp Hematol. 2004;32:622–9. doi: 10.1016/j.exphem.2004.03.012. [DOI] [PubMed] [Google Scholar]