Abstract
Common variable immunodeficiency disease (CVID) is a heterogeneous syndrome characterized by low immunoglobulin serum levels and recurrent bacterial infections. Several studies suggest that CVID patients have a polarized immune response towards a T helper type 1 phenotype (TH1). However, the factors causing the TH1 polarization remain to be determined in this disease. In the present study, serum interleukin (IL)-12, interferon (IFN)-γ levels and the IL-12p40 and IFN-γ gene were studied in CVID patients. Furthermore, we evaluate dendritic cells (DCs) compartment, myeloid dendritic cells (mDCs) and plasmocytoid dendritic cells (pDCs), which help to differentiate naive T cells preferentially into TH1 and TH2, respectively. The serum IL-12p40 subunit levels were increased significantly in CVID patients compared to healthy controls. We examined whether these elevated serum IL-12p40 levels are associated with IFN-γ or IL-12p40 gene polymorphisms, or with new mutations in the IL-12p40 promoter gene. In our hands, no new mutations were found and gene polymorphisms frequencies in CVID patients were similar to the control population. In conclusion, the elevated serum levels of IL-12p40 found in our CVID patients were not related to these genetic variations. The DC compartment analysis did not show an imbalance between pDCs and mDCs, but revealed the presence of low numbers and percentage of both DC populations in CVID.
Keywords: common variable immunodeficiency disease, dendritic cells, gene polymorphism, IFN-γ, IL-12
Introduction
Common variable immunodeficiency disease (CVID) is a primary immunodeficiency disease characterized by hypogammaglobulinaemia and recurrent bacterial infections. Inflammatory, autoimmun e and/or granulomatous diseases are present in some patients. A defect in B cell differentiation and maturation was postulated originally as the main cause of this disorder. Some studies showed a failure of B cells from CVID patients to differentiate into plasma cells and to produce high-affinity antibodies of different isotypes [1]. However, other studies showed restoration of immunoglobulin secretion after the correct stimulation of B cells from CVID patients [2]. T cell abnormalities are also common in CVID patients and include decreased lymphocyte proliferation to different mitogens and antigens, reduced number of antigen-primed T cells [3,4], alterations in interleukin (IL) secretion [5] and increased apoptosis of the CD45RA+ T cell subpopulation [6].
A perfect balance between T helper type 1 phenotype (TH1) and TH2 responses is essential for the achievement of an optimal immune response against most pathogens. Increased secretion of IL-12 [7,8], inducing a TH1 cytokine response, has been described in CVID patients. Conversely, the TH2 response necessary for normal antibody secretion is diminished. The IL-12 cytokine is critical in the initiation of innate and adaptive immune responses to many infections. It promotes cell-mediated responses, inducing natural killer (NK) and T cells to secrete interferon (IFN)-γ and favouring the differentiation of precursor TH0 cells into TH1 effector cells [9].
The implication of professional antigen-presenting cells (APCs) in the alterations found in CVID patients and in the disease pathogenesis remains unclear [10,11]. Denditric cells (DCs) are critical APCs, capable of stimulating naive T cells and initiating primary immune responses [12,13]. Two distinct lineages of dendritic cells have been defined [14]: myeloid dendritic cells (mDCs) that produce large amounts of IL-12 and induce strong TH1 and cytotoxic T lymphocyte responses [15,16], and plasmocytoid dendritic cells (pDCs) that induce TH2 responses.
Cytokine production may be up-regulated and/or down-regulated, like most of the immune system responses, although it differs significantly among individuals. Cytokine gene polymorphisms may determine the individual differences. In general, cytokine genes are highly conserved in terms of exon sequences but silent mutations may alter cytokine production [17,18]. Given the importance of the IL-12p40 subunit in the immune system, genetic variants of this cytokine may be important in causing immunoregulatory abnormalities.
The objective of the present study was to analyse serum concentrations of cytokines that prime the TH1 response and to evaluate the possible association of allelic regions of the IL-12p40 and IFN-γ genes, having gene-regulatory potential, with the IL-12p40 elevated serum levels found in CVID patients. The contribution of the different peripheral blood DCs populations to the TH1 shift present in CVID patients was also studied.
Materials and methods
Subjects
All patients were diagnosed according to the diagnostic criteria of the International Union for Immunological Societies Scientific Group (IUIS) for primary immunodeficiency diseases and were receiving monthly regular substitution therapy with intravenous gammaglobulin (IVIG). In the study we included two X-linked agammaglobulinaemia and three IgG2-deficient patients receiving monthly regular IVIG therapy as a treatment control group. All patients included in the study were free of infections for at least 3 months before sample collection. After informed consent, samples were collected immediately before IVIG infusion. The study was approved by the Hospital Son Dureta institutional human research committee. Patients were classified according to the quantification of switched memory B cells [19] into three groups: MB2- patients with normal number of naive/memory B cells (IgD– CD27+), MB1– patients with a reduced number of switched memory B cells (IgD– CD27+) but normal non-switched memory B cells (IgD+ CD27+) and MB0– patients with a defect in both switched and non-switched memory B cells. The most frequent phenotype was MB0 (53%). The second group was MB1 (33%), and MB2 was 14%. All patients included in the study were free of granulomas.
Serum IL-12 cytokine concentrations were measured in 27 CVID patients and 45 healthy controls. The cytokine gene study included 35 CVID patients and 56 controls. The peripheral blood phenotypic study included 19 CVID patients and 19 healthy non-atopic sex- and age-matched controls.
IL-12 and IFN-γ protein analysis
Serum samples were drawn from each subject and stored at −70°C until use. The concentration of serum IFN-γ, IL-12 p70 was measured by a commercially available enzyme-linked immunosorbent assay (ELISA) kit from Beckman Coulter Company (Marseille, France) and total IL-12 (p40 and p70) from Pierce Endogen (Rockford, IL, USA).
IL-12 p40 and IFN-γ genotyping
All samples were genotyped with DNA extracted from peripheral blood. The final concentration of reaction components were: 200 µM of each dNTP, 2 mM MgCl2, 67 mM Tris base pH 8·8, 16·6 mM ammonium sulphate, 0·01% (v/v) Tween 20, 200 ng DNA and 0·2 units of Taq polymerase (Ecogen, Barcelona, Spain). DNA was amplified using the polymerase chain reaction (PCR) GeneAmp system 9700 (Applied Biosystems, Foster City, CA, USA).
IL-12 p40 gene polymorphism exon 8, 3′UTR A/C (+ 1188)
We performed the studies following the method described by Huang et al. [20].
IL-12p40 promoter polymorphism CTCTAA/GC
We amplified DNA samples with the previous reported primers 5′TCAGACACATTAACCTTGCA-3′ and 5′-ACCT GCCAATGACCACATTA-3′, and PCR was performed as described previously [21]. DNA sequencing was conducted with the Big Dye Terminator Cycle sequencing kit version 3·1 (Applied Biosystems). The products were evaluated on an ABI 3100 DNA sequencer (Applied Biosystems).
IFN-γ polymorphism
Analysis of microsatellite polymorphisms in the first intron of IFN-γ gene was conducted as described previously [22]. Eight µl of the PCR reaction were submitted to electrophoresis on a 6% polyacrylamide sequencing gel. The forward primer was Cy5-labelled, and the results were analysed using the ALFXpress sequencer and Fragment Analyser software (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Mutation analysis of IL-12 p40 gene promoter (HSU89323)
This was performed by PCR amplification of genomic DNA by the use of AmpliTaq Gold polymerase. The forward and reverse primers (numbered: 1, 2, 3, 4) were selected from the PAC sequence (HSU89323) and used to amplify specific segments of IL-12p40 promoter. The forward primer number 4 has a single base change (T→C) in the last nucleotide sequence because we observed that the HSU89323 had a mistake or was a rare polymorphism: (number 1: forward 5′AAGCTTCTTTTGCATAACTGGC-3′ and reverse 5′CTG GCCGTGGGTGGAGAC-3′, product size 548 base pairs (bp); number 2: forward 5′-AGGCCTAGAGGACACAGGG-3′ and reverse 5′AGGTATGCAAAGGTGTACACC3′, product size 568; number 3: forward 5′-ACATGTTCCTGTTCACG TGCA3′ and reverse 5′-CCTGGTTCTTCCCAAGTCAG-3′, product size 549 bp; number 4: forward 5′GATGTACTAAA CCCTTTGCCC-3′ and reverse 5′TTGGGAAGTGCTTAC CTTGCT 3, product size 473 bp. PCR cycling conditions were 7 min at 95°C, 30 cycles of 30 s at 95°C, 30 s at 64°C and 60 s at 72°C, and 5 min at 72°C. DNA sequencing was conducted with the Big Dye Terminator Cycle sequencing kit version 3·1 (Applied Biosystems). The products were evaluated on an ABI 3100 DNA sequencer (Applied Biosystems). The IL-12p40 sequence was compared with the previously described sequences of the promoter (HSU89323) [23].
Flow cytometry analysis
Whole peripheral blood cells were stained following the manufacturer's instructions and analysed on FACScalibur cytometer (BD Pharmingen, San Jose, CA, USA) using CELLQuest software. To detect the mDC (CD11c+) and pDC (CD123+) subsets of peripheral blood dendritic cells, we stained them with a lineage cocktail (Lin 1: fluorescein isothiocyanate (FITC) containing antibodies against CD3, CD14, CD16, CD19, CD20 and CD56), PerCP-conjugated anti-human leucocyte antigen (HLA)-DR and phycoerythrin (PE)-conjugated anti-CD11c or PE-conjugated anti-CD123. Murine immunoglobulins of appropriate isotypes were used as controls. mDCs and pDCs were defined as lin– HLA– DR+ CD11c+ and lin– HLA– DR+ CD123+ cells, respectively. The percentage and absolute number of mDCs and pDCs were calculated from the amount of white blood cells. The ratio of mDCs to pDCs was defined as the quotient between the proportion of mDCs and that of pDCs.
Statistical methods
Data were analysed using the prism statistical package. The Mann–Whitney U-test was used for statistical analysis of DCs populations and serum cytokine levels. Significance level assumed in these comparisons was P < 0·05. All values are expressed as the mean ± s.e.m.
Allelic and genotype frequencies were estimated by direct counting. Case–control association analyses were performed using the Fisher's exact test. When necessary, a Bonferroni correction was applied to obtain the corrected P-value according to the number of comparisons performed, and P < 0·05 was considered statistically significant.
Results
Serum cytokine levels
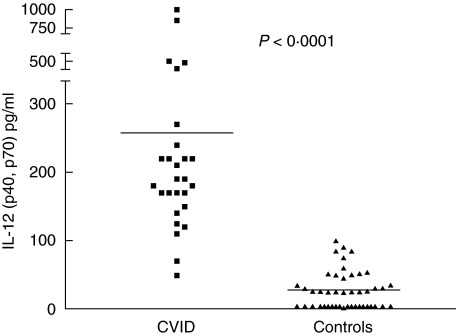
Serum IL-12 (p40 and p70) was significantly increased in CVID compared to controls (257·9 ± 42·0 versus 27·50 ± 4·1, P < 0·001, respectively) (Fig. 1). In this series, three CVID patients who did not receive IVIG treatment also had elevated serum IL-12, whereas in two X-linked agammaglobulinaemic patients receiving IVIG, serum IL-12 concentration was normal (data not shown). Serum IL-12p70 and IFN-γ were barely detectable in patients and controls.
Fig. 1.
Serum interleukin (IL)-12 (p40 and p70) levels in common variable immunodeficiency disease (CVID) patients control group. Total serum IL-12 levels of CVID patients were highly significant (***P< 0·0001). Horizontal lines represent the mean of each group.
IL-12p40 and IFN-γ genotyping
No associations were found between the different IL-12 and IFN-γ polymorphism and the enhanced IL-12p40 production (Table 1). No new mutations in the IL-12 p40 promoter region were found.
Table 1.
Alleles and genotype frequencies of common variable immunodeficiency disease (CVID) patients.
| CVID polymorphism | Control polymorphism | ||||||
|---|---|---|---|---|---|---|---|
| IFN-γ (CA)n intron | |||||||
| Allele n = 64 | Frequency | Genotype n = 32 | Frequency | Allele n = 112 | Frequency | Genotype n = 56 | Frequency |
| 1 (CA)10 | 0 | 2/2 | 7 (21,9%) | 1 (CA)10 | 0 | 2/2 | 13 (23,21) |
| 2 (CA)11 | 34 (53,1%) | 2/3 | 20 (62,5%) | 2 (CA)11 | 57 (50,89) | 2/3 | 27 (48,21) |
| 3 (CA)12 | 28 (43,7%) | 2/4 | 0 | 3 (CA)12 | 48 (42,86) | 2/4 | 2 (3,57) |
| 4 (CA)13 | 1 (1,6%) | 2/5 | 0 | 4 (CA)13 | 5 (4,46) | 2/5 | 2 (3,57) |
| 5 (CA)14 | 1 (1,6%) | 3/3 | 3 (9,4%) | 5 (CA)14 | 2 (1·79) | 3/3 | 10 (17,86) |
| 6 (CA)15 | 0 | 3/4 | 1 (3%) | 6 (CA)15 | 3/4 | 1 (1,79) | |
| 3/5 | 1 (3%) | 3/5 | 0 | ||||
| 4/4 | 0 | 4/4 | 1 (1,79) | ||||
| IL-12 p40 3′UTR +1188 A/C | |||||||
| Allele n = 70 | Frequency | Genotype n = 35 | Frequency | Allele n = 108 | Frequency | Genotype n = 54 | Frequency |
| 1 | 31 (77·5%) | 1/1 | 22 (63%) | 1 | 90 (83,33) | 1/1 | 37 (68,52) |
| 2 | 9 (22%) | 1/2 | 10 (29%) | 2 | 18 (16,67) | 1/2 | 16 (29,63) |
| 2/2 | 3 (9%) | 2/2 | 1 (1,58) | ||||
| IL-12 p40 promoter CTCTAA/GC | |||||||
| Allele n = 44 | Frequency | Genotype n = 22 | Frequency | Allele n = 586 | Frequency* | Genotype n = 293 | Frequency* |
| 1 | 22 (52%) | 1/1 | 5 (24%) | 1 | 286 (49%) | 1/1 | 69 (24%) |
| 2 | 20 (48%) | 1/2 | 12 (57%) | 2 | 300 (51%) | 1/2 | 148 (51%) |
| 2/2 | 4 (19%) | 2/2 | 76 (26%) | ||||
Fisher's exact test was used to compare CVID patients and controls. p >0·05 in all cases.
These control populations frequencies were reported by Morahan et al. 2002 [21]. IFN: interferon; IL: interleukin.
Absolute number and percentage of peripheral blood mDCs and pDCs in CVID patients and healthy subjects
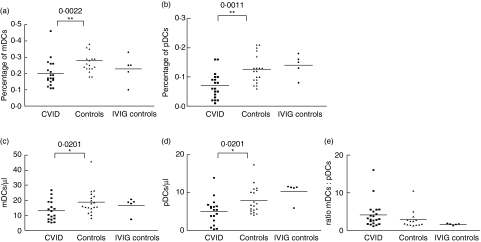
We examined the mDCs and pDCs percentages and absolute numbers in CVID peripheral blood samples. Figure 2 shows the mDC and pDC percentages on total leucocytes from 19 CVID patients, 19 healthy subjects and five IVIG control subjects, and shows the absolute number (cells/µl) and mDCs/pDCs ratio. The percentage and absolute number of peripheral blood pDCs and mDCs were significantly lower in patients than in healthy subjects. In order to assess whether the low leucocyte count observed in some CVID patients could be associated with the lower DCs numbers observed in CVID patients, we performed a correlation analysis between these cell populations and no correlation was found (data not shown). No differences in mDC or pDC percentage or absolute numbers were found between the IVIG control treatment group and the healthy control group (Fig. 2).
Fig. 2.
Myeloid dendritic cells (mDCs) and plasmocytoid DCs (pDCs) percentages, absolute number of mDCs and pDCs and ratio of mDCs/pDCs from peripheral blood. Percentage of (a) mDCs and (b) pDCs of total leucocytes in common variable immunodeficiency disease (CVID) patients, healthy control group and intravenous gammaglobulin (IVIG) treatment control group. The percentage of these DC populations from CVID patients was significantly lower than controls (**P< 0·01). Absolute number (cells/µl) of (c) mDCs, (d) pDCs and (e) ratio of mDCs/pDCs in CVI patients, healthy control group, and IVIG treatment control group. Absolute number of pDCs and mDCs from CVID patients were lower than healthy controls (*P< 0·03) and the mDCs/pDCs ratio of CVID patients was not different to the control group. Horizontal lines represent the mean of each group.
Discussion
Despite extensive studies on CVID, the underlying cause of the disease remains elusive. B cells from CVID patients fail to produce antibodies and hypogammaglobulinaemia is a consistent finding in this syndrome. In vitro studies suggest that B cells are intrinsically normal and there is a failure in the incoming signal [2]. In this sense, several abnormalities have been found in patient T cells that may result in a defective T/B cell cooperation.
We have described previously a significant increase in IL-12p40 serum concentration in a group of CVID patients [7]. Altered monocyte/T cell interaction with an important increase of IL-12 and IFN-γ production was also suggested [8]. Other studies showed an elevated expression of IL-12β1 receptor subunit in CVID patients [24]. Taken together, all these results suggested a bias towards TH1 responses in CVID patients, a diminished TH2 response and low antibody production.
IL-12 is a critical cytokine involved in the initiation of TH1 responses. The IL-12 molecule is a 70 kDa heterodimer that comprises p40 and p35 subunits, encoded by different genes [25–27]. The p35 subunit is expressed constitutively by many cell types, while p40 is an inducible subunit, restricted to several cells of haematopoietic origin that include mDCs but not pDCs. Our analysis of serum IL-12 showed elevated IL-12p40 concentrations in the majority of CVID patients. IVIG administration alters significantly the serum pattern of selected cytokines as IL-6, IL-8, IL-1Rα and tumour necrosis factor (TNF)-α but not IL-1-beta, IFN-γ or IL-2 [28]. This is not the case for IL-12: in our hands, three CVID patients without gammaglobulin treatment presented elevated serum concentrations of IL-12p40, whereas two gammaglobulin-treated X-linked agammaglonulinaemia patients showed normal levels of this cytokine.
The clinical variability of CVID, coupled with the predominant TH1 response described, suggests strongly that genetic variants of some cytokine genes or promoters may be related to the CVID phenotype. We have focused on genes involved in the regulation of TH1 response such as IFN-γ and IL-12p40, but we could not find any relationship between the polymorphisms studied and the altered cytokine pattern secretion, the B cell memory-based classification of the patients, or CVID susceptibility. The sequence of IL-12p40 promoter was normal in CVID patients, although we cannot exclude that other non-analysed cytokine gene regions could be implicated in the IL-12p40 increase.
Recent studies reinforce the role of DCs in the induction of TH1/TH2 polarization in the immune responses [29,30]. Two lineages of DCs have been described: myeloid DCs (mDCs) that prime TH1 responses, and pDCs that polarize immune responses towards TH2 type. In the present study we observed a low percentage and absolute number of peripheral blood DCs compared to healthy subjects. These results corroborate previous studies demonstrating defective dendritric cell function [10,11], and suggest that impairment in the innate immune system may account cumulatively for the inability of CVID patients to eradicate pathogens through conventional immune pathways, resulting in an increased risk for recurrent bacterial infections.
We did not find an altered balance between DC subpopulations that could explain the elevated serum IL-12p40 levels observed in CVID patients. Higher individual IL-12 production by the CVID DCs or the presence of an alternative IL-12 production source as monocytes, suggested by Cambronero et al. [8], must be the cause of these elevated IL-12 serum levels.
Leucopenia is usually a laboratory finding in CVID patients. The lack of correlation between DCs percentage and total leucocytes in our patients excluded that our results were due to the low leucocyte numbers in some of our CVID patients; nor was the diminished number of DCs secondary to IVIG treatment or infections, as demonstrated by the fact that IVIG-treated non-CVID immunodeficient patients had normal numbers of DCs. Neither the elevated IL-12 p40 serum levels nor the studied polymorphisms were related to the classification of the patients.
From our observations the high serum concentration of IL-12p40 is, together with hypogammaglobulinaemia, the most consistent abnormal laboratory finding observed to date in this heterogeneous syndrome.
Acknowledgments
We thank Dr Antoni Gaya and Dr Javier Calvo for expert technical assistance. We would like to thank Dra Aitziber Etxagibel for discussion and proofreading the manuscript. This work was supported by the LAIR Foundation and ISCIII (FIS 01/1015) and (FIS 02/1005).
References
- 1.Agematsu K, Futatani T, Hokibara S, et al. Absence of memory B cells in patients with common variable immunodeficiency. Clin Immunol. 2002;103:34–42. doi: 10.1006/clim.2001.5197. [DOI] [PubMed] [Google Scholar]
- 2.Punnonen J, Kainulainene L, Ruuskanen O, Nikoskelainen J, Arvilommi H. IL-4 synergizes with IL-10 and anti CD40 MoAbs to induce B cell differentiation in patients with common variable immunodeficiency. Sand J Immunol. 1997;45:203–12. doi: 10.1046/j.1365-3083.1997.d01-381.x. [DOI] [PubMed] [Google Scholar]
- 3.Stagg AJ, Funauchi M, Knight SC, Webster AD, Farrant J. Failure in antigen responses by T cells from patients with common variable immunodeficiency (CVID) Clin Exp Immunol. 1994;96:48–53. doi: 10.1111/j.1365-2249.1994.tb06228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondratenko I, Amlot PL, Webster AD, Farrant J. Lack of specific antibody response in common variable immunodeficiency (CVID) associated with failure in production of antigen-specific memory T cells. Clin Exp Immunol. 1997;108:9–13. doi: 10.1046/j.1365-2249.1997.d01-993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrer JM, Iglesias J, Hernandez M, Matamoros N. Alterations in the interleukin secretion (IL-2 and IL-4) by CD4 and CD4CD45RO cells from common variable immunodeficiency patients. Clin Exp Immunol. 1995;102:286–9. doi: 10.1111/j.1365-2249.1995.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iglesias J, Matamoros N, Raga S, Ferrer JM, Mila J. CD95 expression and function on lymphocyte subpobpulations in common variable immunodeficiency: related to increased apoptosis. Clin Exp Immunol. 1999;117:138–46. doi: 10.1046/j.1365-2249.1999.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raga S, Pons J, Iglesias J, Matamoros N. High levels of IL-12 and IL-10 in common variable immunodeficiency. IL-12 neutralization effects. Inmunologia. 1998;17:54. [Abstract]. [Google Scholar]
- 8.Cambronero R, Sewell WA, North ME, Webster AD, Farrant J. Up-regulation of IL-12 in monocytes: a fundamental defect in common variable immunodeficiency. J Immunol. 2000;164:488–94. doi: 10.4049/jimmunol.164.1.488. [DOI] [PubMed] [Google Scholar]
- 9.Aste-Amezaga M, D’Andrea A, Kubin M, Trinchieri G. Cooperation of natural killer cell stimulatory factor/interleukin-12 with other stimuli in the induction of cytokines and cytotoxic cell-associated molecules in human T and NK cells. Cell Immunol. 1994;156:480–92. doi: 10.1006/cimm.1994.1192. [DOI] [PubMed] [Google Scholar]
- 10.Bayry J, Lacroix-Desmazes S, Kazatchkine MD, et al. Common variable immunodeficiency is associated with defective functions of dendritic cells. Blood. 2004;104:2441–3. doi: 10.1182/blood-2004-04-1325. [DOI] [PubMed] [Google Scholar]
- 11.Scott-Taylor TH, Green MR, Eren E, Webster AD. Monocyte derived dendritic cell responses in common variable immunodeficiency. Clin Exp Immunol. 2004;138:484–90. doi: 10.1111/j.1365-2249.2004.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rissoan MC, Soumelis V, Kadowaki N, et al. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–6. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 13.Banchereau J, Seinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 14.Robinson SP, Patterson S, English N, Davies D, Knight SC, Reid CD. Human peripheral blood contains two distinct lineages of dendritic cells. Eur J Immunol. 1999;29:2769–78. doi: 10.1002/(SICI)1521-4141(199909)29:09<2769::AID-IMMU2769>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Macatonia SE, Hosken NA, Litton M, et al. Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J Immunol. 1995;154:5071–9. [PubMed] [Google Scholar]
- 16.Cella M, Scheidegger D, Palmer-Lehmann K, Lane P, Lanzavecchia A, Albert G. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacity: T-T help via APC activation. J Exp Med. 1996;184:747–52. doi: 10.1084/jem.184.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bidwell JL, Wood NA, Morse HR, Olomolaiye OO, Keen LJ, Laundy GJ. Human cytokine gene nucleotide sequence alignments. Eur J Immunogenet. 1998;25(Suppl. 1):83–266. [PubMed] [Google Scholar]
- 18.Bidwell JL, Wood NA, Morse HR, Olomolaiye OO, Keen LJ, Laundy GJ. Human cytokine gene nucleotide sequence alignments. Eur J Immunogenet. 1999;26(Suppl. 1):135–223. doi: 10.1046/j.1365-2370.1999.00143.x. [DOI] [PubMed] [Google Scholar]
- 19.Piqueras B, Lavenu-Bombled C, Galicier L, et al. Common variable immunodeficiency patient classification based on impaired B cell memory differentiation correlates with clinical aspects. J Clin Immunol. 2003;23:385–400. doi: 10.1023/a:1025373601374. [DOI] [PubMed] [Google Scholar]
- 20.Huang D, Cancilla MR, Morahan G. Complete primary structure, chromosomal localisation, and definition of polymorphismos of the gene encoding the human interleukin-12 p40 subunit. Genes Immun. 2000;1:15–520. doi: 10.1038/sj.gene.6363720. [DOI] [PubMed] [Google Scholar]
- 21.Morahan G, Huang D, Wu M, et al. Association of IL-12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet. 2002;360:455–9. doi: 10.1016/S0140-6736(02)09676-9. [DOI] [PubMed] [Google Scholar]
- 22.Pravica V, Asderakis A, Perry C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet. 1999;26:1–3. doi: 10.1046/j.1365-2370.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- 23.Ma X, Chow JM, Gri G, et al. The interleukin 12p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–57. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McQuaid A, Tormey VJ, Trafford B, Webster AD, Bofill M. Evidence for increased expression of regulatory cytokine receptors interleukin-12R and interleukin-18R in common variable immunodeficiency. Clin Exp Immunol. 2003;134:321–7. doi: 10.1046/j.1365-2249.2003.02271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern AS, Podlaski FJ, Hulmes JD, et al. Purification to homogeneity and partial characterization of cytotoxic lymphocyte maturation factor from human B-lymphoblastoid cells. Proc Natl Acad Sci USA. 1989;87:6808–12. doi: 10.1073/pnas.87.17.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf SF, Temple PA, Kobayashi M, et al. Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J Immunol. 1991;146:3074–81. [PubMed] [Google Scholar]
- 27.Gubler U, Chua AO, Schoenhaut DS, et al. Coexpression of two distinct genes is required to genereate secreted, bioactive cytotoxic lymphocyte maturation factor. Proc Natl Acad Sci USA. 1991;88:4143–7. doi: 10.1073/pnas.88.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibanez C, Sune P, Fierro A, et al. Modulating effects of intravenous immunoglobulins on serum cytokine levels in patients with primary hypogammaglobulinaemia. Biodrugs. 2005;19:59–65. doi: 10.2165/00063030-200519010-00007. [DOI] [PubMed] [Google Scholar]
- 29.Pulendran B, Smith JL, Caspary G, et al. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc Natl Acad Sci USA. 1999;96:1036–41. doi: 10.1073/pnas.96.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapsenberg ML, Kalinski P. The concept of type 1 and type 2 antigen-presenting cells. Immunol Lett. 1999;69:5–6. doi: 10.1016/s0165-2478(99)00096-6. [DOI] [PubMed] [Google Scholar]