Abstract
The complement inhibitor Factor H has three distinct binding sites for C3b and for heparin, but in solution uses specifically the most C-terminal domain, i.e. short consensus repeats (SCR) 20 for ligand interaction. Two novel monoclonal antibodies (mABs C14 and C18) that bind to the most C-terminal domain SCR 20 completely blocked interaction of Factor H with the ligands C3b, C3d, heparin and binding to endothelial cells. In contrast, several mAbs that bind to the N-terminus and to the middle regions of the molecule showed no or minor inhibitory effects when assayed by enzyme-linked immunosorbent assay (ELISA) and ligand interaction assays. This paradox between a single functional binding site identified for native Factor H versus multiple interaction sites reported for deletion constructs is explained by a compact conformation of the fluid phase protein with one accessible binding site. On zymosan particles mAbs C14 and C18 blocked alternative pathway activation completely. Thus demonstrating that native Factor H makes the first and initial contact with the C terminus, which is followed by N terminally mediated complement regulation. These results are explained by a conformational hypothetical model: the native Factor H protein has a compact structure and only one binding site accessible. Upon the first contact the protein unfolds and exposes the additional binding sites. This model does explain how Factor H mediates recognition functions during complement control and the clustering of disease associated mutations in patients with haemolytic uraemic syndrome that have been reported in the C-terminal recognition domain of Factor H.
Keywords: complement Factor H, conformational change, haemolytic uraemic syndrome, host recognition, steric effect
Introduction
The innate immune system represents a particularly effective part of immune defence and the immediately acting complement system is important for elimination of microbes and immune complexes [1]. Complement activation occurs directly, particularly on foreign surfaces, and is mediated by three distinct pathways [2,3]. Immune complexes activate the classical pathway (CP), carbohydrates the lectin pathway (LP) and foreign or bacterial surfaces amplify the alternative pathway (AP). Activation of the CP and LP is triggered by C1q or mannose binding lectin, respectively; however, the AP shows intrinsic activation. All three pathways converge at the level of C3 and are controlled by regulators. Activation leads to the formation of a C3 convertase, which − if activation proceeds − generates a potent amplification loop that results in the formation and local deposition of multiple C3b molecules. Because of the high toxicity, complement activation is favoured on foreign surfaces and results in the elimination of bacteria and microbes, but is inhibited and controlled tightly on the surface of self-cells. On host cells C3b deposition is inhibited by a combination of surface attached fluid phase regulators, such as Factor H, FHL-1 and C4 bp and by integral membrane proteins (i.e. DAF/CD55, MCP/CD46, CR1/CD35).
Factor H and FHL-1 are two central human immune regulators that control complement activation at the level of C3b [4]. Both proteins are related structurally and their transcripts are derived from the same gene by means of alternative splicing. The two plasma proteins are composed exclusively of individually folding domains, which are termed complement control protein modules or short consensus repeats (SCR). The 150 kDa Factor H glycoprotein is composed of 20 SCR domains and the 42 kDa protein FHL-1 comprises seven SCRs. The SCRs of FHL-1 are identical to the seven N-terminal SCRs of Factor H; however, the protein has a unique C-terminal extension of four amino acids. The two proteins share complement regulatory activity, which is located within N-terminal SCRs 1–4 [5–7]. However, the proteins differ in recognition function and bind with different affinities to cell surfaces [8]. In addition FHL-1, but not Factor H, displays cell spreading activity via an RGD domain located within SCR 4 [9].
Detailed structure–function studies have localized three distinct binding regions within Factor H for C3b and for glycosaminoglycans or heparin. The three C3 binding regions are located within the N-terminus of the protein, i.e. SCRs 1–4, the middle part, i.e. SCRs 12–14 and the C-terminus, i.e. SCRs 19–20, contained most probably within SCR 20 [5–8,10]. Similarly, the three heparin binding sites have been localized to SCR 7, within the middle region in the vicinity of SCR 13 and to the C-terminal SCRs 19–20 [6,11–15]. The presence of multiple binding sites for a single ligand suggests either a multi-point interaction between two proteins, i.e. between Factor H and C3b, or a single point interaction of one Factor H protein with several ligands (or a combination of the two scenarios).
The contribution of the individual sites in Factor H for ligand interaction is currently unclear. We use novel monoclonal antibodies to dissect the relevance of the individual binding sites in the intact Factor H protein on a functional level. With this set of 11 monoclonal antibodies we show that the C-terminus of Factor H makes the initial contact with C3b, heparin and cell surfaces. Two monoclonal antibodies (mAbs) that bind to the most C-terminal domain of Factor H block ligand interaction of the protein.
Materials and methods
Purification of Factor H from human plasma
Factor H was isolated from human plasma (1000 ml) derived from healthy volunteers by precipitation with polyethylene glycol [30% and 50% (w/v)] at pH 7·4 followed by sequential chromatography steps. All steps were performed at 4°C in the presence of protease inhibitors (20 µM (4-amidinophenyl)-methanesulphonyl fluoride, 2 µM E-64, 1 µM leupeptin and 1 µM pepstatin). After each purification step individual fractions were obtained and assayed for total protein concentration and Factor H content using enzyme-linked immunosorbent assays [16]. Following affinity chromatography on l-lysine–Sepharose 4B columns (GE Health Care, Freiburg, Germany), the eluate was dialysed against 25 mM potassium phosphate, pH 7·0 and applied to a XK 50/60 column (GE Health Care) containing 500 ml of deaminoethylaminoethyl (DEAE)–Sepharose fast flow equilibrated with the same buffer. Bound proteins were eluted with a linear NaCl gradient (0–500 mM), Factor H containing fractions were pooled, concentrated by positive pressure ultrafiltration and loaded onto a 170 ml BioRex-70 column (Bio-Rad, Munich, Germany) equilibrated with 20 mM potassium phosphate, pH 7·0. After elution with a linear NaCl gradient (final conductivity 40 mS) fractions containing Factor H were concentrated and purified further by gel filtration on a Sephacryl-S-300 HR column (100 × 2·6 cm (GE Health Care)). Factor H was finally purified to homogeneity by anion-exchange chromatography using Fractogel TMAE (Merck, Darmstadt, Germany) which was equilibrated with 10 mM Tris-HCl, pH 7·8 and eluted with a linear 0–500 mM NaCl gradient.
Generation of anti-Factor H monoclonal and polyclonal antibodies
Human Factor H-specific mAbs were generated following intraperitoneal immunization of BALB/c mice with 100 µg of purified Factor H in complete Freund's adjuvans (CFA) (Difco, Detroit, MI, USA). After four booster injections at 4-week intervals the fusion was performed according to standard protocols [17] using the myeloma cell line X63-Ag8·653. Positive hybridoma clones were identified by testing enzyme-linked immunosorbent assay (ELISA) reactivity towards Factor H (1 µg/ml) which was bound directly to the solid phase and were subcloned twice by limiting dilution. Hybridoma supernatants or 50% ammonium sulphate precipitates of ascitic fluids were purified by protein A affinity chromatography.
The subtype of the individual monoclonal antibodies was determined by ELISA using subtype-specific antisera according to the manufacturer's instructions (Nordic, Tilburg, the Netherlands). The generation of rabbit polyclonal anti sera with specificity for human Factor H has been described previously [5,9]. Immune sera were precipitated by 50% ammonium sulphate and purified to homogeneity by protein A chromatography.
Recombinant deletion mutants of Factor H
Generation and expression of the various Factor H deletion mutants, i.e. SCRs 1–2, SCRs 1–3, SCRs 1–4, SCRs 1–5, SCRs 1–6, SCRs 1–7, SCRs 8–11, SCRs 11–15, SCRs 15–18, SCRs 15–20 SCRs 8–20 or SCRs 19–20 has been described previously [5,8,18].
Insect cell culture
Spodoptera frugiperda Sf9 cells were grown in monolayers at 27°C in insect express medium (BioWhittaker, Apen, Germany) supplemented with 4% fetal calf serum (FCS), penicillin (100 units/ml), streptomycin (100 µg/ml) and fungizone (250 ng/ml) in 140 mm culture flasks (Nunc, Wiesboden, Germany). For infection with recombinant virus approximately 3 × 106 cells were seeded in 25 ml serum-free medium and infected using a multiplicity of infection of five. Recombinant proteins were isolated from the cell culture supernatant 10 days after infection and purified by nickel chelate chromatography (Pro Bond Resin, Invitrogen, Karlsruhe, Germany), as described previously [19]. Purified proteins were dialysed against phosphate buffer and concentrated using ultrafree-centrifugal filters (Millipore). The protein concentration was measured using the method of Bradford.
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Normal human serum (NHS) or purified recombinant proteins were separated under non-reducing conditions by SDS-PAGE using 8–12% gels. Protein bands were stained either with silver nitrate or transferred to a nitrocellulose membrane using a semidry system [19].
Factor H binding to C3b
A competitive binding assay was established in order to determine the ability of the newly generated mAbs to interfere with Factor H binding to the C3b fragment. Factor H was biotinylated with a sevenfold molar excess of Biotin-X-NHS (Calbiochem, Schwalbuck, Germany) and was incubated [16 h at room temperature (RT)] with the various anti-Factor H monoclonal antibodies at a molar ratio of 1 : 20 in phosphate-buffered saline (PBS)/0·1% bovine serum albumin (BSA). Individual wells of Maxisorp microtitre plates (Nunc) were coated with 100 µl purified C3b (20 µg/ml) [18] in 50 mM carbonate, pH 9·6 for 16 h at 4°C. After washing with PBS/0·2% BSA, 100 µl of Factor H (40 µg/ml) was added in the presence or absence of anti-Factor H mAb to each well and incubated for 2 h at RT. Wells were washed with PBS-T, then streptavidin–peroxidase conjugate (GE Health Care; 1 : 500) was added and incubated for 45 min. The wells were washed four times with PBS-T and 2 mM 2,2-azino-di(3-ethylbenzthiazolinsulphonacid-6) in 0·1 M sodium acetate, 0·05 M sodium dihydrogen phosphate and 2·5 mM hydrogen peroxide (H2O2) as the peroxidase substrate was applied to each well. The optical density was measured photometrically (MR 600, Dynatech, Denkendorf, Germany) and quantified using the manufacturer's software.
Factor H binding to heparin
A solid-phase assay was established in order to assay the inhibitory capacity of the mAbs (N22, M16, C02, C14 and C18) for Factor H binding to heparin. Polymeric heparan sodium salt (100 µg per well, Fluka, Taufkirchen, Germany) diluted in bicarbonate coating buffer (Sigma, Taufkirchen, Germany) was immobilized onto microtitre wells (Nunc Maxisorb; Nalge Nunc Intl., Roskilde, Denmark). After coating overnight (4°C), wells were washed four times with PBS-T and unspecific binding sites were blocked by 3% BSA/PBS-T for 15 min at RT. Factor H (0·2 µg/ml) was preincubated with a fivefold excess of monoclonal antibody for 15 min at RT and these Factor H-antibody complexes were added to individual wells. For detection of bound Factor H, polyclonal goat anti-human Factor H antiserum (Calbiochem) was added in a 1 : 3000 dilution, followed by a horseradish peroxidase (HRP)-conjugated rabbit anti-goat anti-serum (Dako, Hamburg, Germany). OPD (Dako) and H2O2 was added. The colour reaction was stopped by addition of 3 M sulphuric acid after 2·5 min. The optical density was measured photometrically (MR 600, Dynatech).
Inhibition of heparin affinity
Factor H (Calbiochem) treated with various mABs or left untreated was applied to heparin affinity chromatography using a 1 ml HiTrap heparin column and an ÄKTAPrime System (GE Health Care). The flow-through was collected and reloaded. The column was washed extensively and bound proteins were eluted using a linear salt gradient, ranging from 30 to 500 mM NaCl in a total volume of 30 ml. Individual fractions of 400 µl were collected, and the presence of Factor H was assayed upon SDS-PAGE separation and Western blotting.
Epitope mapping
The binding sites of the Group III mAbs C02, C14 and C18 on Factor H were mapped to the amino acid level. 12-mer peptides, covering the complete sequence of SCRs 19 and 20, were synthesized whereby cysteine residues were replaced by serine residues in order to reduce background staining. The panel of peptides was used in a linear scan for the binding epitopes of mAbs (Pepscan Systems, the Netherlands) and binding of antibody to the immobilized peptides was determined photometrically.
Cell binding assay
Human umbilical vein endothelial cells (HUVEC) were cultivated as described [20]. Cells incubated over night in the absence of FCS were treated with human serum (20% diluted in 0·5× PBS) for 20 min at 4°C in the absence or the presence of the various mABs. Following incubation with a polyclonal Factor H specific antiserum, a secondary fluorescein isothiocyanate (FITC)-labelled anti-goat anti serum was added. The cells were processed by flow cytometry using a fluorescence activated cell sorter (FACSCalibur; Becton and Dickinson, Heidelberg, Germany). In parallel experiments cells incubated with serum in the presence or absence of the mAbs were lysed and the cell lysate was separated by SDS-PAGE. Following transfer by electroblotting [19], bound Factor H was visualized using a polyclonal goat Factor H antiserum.
Surface plasmon resonance studies
The binding of the N-terminal fragment of Factor H (i.e. SCRs 1–7) to immobilized intact Factor H was assayed by surface plasmon resonance technique (Biacore 3000; Biacore AB, Uppsala, Sweden) [20,21]. Briefly, Factor H was immobilized via a standard amine-coupling procedure to flow cells of a CM5 sensor chip. Two flow cells were activated and Factor H diluted in coupling buffer (10 mM acetate buffer, pH 5·0) was injected in several cycles (> 20 µg/ml) into one flow cell until an appropriate coupling (> 4000 resonance units) was achieved. A flow cell without protein was used as control. Unreacted groups were inactivated with ethanolamine-HCl (35 µl). After coupling, the flow cells were washed thoroughly by sequential injection of regeneration buffer (3 M NaCl, 10 mM acetate buffer, pH 4·6) and running buffer (1/3× veronal buffered saline (VBS), pH 7·4). SCRs 1–7 were injected at a flow rate of 5 µl/min at 22°C and all binding assays were repeated at least three times.
Complement regulatory activity on zymosan surfaces
Human serum was depleted of Factor H (residual Factor H concentration 26 µg/ml) by immunoaffinity chromatography using mAb C02. Complement activation was assayed using zymosan particles treated with human Factor H depleted serum to allow alternative pathway activation. Alternative pathway of complement activation was initiated by incubation (20 min at 37°C) in the presence of a limited concentration (0·4 mg/ml) of zymosan (Serva, Heidelberg, Germany). The reaction was stopped by addition of 20 mM ethylenediamine tetraacetic aid (EDTA). Activation is dose-dependent and is evidenced by the generation of the Ba activation product, which was determined in the supernatant by ELISA [16]. For this activation assay Factor H-depleted serum was substituted with purified Factor H (500 µg/ml) which had been preabsorbed with a fourfold molar excess of the different anti-Factor H or control mAbs. Activation correlates inversely with Factor H concentration (data not shown). The effect of the individual mAbs to interfere with co-factor activity on this zymosan surface was assayed.
Results
We generated and used a set of novel mAbs to determine a hierarchy of the individual binding sites in the native Factor H protein for immune recognition.
Mapping the binding domains of novel mAbs within Factor H
Reactivity with human serum
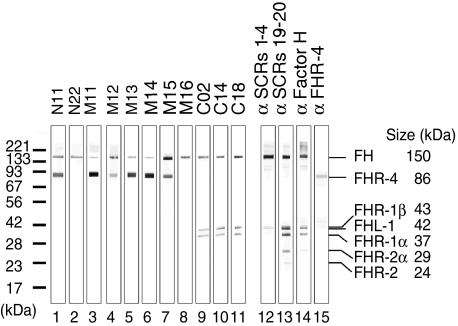
The reactivity of 11 novel mAbs with Factor H was tested with human serum separated by SDS-PAGE and blotted onto nitrocellulose. The 150 kDa Factor H protein was detected by each mAb (Fig. 1, lanes 1–11). The three mAbs M11, M13 and M14 reacted weakly and N22 and mAb M16 reacted exclusively with Factor H (Fig. 1, lanes 3, 5–6, 2 and 8). The other mAbs recognized additional bands, which represent members of the Factor H protein family: mAbs N11, M11, M12, M13, M14 and M15 identified a band of 86 kDa that represents the FHR-4A protein (Fig. 1, lanes 1 and 3–7). Three antibodies (C02, C14 and C18) identified the 42 and 37 kDa FHR-1 proteins (Fig. 1, lanes 9–11). The two forms of FHR-2 reacted with antiserum raised against the C-terminal SCRs 19–20 of Factor H, but under the given conditions no mAbs identified these proteins (Fig. 1, lanes 13 and 14). The different reactivity of the various antibodies indicates binding to distinct domains of Factor H.
Fig. 1.
Reactivity of 11 novel monoclonal antibodies with normal human serum proteins. Normal human serum was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% gel under non-reducing conditions, blotted to a nitrocellulose membrane, which was cut into stripes. Each stripe was incubated with a single monoclonal antibody (mAb) (lanes 1–11) or various polyclonal antisera used as controls (lanes 12–15). The 150 kDa band of the factor H protein is detected by all mAbs. Six mAbs (lanes 1, 3–7) identify a 86 kDa band and three mAbs (lanes 9–11) detect the two differently glycosylated forms of FHR-1. In addition to Factor H, polyclonal antisera identify the 42 kDa Factor H related protein (FHL)-1 (lanes 12 and 14) and both FHR-2 proteins (lanes 13 and 14). Antiserum raised against FHR-4 reacts with the 86 kDa protein (lane 15).
Mapping the binding domains of the novel mAbs
In order to map the binding domains to the level of SCRs, the reactivity of individual mAbs to recombinant deletion mutants of Factor H was tested using a panel of mutants, which together cover the complete Factor H protein. In the first approach mAbs N11 and N22 reacted with SCRs 1–7 but not with SCRs 8–20. All other mAbs detected SCRs 8–20 but not SCRs 1–7 (Table 1; and data not shown).
Table 1.
Localization of binding domain of monoclonal antibodies (mAbs) raised against Factor H; reactivity of anti-Factor H mAbs with Factor H deletion mutants as determined by dot blot and Western blot analyses.
| SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | 1–7 | 8–20 | 1–2 | 1–3 | 1–4 | 1–5 | 1–6 | 3–5 | 8–11 | 11–15 | 15–20 |
| Group I | |||||||||||
| N11 | + | – | – | + | + | + | + | + | – | – | – |
| N22 | + | – | – | – | – | + | + | + | – | – | – |
| SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | |
|---|---|---|---|---|---|---|---|
| (b) | 1–7 | 8–20 | 8–11 | 11–15 | 15–20 | 15–18 | 19–20 |
| Group II | |||||||
| M11 | – | + | – | – | – | – | – |
| M12 | – | + | – | + | – | – | – |
| M13 | – | + | – | – | + | + | – |
| M14 | – | + | + | + | – | – | – |
| M15 | – | + | – | + | + | + | – |
| M16 | – | + | – | – | + | + | – |
| SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | SCRs | |
|---|---|---|---|---|---|---|---|
| (c) | 1–7 | 8–20 | 8–11 | 11–15 | 15–20 | 15–18 | 19–20 |
| Group III | |||||||
| C02 | – | + | – | – | + | – | + |
| C14 | – | + | – | – | + | – | + |
| C18 | – | + | – | – | + | – | + |
+: Positive reactivity. + reactivity; – no reactivity. SCR: short consensus repeats.
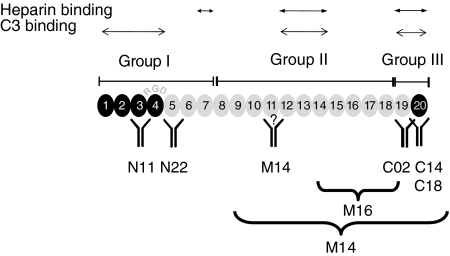
The use of additional recombinant deletion mutants of Factor H allowed mapping of the binding domains for most antibodies: Group I mAbs, i.e. N11 and N22 bind to SCR 3 or SCR 5 of Factor H, respectively (Fig. 2 and Table 1a).
Fig. 2.
Binding domains and epitopes of the various monoclonal antibodies (mABs). The binding domains of the various mAbs is indicated in an extended, structural model of Factor H. The three heparin and three C3 binding sites are shown. The N-terminal complement regulatory domain, represented by short consensus repeats (SCRs) 1–4, is shown by black modules and the central recognition domain at the C-terminus (SCR 20) is also shown in black.
Six antibodies termed group II, i.e. mAbs M11, M12, M13, M14, M15 and M16, bind to the middle region of Factor H. These antibodies reacted with the mutant representing SCRs 8–20, but not with the N-terminal SCRs 1–7, nor with the most C-terminal SCRs 19–20. The binding region of three of these mABs has been mapped to SCR 11 (i.e. M14), to the middle region (i.e. M15) and to a stretch covered by SCRs 15–18 (i.e. M16) (Fig. 2 and Table 1b).
The three group III antibodies, i.e. C02, C14 and C18, bind to the C-terminus of Factor H (SCRs 19 and 20). These mAbs reacted with SCRs 8–20, SCRs 15–20 and SCRs 19–20, but not with SCRs 15–18 (Fig. 2, Table 1c and data not shown). Given the importance of the C-terminus for Factor H function, the binding epitope of all three group III antibodies was characterized to the amino acid level. For mAb C02 this approach revealed the binding epitope DITSFPLSVYAP (position 1119–1130) within SCR 19 and for mAbs C14 and C18, overlapping binding epitopes with the sequence RTGESVEFVCKR (position 1192–1203) in SCR 20.
Characterization of mAbs in terms of Factor H function
Factor H has three binding sites for C3b as well as for heparin. It was therefore of interest to assay whether in the native protein all three sites are simultaneously accessible or − as indicated previously by diseases associated with clustering of mutations in the C-terminus − if a hierarchy exists among the individual sites [22]. Consequently, the capacity of these domain mapped mAbs to block Factor H ligand interaction was assayed with the intact native protein.
Inhibition of Factor H binding to C3b
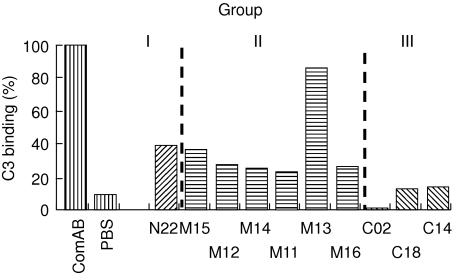
An ELISA-based assay was used to test how the individual mAbs affect Factor H binding to C3b. The group I antibodies N11 and N22 affected binding and showed 50 and 61% inhibition. The group II antibodies M11, M12, M13, M14, M15 and M16 showed moderate to strong inhibition, ranging from 69, 68, 17, 69, 62 and 71%, respectively. The three group III antibodies mAbs C02, C14 and C18 showed strong blocking activity and inhibited binding of Factor H to C3b by 98, 87 and 88%, respectively (Fig. 3 and Table 2). Thus the strong inhibitory activity of all three group III mABs demonstrates that native Factor H uses primarily the C-terminal site for C3b interaction.
Fig. 3.
Factor H binding to C3b. The ability of antibodies to interfere with Factor H binding to its ligand C3 was tested by enzyme-linked immunosorbent assay (ELISA). For evaluation, binding of wild-type Factor H was set to 100% and the inhibitory effect of various monoclonal antibodies (mAbs) was assayed and compared to the wild-type protein. Similar results were obtained in four different experiments.
Table 2.
Inhibitory effects of anti-Factor H monoclonal antibodies (mAbs).
| Inhibition (%) | |||||
|---|---|---|---|---|---|
| C3b binding | Heparin binding | Cellbinding | Co-factor activity1 | ||
| Group I | |||||
| N11 | IgG1 | 50 | n.d. | – | n.d. |
| N22 | IgG1 | 61 | 35 | 50 | 45 |
| Group II | |||||
| M11 | – | 69 | n.d. | Enhance | n.d. |
| M12 | IgG2a | 68 | n.d. | Enhance | 12 |
| M13 | IgG1 | 17 | n.d. | – | n.d. |
| M14 | – | 69 | n.d. | – | n.d. |
| M15 | IgG1 | 62 | n.d. | Enhance | 15 |
| M16 | IgG1 | 71 | 19 | – | 16 |
| Group III | |||||
| C02 | IgG1 | 98 | 18 | 10 | 45 |
| C14 | IgA | 87 | 98 | 99 | 86 |
| C18 | IgG1 | 88 | 99 | 98 | 74 |
Inhibitory effect of anti-Factor H mAbs determined for C3b and for heparin binding using specific enzyme-linked immunosorbent assay (ELISA) systems, Cell binding as determined by flow cytometry and Western blotting and for surface dependent co-factor activity using zymosan particles and Factor H depleted human serum as a model for non activator surfaces. n.d. not determined.
Co-factor activity was determined using zymosan and Factor H depleted human serum. The newly generated Ba activation fragment was quantified by enzyme-linked immunosorbent assay (ELISA). The activity observed with Factor H depleted human plasma was set to 100%. Italic type and bold characters: more than 75% inhibition.
Heparin binding
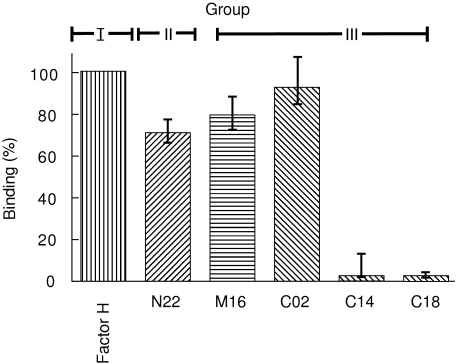
An ELISA system was used to test how each mAb affects heparin binding. Binding of native Factor H to heparin was set to 100%. The group I mAb N22, as well as group II mAb M16, showed little effect (35 and 19%, respectively) (Fig. 4 and Table 2). The pattern of the group III was diverse. mAb C02 showed no significant inhibitory activity (18%), but mAb C14 and C18 blocked binding completely (Fig. 4). These results show that native Factor H binds heparin almost exclusively with its C terminus, i.e. SCR 20.
Fig. 4.
Factor H binding to heparin. The ability of antibodies to interfere with Factor H binding to heparin was tested by enzyme-linked immunosorbent assay (ELISA). Monoclonal antibodies (mAbs) were mixed with Factor H before addition to the heparin coated wells. The absorbency was measured and compared to that of a probe of Factor H with no antibody added (Factor H), which was set to 100%. Results obtained with mAbs N22, M16 and C02 (65, 81 and 82%, respectively) and that of the inhibitory mAbs C14 and C18 are shown. Standard deviation between individual values (n = 4) is indicated. Similar results were obtained in four different experiments.
In addition, Factor H binding was assayed by heparin chromatography. Factor H was incubated with the various antibodies, applied to heparin chromatography and the maximum elution peak was determined (Table 3). Wild-type Factor H eluted at 28·51 min (corresponding to 241 nM NaCl). The N terminal mAB N22 and the group II mAB M16 had no or moderate effects with 28·61 and 28·31 min, corresponding to 242 and 237 mM NaCl, respectively. The group III mAB C02 apparently increased the interaction reaching 29·89 min, corresponding to 260 nM NaCl. Both C-terminal binding antibodies C14 and C18 reduced heparin binding strongly. Retention time was 27·1 and 26·44 min, which corresponded to an elution at 206 mM and 197 mM NaCl, respectively.
Table 3.
Inhibition of various monoclonal antibodies (mABs) on Factor H binding to C3b.
| mABs added | RT (min) | NaCl (mM) | |
|---|---|---|---|
| Factor H | None | 28·51 | 241 |
| N22 | 28·61 | 242 | |
| M16 | 28·31 | 237 | |
| C02 | 29·89 | 260 | |
| C14 | 27·10 | 206 | |
| C18 | 26·44 | 197 |
Factor H was applied to heparin affinity chromatography and eluted with a linear salt gradient. Following preincubation with the indicated mABs the elution time (peak maximum) and the corresponding NaCl concentration required for maximal elution were recorded. RT: retention time.
Cell binding
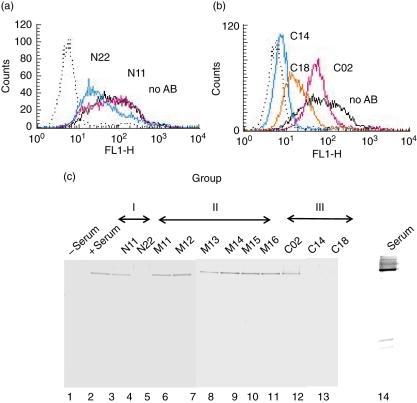
The inhibitory effect of the various mAbs on Factor H binding to endothelial cells (HUVEC) was assayed by flow cytometry. The group I mAb N11 affected binding weakly, and mAb N22 showed 50% inhibition (Fig. 5a). Among the three group III mAbs, mAB C02 showed minor effects, but mAbs C14 and C18 inhibited binding of Factor H to endothelial cells strongly (Fig. 5b).
Fig. 5.
Factor H binding to endothelial cells. Human umbilical vein endothelial cells (HUVECs) cultivated in the absence of serum were incubated with human serum in the absence or presence of the indicated group I (a), group II or group III (b) monoclonal antibodies (mAbs). Binding was assayed by flow cytometry. (c) HUVECs cultivated in the absence of serum were treated with serum in the absence or presence of the indicated group I, group II or group III mABs. Following this treatment, cells were lysed, the lysate was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a membrane and assayed by Western blotting using a Factor H specific goat antiserum.
The inhibitory effect of the various mAbs on cell binding of Factor H was tested further by identifying cell surface attached Factor H in the cell lysate using a combination of SDS-PAGE and Western blotting. When HUVEC cells were cultivated in Factor H containing serum, surface attached Factor H was identified (Fig. 5c, lane 2). The group I mAB N11 showed no effect; however, mAb N22 showed an inhibitory effect (lanes 3 and 4). All six group II mABs and also group III mAB C02 did not affect binding. However, both group III mABs C14 and C18 inhibited binding of Factor H to endothelial cells (Fig. 5c, lanes 12, 13), thus confirming that Factor H binding to endothelial cells is inhibited by blocking domain 20.
Characterization of mAbs for Factor H-mediated complement control on zymosan particles
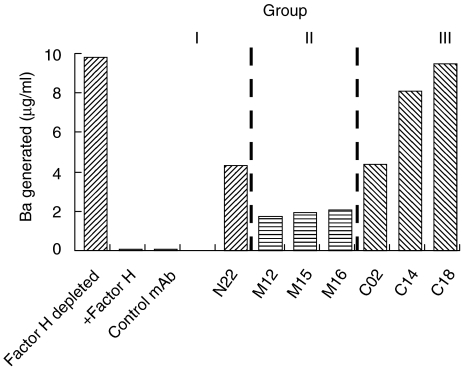
Factor H controls alternative pathway activation by regulating the C3 convertase and by acting as a co-factor for Factor I in cleaving C3b. Complement activation via the alternative pathway on zymosan particles with Factor H-depleted serum was followed by measuring the Ba activation product (Fig. 6, lane 1). In this system addition of Factor H blocked complement activation in a dose-dependent manner (Fig. 6, lane 2; and data not shown).
Fig. 6.
Inhibitory potential of the monoclonal antibodies (mAbs) to Factor H complement regulatory activity. Complement activation on activator zymosan particles using Factor H depleted human serum was measured by the generation of the Ba product in an enzyme-linked immunosorbent assay (ELISA). Factor H-depleted serum, reconstituted with Factor H treated with an irrelevant mAb, showed inhibition of complement activation as determined by the absence of the Ba activation product (control antibody) and phosphate-buffered saline (PBS) in the absence of depleted serum showed no effect. The inhibitory role of Factor H treated with the indicated mAbs on complement activation was tested. Factor H, which was treated with mAbs N22, M12, M15, M16 and C02, showed moderate effects. In contrast, the two group III mAbs C14 and C18 which bind to the C-terminus of the protein blocked the regulatory activity of the Factor H. A representative experiment out of five independent experiments is shown.
The inhibitory activity of the individual mABs was assayed upon substituting the serum with Factor H. The group I mAb N22, which binds to the N-terminus, showed a weak inhibitory effect (45%). The group II mABs, i.e. M12, M15 and M16, and similarly an unrelated control mAb, displayed mild or no inhibitory effects (12%, 15%, and 16%) (Fig. 6 and Table 2). The group III mAb C02 had a moderate (45%) and the two mABs C14 and C18, that bind to SCR 20, showed strong inhibitory activity on Factor H function (86 and 74%). These results demonstrate that blocking of the C-terminal recognition domain affects the N-terminal-mediated complement regulatory activity.
Intramolecular interaction of Factor H
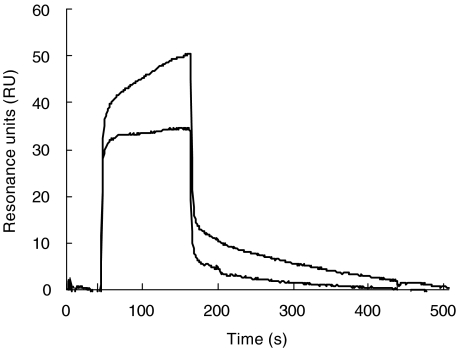
Intramolecular interaction might lead to a masking of binding sites and might explain the difference between a single accessible site of the native Factor H protein and the presence of multiple binding sites. Therefore, binding of the N-terminal domain (i.e. SCRs 1–7) to the intact Factor H protein was assayed by surfacing plasmon resonance. A deletion fragment of Factor H represented the N-terminal SCRs 1–7 bound to immobilized intact Factor H, as demonstrated by the association profile which shows a complex formation and the dissociation profile which shows that the complex is stable (Fig. 7). This assay shows intramolecular interaction of Factor H and demonstrates that the N-terminus binds to the intact Factor H protein.
Fig. 7.
Binding of the N-terminus of Factor H [short consensus repeats (SCR) 1–7)] to immobilized intact Factor H. Factor H was immobilized to the surface sensor chip and interaction of the N-terminus (i.e. SCR 1–7) was followed. The association and dissociation profile provide evidence for a intramolecular interaction.
Discussion
The alternative pathway inhibitor Factor H uses the C-terminus for initial contact with its ligands C3b, C3d, heparin and for binding to cell surfaces. This initial interaction of the native protein precedes regulation, i.e. co-factor activity for the cleavage of C3b which is mediated by the N-terminal regulatory region. We show that two novel mAbs that bind to SCR 20 the C-terminal recognition and cell binding region of Factor H block completely ligand interaction and cell binding. We interpret these results that native, fluid phase Factor H has exclusively the C-terminus binding site accessible for ligand recognition and that the additional sites in the middle region and in the N-terminus are masked or hidden. The apparent paradox between a single binding site demonstrated experimentally for fluid phase Factor H and multiple binding sites, determined with recombinant deletion mutants, is explained with a conformational model, in which native Factor H protein has a compact form which has exclusively the C-terminal recognition region exposed and accessible. Upon initial ligand interaction, e.g. with surface attached C3b, the protein unfolds and exposes the additional binding sites. This conformation model suggests that target recognition by the C-terminus precedes complement regulation by the N-terminus.
Factor H has two central functional regions, i.e. the N-terminal complement regulatory region located within domains 1–4 and the C-terminal recognition region in domains 19–20 (Fig. 2). Multiple binding sites for the ligands C3b, heparin have been identified using recombinant deletion mutants and proteolytic protein fragments. The three C3b binding sites have been mapped to SCRs 1–4, SCRs 8–15 and SCRs 19–20 and the three heparin binding sites were localized to SCR 7, to or near SCR 13 and SCR 20 [5–8,10,11,23,24]. So far it has been unclear whether the native protein has all these sites accessible simultaneously. Here we demonstrate a hierarchy among the multiple binding sites and demonstrate that the C-terminus of Factor H, which represents the major recognition and cell binding sites, is the single accessible binding site in the native Factor H protein.
Native Factor H uses the C-terminus for initial contact with the ligands C3b, C3d, heparin and for binding to cell surfaces. The C-terminus, i.e. SCR 20, which represents overlapping binding sites for C3b, C3d, heparin and for cell surfaces, is essential for Factor H activity particularly on surfaces [25,26].
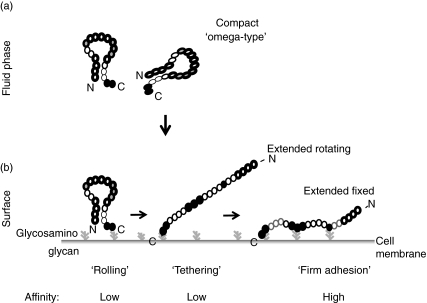
This conformational model explains the discrepancy between a single accessible domain and the multiple-mapped binding sites. Native fluid phase Factor H has a compact form, the protein bends and the N-terminus binds to the C-terminus. Interaction of the N-terminus with Factor H was shown experimentally (Fig. 7). We suggest that the compact form has an ‘omega type’ structure (Fig. 8), which has the C-terminal binding sites accessible and thus forms the major interaction sites for C3b, C3d, glycosaminoglycans and cell surfaces. The other binding sites and also the complement regulatory domain in the N-terminus are hidden or masked. Upon ligand interaction the protein unfolds and forms the extended rod-like conformation; consequently the additional binding sites and the complement regulatory domain become accessible (Fig. 8). This model explains how Factor H controls complement activation: the C-terminus makes the initial contact and is essential for the discrimination between activator and non-activator surfaces.
Fig. 8.
A conformational hypothetical model of native Factor H. (a) Native Factor H has a compact, omega-type structure, which has the C-terminal site accessible and the additional binding sites and also the functional complement regulatory region masked and inaccessible. (b) Initial contact of the C-terminal recognition region occurs with C3b/C3d, glycosaminoglycan or cell surfaces. This interaction induces a conformational change; the protein unfolds and exposes the additional binding sites for C3b (located within domains 1–4 and 8–15) and heparin (i.e. domains 7 and 11–13) and also the complement regulatory region in domains 1–4. Thus on surfaces recognition mediated by the C-terminus precedes regulation, mediated by the N-terminus. The structure and conformation of fluid phase Factor H is shown by the individual short consensus repeat (SC) domains. The three C3 binding sites have been mapped to domains SCRs 1–4, SCR 8–15 and SCR 19–20. The C3 binding sites are indicated by thick lines. Inactive sites are shown stippled, and active sites are shown in black. Heparin binding domains have been mapped to domains SCR 7, SCR 11–13 and SCR 19–20. Active heparin binding sites are filled black and inactive sites are filled with grey colour.
This model of a compact ‘omega-type’ form of native Factor H is based on results presented in this work and does also explain several published data, which so far have been in contrast to the ‘bead-on-a-string model’ [9,14,27–32]. Bending of Factor H, with the N- and C-terminal ends touching each other, has been observed by electron microscopy [23]. In addition, (i) similar to the results shown here, mAb VIG8 which binds within SCR 20 blocks heparin binding of Factor H completely [24]; (ii) the C-terminus of Factor H mediates host recognition functions [25]; (iii) cell spreading assays show that in native Factor H the RGD domain in SCR 4 is hidden and inaccessible [9]; (iv) most Factor H mAbs, which were raised with native purified protein, bind preferentially to the C-terminus, and not (or very rarely) to the middle or N-terminal regions (Fig. 2a) [24,25]; (v) several monoclonal antibodies recognize conformational epitopes of Factor H protein, e.g. in ELISA, but do not or very poorly react with linear Factor H following SDS-PAGE separation or do not react with individual SCR domains (e.g. mAbs M11, M15 or M16) (Figs 1 and 2); (vi) native Factor H, which has three binding sites, and the recombinant fragment SCRs 15–20 of Factor H with one binding site show comparable affinity to C3b and to cells (data not shown); (vii) two forms of Factor H: phi1 and phi2 have been described, which show different biological activities in terms of activation of human monocytes and receptor binding [27–30]; (viii) X-ray and neutron scattering assays show for native Factor H a length of ca. 40 nm, which is about 50% of the size predicted for the extended protein consisting of 20 SCR domains [31]; and (ix) the sigmoid binding of Factor H to C3b versus the hyperbolic binding of Factor B to sepharose coated with different densities of C3b [32].
The conformational model explains how Factor H mediates recognition functions and alternative complement control, particularly at cell surfaces, and in addition highlights the central role of the C-terminus for Factor H-mediated surface recognition. The model also explains the clustering of the multiple gene mutations within domains 19 and 20 that have been reported for patients with the Factor H-associated form of haemolytic uraemic syndrome (HUS) [33–41]. Recent mutagenesis and functional, as well as structural analyses of HUS-associated Factor H gene mutations occurring in SCR domain 20 (i.e. E1172Stop, R1210g and R1215G), showed severely reduced binding to C3b, to heparin and to endothelial cells for each of the three mutant proteins. In fluid phase the complement regulatory activities of the mutant proteins were intact and comparable to wild-type Factor H, but were abrogated on cell surfaces [20,41] (Stefan Heinen, Andrea Hartman and PFZ, unpublished data). In summary, the data presented here explain how defective recognition results in endothelial cell damage that is considered a hallmark of HUS.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and the Thüringer Ministerium für Wissenschaft, Forschung und Kunst.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Walport MJ. Complement. N Engl J Med. 2001;344:1140–4. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 3.Morgan BP, Harris CL. Complement regulatory proteins. San Diego: Academic Press; 1999. [Google Scholar]
- 4.Zipfel PF, Jokiranta ST, Hellwage J, Koistinen V, Meri S. The factor H-protein family. Immunopharmacology. 1999;42:53–60. doi: 10.1016/s0162-3109(99)00015-6. [DOI] [PubMed] [Google Scholar]
- 5.Kühn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H. J Immunol. 1995;155:5663–70. [PubMed] [Google Scholar]
- 6.Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc Natl Acad Sci USA. 1996;93:10996–1001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin D. Identification of complement regulatory domains in human factor H. J Immunol. 1995;155:348–6. [PubMed] [Google Scholar]
- 8.Kühn S, Zipfel PF. Mapping of the domains required for co-factor activity of complement factor H. Eur J Immunol. 1996;6:2383–7. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- 9.Hellwage J, Kühn S, Zipfel PF. The human complement regulatory factor H-like protein 1, which represents a truncated form of factor H displays cell spreading activity. Biochem J. 1997;326:321–7. doi: 10.1042/bj3260321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on complement factor H interacts with a distinct site on C3b. J Biol Chem. 2000;275:27657–62. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- 11.Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J Immunol. 1996;157:5422–7. [PubMed] [Google Scholar]
- 12.Pangburn MK, Atkinson MA, Meri S. Localization of the heparin-binding site on complement factor H. J Biol Chem. 1991;266:16847–53. [PubMed] [Google Scholar]
- 13.Blackmore TK, Hellwage J, Sadlon TA, et al. Identification of the second heparin-binding domain in human complement factor H. J Immunol. 1998;160:3342–8. [PubMed] [Google Scholar]
- 14.Hellwage J, Jokiranta TS, Friese MA, et al. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol. 2002;169:6935–44. doi: 10.4049/jimmunol.169.12.6935. [DOI] [PubMed] [Google Scholar]
- 15.Pangburn MK. Localization of the host recognition functions of complement factor H at the carboxyl-terminal: implications for hemolytic uremic syndrome. J Immunol. 2002;169:4702–6. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- 16.Peters JH, Baumgarten H. Monoclonal antibodies. Berlin: Springer Verlag; 1992. [Google Scholar]
- 17.Würzner R, Oppermann M, Zierz R, Baumgarten H, Gotze O. Determination of the epitope specificities of monoclonal antibodies using unprocessed supernatants of hybridoma cultures. J Immunol Meth. 1990;126:231–7. doi: 10.1016/0022-1759(90)90155-o. [DOI] [PubMed] [Google Scholar]
- 18.Kühn S, Zipfel PF. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene. 1995;162:225–9. doi: 10.1016/0378-1119(95)00360-i. [DOI] [PubMed] [Google Scholar]
- 19.Skerka C, Hellwage J, Weber W, et al. The human factor H related protein 4: a novel short consensus repeat-containing protein is associated with human triglyceride-rich lipoproteins. J Biol Chem. 1997;272:5627–34. doi: 10.1074/jbc.272.9.5627. [DOI] [PubMed] [Google Scholar]
- 20.Manuelian T, Hellwage J, Meri S, et al. Factor H gene mutations in hemolytic uremic syndrome: single amino acid exchanges reduce binding affinity to C3b, heparin and surface attachment to endothelial cells. J Clin Invest. 2003;111:1181–90. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieland GD, Nehmann N, Muller D, et al. Early growth response proteins EGR-4 and EGR-3 interact with immune inflammatory mediators NF-kappaB p50 and p65. J Cell Sci. 2005;118:3203–12. doi: 10.1242/jcs.02445. [DOI] [PubMed] [Google Scholar]
- 22.Zipfel PF, Heinen S, Jozsi M, Skerka C. Complement and diseases: defective alternative pathway control results in kidney and eye diseases. Mol Immunol. 2006;43:97–106. doi: 10.1016/j.molimm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 23.DiScipio RG. Ultrastructures and interactions of complement factors H and I. J Immunol. 1992;149:2592–9. [PubMed] [Google Scholar]
- 24.Prodinger W, Hellwage J, Spruth M, Dierich MP, Zipfel PF. The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochem J. 1998;331:41–7. doi: 10.1042/bj3310041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jokiranta ST, Zipfel PF, Hakulinen J, et al. Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal anti-factor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett. 1996;393:297–302. doi: 10.1016/0014-5793(96)00905-2. [DOI] [PubMed] [Google Scholar]
- 26.Jokiranta TS, Cheng ZZ, Seeberger H, et al. Binding of complement Factor H to endothelial cells is mediated by the carboxy terminal gylcosaminglycan binding site. Am J Pathol. 2005;167:1173–81. doi: 10.1016/S0002-9440(10)61205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripoche J, Erdei A, Gilbert D, Al Salihi A, Sim RB, Fontaine M. Two populations of complement factor H differ in their ability to bind to cell surfaces. Biochem J. 1988;53:475–80. doi: 10.1042/bj2530475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripoche J, Al Salihi A, Rousseaux J, Fontaine M. Isolation of two molecular populations of human complement factor H. Biochem J. 1984;221:89–96. doi: 10.1042/bj2210089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtsuka H, Imamura T, Matsushita M, et al. Thrombin generates monocyte chemotactic activity from complement factor H enerates monocyte chemotactic activity from complement factor H. Immunology. 1993;80:140–5. [PMC free article] [PubMed] [Google Scholar]
- 30.Imamura T, Ohtsuka H, Matsushita M, Tsuruta J, Okada H, Kambara T. A new biological activity of the complement factor H. Identification of the precursor of the major macrophage-chemotactic factor in delayed hypersensitivity reaction sites of guinea pigs. Biochem Biophys Res Commun. 1992;185:505–9. doi: 10.1016/0006-291x(92)91653-8. [DOI] [PubMed] [Google Scholar]
- 31.Perkins SJ, Gilbert HE, Aslam M, Hannan J, Holers VM, Goodship THJ. Solution structures of complement components by X-ray and neutron scatter and analytical ultracentrifugation. Biochem Soc Trans. 2002;30:996–1001. doi: 10.1042/bst0300996. [DOI] [PubMed] [Google Scholar]
- 32.Koistinen V. Effect of complement-protein-C3b density on the binding of complement factor H to surface-bound C3b. Biochem J. 1991;280:255–9. doi: 10.1042/bj2800255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Józsi M, Manuelian T, Heinen S, Oppermann M, Zipfel PF. Attachment of the soluble complement regulator factor H to cell and tissue surfaces: relevance for pathology. Histol Histopathol. 2004;19:251–8. doi: 10.14670/HH-19.251. [DOI] [PubMed] [Google Scholar]
- 34.Taylor CM. Complement factor H and the haemolytic uraemic syndrome. Lancet. 2001;358:1200–2. doi: 10.1016/s0140-6736(01)06339-5. [DOI] [PubMed] [Google Scholar]
- 35.Zipfel PF. Hemolytic uremic syndrome: how do factor H mutants mediate endothelial damage? Trends Immunol. 2001;22:345–8. doi: 10.1016/s1471-4906(01)01972-x. [DOI] [PubMed] [Google Scholar]
- 36.Caprioli J, Bettinaglio P, Zipfel PF, et al. The molecular basis of familial hemolytic uremic syndrome. mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol. 2001;12:297–307. doi: 10.1681/ASN.V122297. [DOI] [PubMed] [Google Scholar]
- 37.Richards A, Buddles MR, Donne RL, et al. Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. Am J Hum Genet. 2001;68:485–90. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Caballero D, Gonzalez-Rubio C, Gallardo ME, et al. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet. 2001;68:478–84. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann HPH, Bohnert-Iwan B, Manuelian T, et al. Frequency of factor H gene mutations in hemolytic uremic syndrome. J Med Genet. 2003;40:676–81. doi: 10.1136/jmg.40.9.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins SJ, Goodship TH. Molecular modeling of the C-terminal domains of factor H of human complement: a correlation between haemolytic uraemic syndrome and a predicted heparin binding site. J Mol Biol. 2003;2316:217–24. doi: 10.1006/jmbi.2001.5337. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Corral P, Perez-Caballero D, Huarte O, et al. Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am J Hum Genet. 2002;71:1285–95. doi: 10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]