Abstract
For most bacteria, adherence to human cells is achieved by bacterial lectins binding to mammalian surface glyconjugates. 6-Phosphogluconate dehydrogenase (6PGD) was identified by us as one of Streptococcus pneumoniae cell wall lectin proteins, which elicits an age-dependent immune response in humans. This study assesses the role of 6PGD in S. pneumoniae pathogenesis as an adhesin and its ability to elicit a protective immune response in mice. Recombinant 6PGD (r6PGD) was cloned from S. pneumoniae serotype 3 (strain WU2). r6PGD interference in adhesion of three genetically unrelated unencapsulated pneumococcal strains (3·8, 14·8 and R6) and two genetically unrelated encapsulated pneumococcal strains (WU2 and D39) to A549 type II lung carcinoma cell was tested. BALB/c mice were immunized with r6PGD and boosted after 3 weeks. Immunized mice were challenged intranasally with a lethal dose of S. pneumoniae. r6PGD inhibited 90% and 80% of pneumococcal adhesion to the A549 cells of three unencapsulated S. pneumoniae strains and two encapsulated S. pneumoniae strains, respectively, in a concentration-dependent manner (P< 0·05). Antibodies to r6PGD produced in mice significantly inhibited bacterial adhesion to A549 cell (P< 0·05). Immunization of mice with r6PGD protected 60% (P< 0·001) of mice for 5 days and 40% (P< 0·05) of the mice for 21 days following intranasal lethal challenge. We have identified 6PGD as a surface-located immunogenic lectin protein capable of acting as an adhesin. 6PGD importance to bacterial pathogenesis was demonstrated by the ability of r6PGD to elicit a protective immune response in mice.
Keywords: immunoproteomics, proteomics, Streptococcus pneumoniae, vaccine
Introduction
Streptococcus pneumoniae is a major cause of morbidity and mortality worldwide. It colonizes asymptomatically the upper respiratory tract in approximately 10% of the adult population and in >50% of young children under 2 years of age [1]. However, in the minority of the carriers this pathogen may cause otitis media, pneumonia bacteraemia and meningitis [2]. The mechanisms that turn the benign carriage into disease are unclear.
An understanding of the sequential molecular interactions of S. pneumoniae with the host is emerging. Pneumococci have been shown to bind to upper and lower respiratory tract cells. The attachment is characterized by a broad area of contact between the bacterial surface and the host cells, suggesting multiple receptor interactions [3].
For most bacteria, adherence to human cells is achieved by bacterial lectins binding to the mammalian surface glyconjugates. Pneumococci display at least five lectin specificities, depending on target cells. Inhibition of pneumococcal adherence to Buccal epithelial cells could be achieved by N-acetylglucosamine (GlcNAc) (β1–3)Gal [4]. Inhibition of pneumococcal adherence to conjunctival or primary bronchial cells could be achieved by the glyconjugates of Gal(β1–4) GlcNAc, particularly if the sugars were sialylated [5]. Attachment of pneumococci to non-inflamed pulmonary epithelial and endothelial cells could be disrupted by the disaccharides GlcNAc (β1–4)Gal [6] and GlcNAc (β1–3)Gal [7]. A combination of these two sugars, as represented by asialo_GM2 and globoside, eliminates in vitro adhesion.
Activation of cells by proinflammatory cytokines up-regulates several cellular receptors, including the platelet-activating factor (PAF) receptor [8]. The enhanced adhesion of pneumococci to activated cells could be inhibited by the PAF receptor antagonist and the sugars GlcNAc and lacto-N-neotetraose. However, GlcNAc and lacto-N-neotetraose did not inhibit pneumococci adherence to resting cells. It appears that PAF receptors enhance invasion of endothelial cells by pneumococci, although no signal transduction downstream to this receptor could be observed [9].
A further attempt to identify the pneumococcal ligands involved in adhesion to the host cells was performed using a genetic approach. A library of phoA fusion mutants, in which targeted insertion duplication mutagenesis yielded loss of function in an array of pneumococcus surface molecules, showed that among the molecules involved in adhesion, structural, regulatory and accessory proteins can be found [10]. Another pneumococcal surface protein PspC/CbpA was identified as an adhesin [11]. Recently the target receptor on human cells to this adhesin has been shown to be the human polymeric immunoglobulin receptor [12,13].
To further identify proteins involved in S. pneumoniae interaction with the host cells and host immune system we have used proteomics and immunoproteomics of the pneumococcus cell wall. Cell wall proteins were fractionated by fetuin affinity chromatography in attempt to identify lectin proteins [14]. Using proteomic analysis, several proteins were identified in the fetuin-binding fraction. We have analysed these proteins further using immunoproteomics [15]. One of the proteins that exhibited lectin characteristics and age-dependent immunogenicity in humans has been found to be 6-phosphogluconate dehydrogenase (6PGD). Cytoplasmic 6PGD is responsible for the conversion of 6-phosphogluconate to ribose 5 phosphate. However, the existence of bacterial cytoplasmic proteins in the cell wall has been described for S. pneumoniae [16] and for other bacteria such as S. pyogenes [17].
In the current study we present our results demonstrating that 6PGD may function as an adhesin. 6PGD's abilities to be a virulence factor and to elicit protective immune responses were also assessed. The present study could contribute to the understanding of the sequential molecular interactions of S. pneumoniae with the host.
Materials and methods
Reagents
Unless otherwise stated, all chemicals and biochemicals of the highest purity available were purchased from Sigma-Aldrich (St Louis, MO, USA).
Bacterial strains, growth conditions and growth medium
Six capsulated S. pneumoniae strains and three unencapsulated strains are shown in Table 1. In this study we have used the unencapsulated derivative of three of the capsulated strains. The unencapsulated S. pneumoniae strain R6 is spontaneously derived unencapsulated strain from the encapsulated Avery strain D39 [18]. Strains 3·8 [19] and 14·8 [20] are unencapsulated mutants derived from the encapsulated strains WU2 [21] and 14DW, respectively. The last two pairs and their parental strains were kindly obtained from Professor Watson (Dallas, TX, USA). In addition two Escherichia coli strains were used in this study (Table 1). Except for the strains 14R and 9 V, all strains of S. pneumoniae used in this study were genetically unrelated, as described previously [22]. E. coli were grown in Luria–Bertani (LB) broth. Pneumococci were grown to mid-logarithmic growth phase as determined by optical density (OD) in Todd–Hewitt broth (Difco Laboratories, Detroit, MI, USA) supplemented by yeast extract (Difco Laboratories).
Table 1.
Strains used in this study.
| Sourcea | Capsule type | Strain |
|---|---|---|
| Streptococcus pneumoniae capsulated | ||
| types | ||
| ATCC | 2 | D39 |
| D. Watson | 3 | WU2 |
| R. Dagan | 6B | 6BR |
| R. Dagan | 9V | 9VR |
| D. Watson | 14 | 14DW |
| R. Dagan | 14 | 14R |
| S. pneumoniae unencapsulated types | ||
| ATCC | 2 | R6 |
| D. Watson | 3 | 3·8 |
| D. Watson | 14 | 14·8 |
| Escherichia coli | ||
| Invitrogen Corp, Carlsbad, CA, USA | DH5α UltraMAX | |
| Promega Corp, Madison, WI, USA | BL21(DE3)pLysS | |
Pneumococcal isolates were obtained from the collections of Prof. D. Watson (Dallas, TX, USA), Prof. R. Dagan (Pediatric Infectious Disease Unit, Soroka University Medical Center, Beer Sheva, Israel) and American Type Culture Collection (ATCC) (Manassas, VA, USA). It should be noted that the parental strains for the unencapsulated strains R6, 3.8 and 14.8 are D39, WU2 and 14DW, respectively.
Isolation of S. pneumoniae cell wall (CW) proteins
The proteins were isolated by the method of Siegel et al. [23]. Briefly, bacterial cells were harvested by centrifugation at 4700 g for 15 min, washed with phosphate buffered saline (PBS) and incubated with mutanolysin for 1 h at 37°C. The supernatant containing the soluble proteins released from bacteria were collected after centrifugation and stored at −70°C. Less than 10% of cytoplasmic proteins are reported to be found in the supernatant due to leakage using this procedure. We have confirmed these data in comparing two-dimensional polyacrylamide gel electrophoresis (PAGE) of CW proteins and cytoplasmic proteins showing a different protein distribution [15].
Protein gel electrophoresis and staining
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the Hoefer mini VE vertical electrophoresis system (Amersham Biosciences, San Francisco, CA, USA). Isoelectric focusing (pI 4–6·5) was carried out in a Hoefer SE 220 Mighty Small Tube Gel Adaptor (Amersham Biosciences). The proteins were then separated in the second dimension using 10% polyacrylamide gels and stained using Coomassie brilliant blue [24].
Cloning, expressing and purification of recombinant proteins
The 6PGD accession code-NP 344902 gene was amplified from S. pneumoniae strain WU2 genomic DNA by PCR with the following primers: forward: (26·3 nmol, Mm 8225, 7·5 OD, Tm 63·4) 5′-TCG AGC TCT TTT TCG TCA TAC CAA GAG-3′; reverse: (254·8 nmol, Mm 8984, 8·9 OD, Tm 65·3) 5′-TCG GAT CCA TGA AAC AAG AGG AGT GTC AA-3′. The forward and reverse primers contain BamHI and SacI recognition sequences, respectively, and all primers contain 5′-TC spacers. The primers flank the entire open reading frames. The amplified and BamHI-SacI (Takara Bio Inc, Shiga, Japan)-digested DNA-fragments were cloned into the pHAT expression vector (HAT epitope is 19-amino-acid sequence with six non-consecutive His residues (BD Biosciences Clontech, Palo Alto, CA, USA) and transformed in DH5a UltraMAX ultracompetent E. coli cells. Ampicillin-resistant transformants were cultured and plasmid DNA was analysed by PCR. The pHAT-6PGD vector was purified from DH5α UltraMAX cells using the Qiagen High Speed Plasmid Maxi Kit (Qiagen GMBH, Hilden, Germany) and transformed in E. coli host expression strain BL21(DE3)pLysS (Stratagene, La Jolla, CA, USA). The identity of the insert was confirmed by sequencing. Bacteria were grown overnight and expression of the recombinant HAT-tagged proteins was induced by the addition of 1 mM IPTG to BL21(DE3)pLysS+6PGD cells for 5 h. The cells were harvested by centrifugation and lysed in lysis buffer containing urea (8 M urea, 0·1 M NaH2PO4, 0·01 M Tris-Cl pH 8·0), as the r6PGD protein was found previously in inclusion bodies. The HAT-tagged recombinant proteins were purified using a Ni-NTA column (Qiagen GMBH) binding for 1 h at room temperature. The column was then washed with wash buffer (8 M urea, 0·1 M NaH2PO4, 0·01 M Tris-Cl pH 6·3), and the recombinant proteins were recovered from the column using elution buffer (8 M urea, 0·1 M NaH2PO4, 0·01 M Tris-Cl, pH 5·9). After purification recombinant protein was dialysed for partial refolding against PBS buffer for 48 h with three exchanges of the buffer. Isolation of the proteins was confirmed by Western blot analysis using anti-HAT antibodies (BD Biosciences Clontech) and by MALDI-TOF mass spectrometry sequencing.
Inhibition of S. pneumoniae adhesion to A549 cells
A549 [25] cells (type II epithelial lung carcinoma cells; American Type Culture Collection, Rockville, MD, USA) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) with penicillin and streptomycin (100 µg/ml each) at 37°C in a humidified incubator. A549 cells (2·5 × 104/well) were cultured on fibronectin-coated plates to increase A549 cell adhesion to the wells and prevent their detachment during washings in the absence of antibiotic for 24 h, at which time the cells reached confluence (5 × 104 cells/well on average). The plates were blocked with 1% gelatine for 1 h and 4 × 105 colony-forming units (CFU) bacteria were added per well. The adhesion of S. pneumoniae to uncoated plates was negligible (< 80 CFU/ml). The number of bacteria that adhered to fibronectin-coated plates (8000 ± 100 s.d. CFU/ml on average) was subtracted afterwards to determine the net number of bacteria that adhered to the A549 cells only. r6PGD at the denoted concentrations (ranging from 0·2 to 25 µg/ml) was added to the cultured cells and incubated for 1 h. Following extensive washings S. pneumoniae (106 CFU) were added for 1 h incubation. The wells were washed extensively (× 5) for the removal of non-adherent bacteria. The A549 cells were liberated by 0·25% trypsin-ethylenediamine tetraacetic acid (EDTA) for 5 min at 37°C and plated, in serial dilutions, onto blood agar plates. The blood agar plates were incubated for 18 h at 37°C.
Immunization of mice with 6PGD
Six-week-old BALB/c female mice (Harlan Laboratories, Israel) were housed in sterile conditions under 12-h light/dark cycles and fed Purina chow and tap water ad libitum. Animal experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Ben-Gurion University of the Negev, Beer Sheva, Israel. Mice were immunized intraperitoneally with 25 µg of r6PGD and 75 µl of Inject Alum adjuvant (Pierce Biotechnology Inc., Rockford, IL, USA) on days 0 (primary immunization) and 21 (booster). Control mice were sham-immunized with adjuvant only. Blood samples were collected from mice 1 week prior to immunization and 1 week after booster immunization. The sera were pooled for immunological assays.
For inhibition of adhesion experiments S. pneumoniae (106 CFU) were added to A549 cells, as described above, prior to or after 30 min incubation with serum obtained from r6PGD immunized mice. The bacteria were spun and resuspended in culture media prior to their addition to the cultured A549 cells. Incubation and results evaluation was performed as described in the previous section.
Bacterial challenge of mice
For respiratory challenge r6PGD immunized (n = 29) and control (n = 14) mice were anaesthetized with pentobarbital sodium (0·6 mg/kg) and inoculated intranasally with 1 × 108 S. pneumoniae strain WU2 (in 25 µl PBS). This inoculum's size was used as it was found to be the lowest that causes 100% mortality in our mouse model system within 96 h. Survival was monitored daily. The experiments were conducted on three different occasions and the results were pooled.
Bacterial load was determined in r6PGD immunized (n = 3) and control mice (n = 3). The nasopharynx and lungs were excised and homogenized and blood was withdrawn. Bacterial load in the nasopharynx, lungs and the blood was determined 48 h following intranasal challenge with S. pneumoniae strain WU2.
Flow cytometry
Bacteria were incubated with titrated amounts of anti-r6PGD serum or control mouse serum, washed and stained with fluorescein isothiocyanate (FITC)-conjugated-F(ab′)2 goat-anti-mouse-IgG + IgM (Jackson ImmunoResearch, West Grove, PA, USA). Staining and washing buffer consisted of 2% (v/v) FCS and 0·05% sodium azide in PBS. Flow cytometry was performed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA). Data files were acquired and analysed using BD CellQuestTM 3·3 software. Staining results shown are with serums diluted at 1 : 15 and are presented as overlays of staining histograms (the x-axis represents fluorescence intensity and the y-axis represents cell counts).
Statistical analysis
Pearson's regression analysis was used to verify the ability of r6PGD and anti-r6PGD antibodies to inhibit bacterial adhesion to A549 lung epithelial cells in a concentration-dependent manner. Further verification was performed using two-way analysis of variance (anova) for repeated measurements. Kaplan–Meier survival analysis was used for the vaccination studies.
Bioinformatic analysis
The nucleotide and amino acid sequences of 6PGD were analysed for functional domains using the TIGR4 and R6 sequences available on databases. The nucleotide sequence of S. pneumoniae 6PGD sequence was compared to the entire human genome at NCBI and UCSC databases.
Results
Cloning and expression of r6PGD
The amplified and BamHI-SacI digested DNA fragments were cloned into the pHAT expression vector and the out-coming plasmid was used to transform the E. coli host expression strain DHα followed by plasmid isolation and transformation of BL21(DE3)pLysS. The insert was amplified and the existence of the expected 1440 base pairs (bp) size band was confirmed (data not shown). The identity of the amplified 1440 bp band as the 6pgd gene was confirmed further by sequencing. The HAT-tagged r6PGD fusion protein was purified by Ni+ NTA affinity chromatography. Resolution of the eluted protein by one-dimensional PAGE revealed a single band following staining of the gel with Coomassie brilliant blue (53·4 kDa band; data not shown). Further confirmation of the identity of the band as the expected HAT-tagged-r6PGD was performed by Western blot analysis using anti-HAT antibodies (data not shown). MALDI-TOF sequencing of the purified r6PGD protein separated on two-dimensional PAGE gave the final confirmation of the identity of the cloned protein as 6PGD.
Inhibition of S. pneumoniae adhesion to cultured A549 epithelial cells by r6PGD
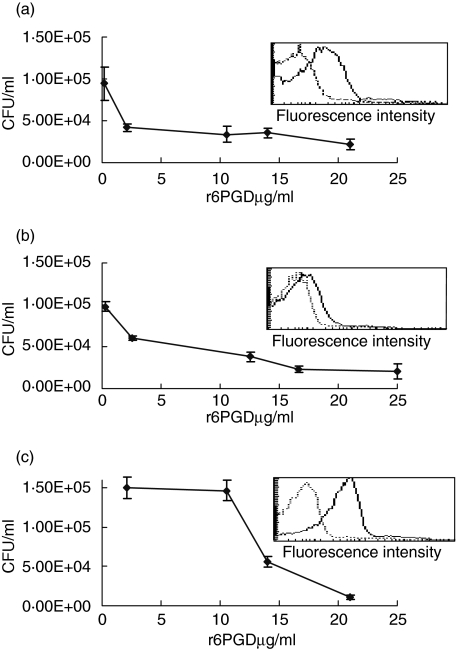
To determine whether 6PGD is involved in bacterial adhesion to respiratory epithelial cells we tested its ability to inhibit bacterial adhesion to A459 type II lung carcinoma cells in a concentration-dependent manner. The ability of r6PGD to interfere with the adhesion of S. pneumoniae was tested with three genetically different unencapsulated strains of S. pneumoniae (3·8, 14·8 and R6; Fig. 1a–c). r6PGD inhibited S. pneumoniae adhesion to A549 cells in a concentration-dependent manner to all tested unencapsulated strains of the bacteria tested (Pearson's regression analysis 3·8; r =−0·754, P < 0·001, 14·8; r = −0·626, P < 0·05, R6; r = −0·782, P < 0·001). Inhibitory concentration of 50% (IC50) of S. pneumoniae strains 3·8, 14·8 and R6 adhesion to A549 cells was at 2, 6·5 and 13 µg/ml, respectively. The experiment was performed in triplicate and was repeated on three different occasions.
Fig. 1.
Inhibition of unencapsulated Streptococcus pneumoniae adhesion to A549 epithelial cells. S. pneumoniae strains were added to the cultured A459 type II lung carcinoma cells prior and following treatment with increasing concentration of purified recombinant 6-phosphogluconate dehydrogenase (r6PGD). r6PGD interfered with the adhesion of unencapsulated S. pneumoniae strains, 3·8 (a), 14·8 (b) and R6 (c) in a concentration-dependent manner (3·8; r = −0·754, P < 0·001, 14·8; r = −0·626, P < 0·05, R6; r = −0·782, P < 0·001). Insert in each panel: primary fluorescence activated cell sorter (FACS) histogram overlays showing staining of the corresponding bacteria with anti-6PGD serum (plain line) and control mouse serum (dotted line).
To further verify the surface location of 6PGD, flow cytometry analysis was performed using antibodies to r6PGD obtained from immunized mice as the primary antibodies. Fluorescence activated cell sorter (FACS) analysis revealed the existence of 6PGD on S. pneumoniae strains 3·8, 14·8 and R6 (geometric means 8·95, 6·03 and 26·26, respectively), in comparison to bacteria stained with negative serum taken from control mice (geometric means 3·35, 3·91 and 3·83). The existence of 6PGD on the bacterial cell wall, as analysed by flow cytometry (Fig. 1, inserts), was found to be higher on the R6 S. pneumoniae strain. This correlated with the higher amounts of r6PGD needed to inhibit bacterial adhesion. r6PGD could be found on the 14DW encapsulated S. pneumoniae strain (data not shown).
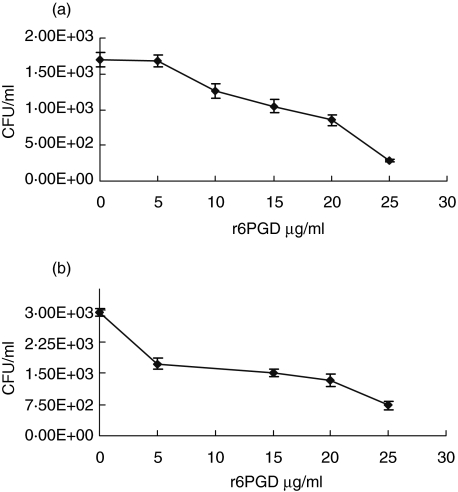
In addition, 6PGD inhibited the adhesion of two genetically different encapsulated strains (WU2 and D39; Fig. 2a,b) to A549 lung cells in a concentration-dependent manner (Pearson's regression analysis WU2; r = −0·921, P < 0·001, D39; r = −0·771, P < 0·001). IC50 of adhesion of S. pneumoniae strains WU2 and D39 to A549 cells was 16·5 and 20 µg/ml, respectively. It should be noted that 3·8 and 14·8 strains are unencapsulated mutants of WU2 and 14DW S. pneumoniae strains. Flow cytometry analysis of 6PGD on WU2 and D39 encapsulated strains was below the level of detection (data not shown).
Fig. 2.
Inhibition of encapsulated Streptococcus pneumoniae adhesion to A549 epithelial cells. S. pneumoniae encapsulated strains WU2 (a) and D39 (b) were added to the cultured epithelial cells prior and following treatment with purified recombinant 6-phosphogluconate dehydrogenase (r6PGD). r6PGD inhibited S. pneumoniae adhesion in concentration-dependent (WU2; r = −0·921, P < 0·001, D39; r = −0·771, P < 0·001).
Specificity of anti-r6PGD antibodies
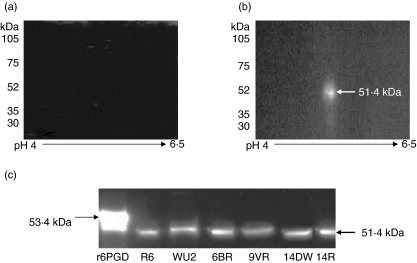
Cell wall-extracted proteins from S. pneumoniae strain WU2 were resolved by two-dimension PAGE, transferred onto nitrocellulose membrane and probed with sera obtained prior to and following immunization of mice with r6PGD. No proteins were identified with sera obtained prior to immunization (Fig. 3a), while a single spot of 51·4 kDa was detected with sera obtained from immunized mice (Fig. 3b). r6PGD and cell wall proteins obtained from six genetically or capsularly unrelated strains of S. pneumoniae (R6, WU2, 6B, 9 V, 14DW and 14R) were tested by one-dimension PAGE Western blots and probed with sera obtained from r6PGD immunized mice. The sera identified the HAT-tagged-r6PGD fusion protein as a 53·4 kDa protein. The native protein found in the cell wall extracts from all the bacterial strains used in this experiment was identified as 51·4 kDa protein (Fig. 3c). The difference in molecular weight between the recombinant and native proteins in the cell wall extract is due to the addition of HAT sequences in the r6PGD-HAT fusion protein.
Fig. 3.
The specificity of the serum obtained from the immunized mice. The specificity of the antibodies produced in mice immunized with recombinant 6-phosphogluconate dehydrogenase (r6PGD) was tested by two-dimensional polyacrylamide gel electrophoresis (PAGE) Western blot of total cell wall proteins. (a) Two-dimensional PAGE, transferred onto nitrocellulose membrane and probed with serum obtained from mice before immunization with r6PGD protein. (b) Two-dimensional PAGE, transferred onto nitrocellulose membrane and probed with serum obtained from mice after immunization with r6PGD protein. (c) r6PGD and total cell wall proteins of Streptococcus pneumoniae strains R6, WU2, 6BR, 9VR, 14DW and 14R were separated on PAGE, transferred onto nitrocellulose membranes and probed with sera obtained from mice immunized with r6PGD protein.
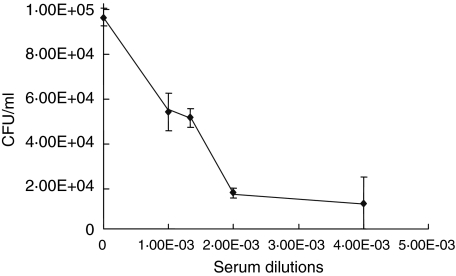
To determine whether the antibodies produced in r6PGD immunized mice have a functional significance, we tested their ability to interfere with S. pneumoniae adhesion to A549 cells. S. pneumoniae strain 3·8 bacteria were treated for 30 min with diluted serum obtained from r6PGD-immunized mice and then centrifuged, resuspended in PBS and added to the cultured A549 cells. Following incubation the trypsin-liberated cells were plated onto blood agar plates for colony number determination. The anti-r6PGD-antibodies inhibited 90% of S. pneumoniae adhesion to the cultured cells in a concentration-dependent manner, as determined by Pearson's correlation regression analysis (r = –0·886; P < 0·05; Fig. 4).
Fig. 4.
Inhibition of Streptococcus pneumoniae adhesion to A549 lung cells by anti-r6PGD antibodies. Antibodies to r6PGD were produced after immunization of mice with recombinant 6-phosphogluconate dehydrogenase (r6PGD) protein. S. pneumoniae (strain 3·8) bacteria were added to A549 cells in the presence of pre-immune mouse serum (control) or serum obtained from r6PGD immunized mice. The serum obtained from immunized mice inhibited S. pneumoniae adhesion in a concentration-dependent manner (r = −0·886, P < 0·05).
Vaccination with r6PGD, bacterial challenge of mice and bacterial load
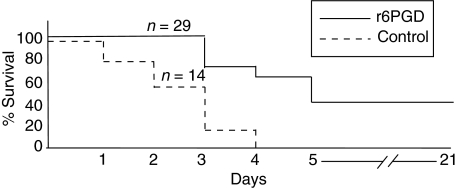
To determine whether immunization with r6PGD can elicit protective immunity, BALB/c mice were immunized as described in Methods. Immunization of mice with r6PGD delayed mortality and protected 60% of the mice for 5 days (P< 0·001) and 40% for 21 days (P< 0·05) following intranasal lethal challenge with S. pneumoniae WU2, while none of the control mice survived (P< 0·001; Fig. 5).
Fig. 5.
Survival of 6-phosphogluconate dehydrogenase (6PGD) immunized mice following S. pneumoniae challenge. Six-week-old BALB/c female mice were immunized intraperitoneally with 25 µg of r6PGD (n = 29) on day 0 (primary immunization) and day 21 (booster). Control mice (n = 14) were sham-immunized with adjuvant. r6PGD protected 60% and 40% of the mice after 5 (P < 0·001) and 21 (P < 0.05) days, respectively, after intranasal lethal challenge with S. pneumoniae WU2.
In addition, bacterial load was determined in r6PGD immunized and control mice. Bacterial load in the nasopharynx, lungs and the blood was determined 48 h following intranasal challenge with S. pneumoniae strain WU2. The bacterial load results were in agreement with the vaccination results being lower in the immunized mice in comparison to the unimmunized mice, as expected (Table 2).
Table 2.
Bacterial load after 6-phosphogluconate dehydrogenase (6PGD) immunization.
| Blood colonization (CFU/ml)a | Lung colonization (CFU/ml) | Nose colonization (CFU/ml) | |
|---|---|---|---|
| Immunized mice | 226 000 ± 2878 | 138 500 ± 36 062 | 35 766 ± 1328 |
| Control mice | 1130 000 ± 183 827 | 1023 500 ± 97 936 | 135 000 ± 35 355 |
Each box represents average results (with standard deviation) of three mice. CFU: colony-forming units.
Bioinformatic analysis
S. pneumoniae 6PGD does not contain a signal peptide, choline-binding domain or LPXTG sequences. S. pneumoniae 6PGD protein has 40% homology to its human orthologue, as revealed by blast database search. The protein sequence of the cloned 6PGD from WU2 was confirmed by MALDI-TOF analysis against the database of S. pneumoniae TIGR4 (http://www.matrixscience.com).
Discussion
We have initiated biochemical and proteomic studies to identify surface molecules involved in S. pneumoniae pathogenicity [15]. In the current study we describe an immunogenic cell wall-derived lectin, 6PGD, as a putative adhesin. S. pneumoniae adhesion to cultured A549 type II lung epithelial cells was inhibited significantly by r6PGD. Similarly, antibodies elicited against r6PGD in mice inhibited bacterial adhesion to the cultured A549 cells. The reduction in bacterial load in the nasopharynx, lungs and blood suggests that the protective immune response elicited by r6PGD affected all stages of disease development. We found further that r6PGD can elicit a partially protective immune response against an intranasal lethal challenge of S. pneumoniae in the mouse model system, signifying its involvement in S. pneumoniae pathogenicity
Bacterial attachment is a prerequisite for disease development and may serve in a number of basic functions. The attached state of the bacteria is thought to promote nutrient uptake and to allow the bacteria to multiply. In certain cases it was demonstrated that adherence promotes the delivery of toxins [26]. Moreover, it was demonstrated that adherence of a pathogen initiates the local response to infection by activating a signal transduction cascade that result in the production of proinflammatory mediators [27], probably through nuclear factor (NF)κB-dependent activation cascades [28].
In the case of uropathogenic pyelonephritic E. coli the initial interaction involved the recognition of a glycosylated receptor by the tip of the P fimbriae [29] and G fimbriae, respectively (fibriae are filamentous polymers of protein subunits called fimbrillins) [30]. In the case of S. pneumoniae, administration of sialyated oligosaccharides was shown to prevent the development of pneumococcal pneumonia in mice, presumably by interfering with adhesion of the pathogen to host cells [31], suggesting the involvement of pneumococcal lectins (carbohydrate-binding proteins) in the process. Furthermore, the ability of sialylated and non-sialylated oligosccharides and other oligosaccharides [5–8] to inhibit S. pneumoniae adhesion to cultured cells encouraged us to use proteomics for the identification of cell wall lectins and study their putative role in S. pneumoniae adhesion to epithelial cells.
We have demonstrated previously that cell wall lectins inhibit pneumococcal adhesion to cultured epithelial cells more efficiently than cell wall non-lectin proteins [14]. Furthermore, we found that the lectin fraction is capable of eliciting a partial protective immune response against intranasal bacterial lethal challenge but not to the intraperitoneal lethal challenge [32]. We have used MALDI-TOF sequencing to identify proteins from the lectin fraction with age-dependent immunogenicity in infants [14]. 6PGD was found to belong to this group of proteins [15], and as such was selected for further studies.
It should be mentioned that cytoplasmic 6PGD is responsible for the conversion of 6-phosphogluconate to ribose-5-phosphate, which might explain its ability to bind sugars, although the saccharidic entity to which this protein binds in a receptor molecule may differ.
The finding that 6PGD could be identified by flow cytometry on the surface of the bacteria further confirmed 6PGD surface location. A dual function for housekeeping proteins in prokaryotes has been demonstrated, among others, in S. agalactia [33], S. pyogenes [34] and S. pneumoniae [35]. The mechanism of export of these proteins to the cell wall is unknown [36], because many of them do not carry recognizable export domains such as signal peptides [37] or LXPTG [38] sequences.
In the current study we have evaluated 6PGD involvement in S. pneumoniae interaction with human cells in culture. r6PGD and antibodies produced against this protein in mice demonstrated the ability to inhibit bacterial adhesion to cultured type II lung cells derived from a lung carcinoma. We have chosen human lung carcinoma cells (A549), which resemble type II pneumocytes, as our model system. These cells retained many characteristics of the type II lung carcinoma cells such as human karyotype, type II multi-lamellar cytoplasmic inclusions [25] and phospholipid biosynthesis [39]. In addition to these morphological and biochemical characteristics they also demonstrate immunological characteristics resembling type II lung cells, such as secretion of surfactants [40] and constitutive and inducible nitric oxide activity [41]. The A549 lung carcinoma cells have been used widely as a model to study S. pneumoniae interaction with human cells [42,43].
Recombinant 6PGD inhibited the adhesion of several genetically unrelated strains of S. pneumoniae, suggesting the conservation of this protein in the tested bacterial strains. Furthermore, we have used an encapsulated strain (WU2 [21]) and its unencapsulated mutant (3·8 [18]). It is of interest to note that the IC50 of r6PGD inhibition of S. pneumoniae strain 3·8 adhesion to A549 cells was the lowest (2 µg/ml). This could be the result of a lack of not only the capsule, but also other virulence factors [44]. Specifically, this strain was described as lacking PspA [45] and its parental strain, WU2, was shown to lack PspC/CbpA [46]. We have demonstrated further that r6PGD was also capable of inhibiting the adhesion of other encapsulated pneumococcal strains. Interestingly, although the unencapsulated strains adhere more readily to the cultured A549 cells, inhibition of adhesion by r6PGD could be achieved at lower IC50 for the unencapsulated strains than to the encapsulated strain (2, 6 and 13 µg/ml versus 16·5 and 20 µg/ml, respectively). A possible explanation could be the higher sensitivity of the unencapsulated strains to oxidative activity that occurs in the A549 cells upon activation similar to that occurring in primary alveolar cells.
The ability of 6PGD to inhibit S. pneumoniae adhesion to A549 pneumocytes suggests the existence of binding sites specific to 6PGD. Furthermore, surface location of 6PGD on unencapsulated strains of bacteria was confirmed by FACS analysis. The same analysis with encapsulated strains revealed staining below the level of detection, except for encapsulated strain 14DW. We have described previously that the amount of carbohydrates on the 14DW strain is lower than on WU2 and D39 strains [21]. Thus it is possible that the capsule masks 6PGD to below FACS detection level, but not below functional involvement in adhesion, as has been found in this study.
Several pneumococcal cell wall proteins were described which could elicit a protective immune response. Among these proteins are PspA [47], presumably a protein involved in factor H binding, PsaA [48], a lipoprotein belonging to the ABC transporters involved in Mn+2 transport and possibly in cell adhesion [49,50]. Additional pneumococcal surface proteins capable of eliciting a protective immune response are CbpA/PspC (a major pneumococcal adhesion) [51], pneumolysin [52] and BVH (a surface exposed accessible protein) [53].
r6PGD could also potentially be a candidate for vaccine development. However, the high homology with the human orthologue is discouraging. Currently, T and B cell epitopes are being looked for in the 6PGD for the identification of sequences lacking homology to humans that may enable the development of a putative vaccine from this protein. Nevertheless, understanding the molecular mechanisms underlying S. pneumoniae interaction with the host and their target molecules may identify new therapeutic targets.
Acknowledgments
This work was supported by grants from the Israeli Ministry of Health nos 4476 and 5540, BGNegev Biotechnology, BGU seed money no. 80904101, a grant from the Center of Emerging Diseases no. 2506, Israeli Science Foundation G13/04 to Y. M. N.
References
- 1.Centers for Disease Control and Prevention. Prevention of pneumococcal disease. Recommendations of the Advisory Committee on Immunization Practices. MMRW. 1997;46:1–24. [PubMed] [Google Scholar]
- 2.Musher DM. Infection caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity and treatment. Clin Infect Dis. 1992;14:801–9. doi: 10.1093/clinids/14.4.801. [DOI] [PubMed] [Google Scholar]
- 3.Tuomanen EI, Masure HR. Molecular and cellular biology of pneumococcal infection. In: Tomasz A, editor. Streptococcal pneumoniae. New York: Mary Ann Liebert, Inc.; 2000. pp. 295–308. molecular biology, mechanism of disease. [Google Scholar]
- 4.Andersson B, Dahmen J, Frejd T, et al. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J Exp Med. 1983;158:559–70. doi: 10.1084/jem.158.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthelson R, Mobasseri A, Zopf D, Simon P. Adherence of Streptococcus pneumoniae to respiratory epithelial cells is inhibited by sialylated oligosaccharides. Infect Immun. 1998;66:1439–44. doi: 10.1128/iai.66.4.1439-1444.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krivan HC, Roberts DD, Ginsburg V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc beta 1-4Gal found in some glycolipids. Proc Natl Acad Sci USA. 1988;85:6157–61. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundell DR, Tuomanen EI. Receptor specificity of adherence of Streptococcus pneumoniae to human type-II pneumocytes and vascular endothelial cells in vitro. Microb Pathog. 1994;17:361–74. doi: 10.1006/mpat.1994.1082. [DOI] [PubMed] [Google Scholar]
- 8.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 1995;377:435–8. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 9.Geelen S, Bhattacharyya C, Tuomanen E. The cell wall mediates pneumococcal attachment to and cytopathology in human endothelial cells. Infect Immun. 1993;61:1538–43. doi: 10.1128/iai.61.4.1538-1543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce BJ, Yin YB, Masure HR. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–50. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 11.Rosenow C, Ryan P, Weiser JN, et al. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol. 1997;25:819–29. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JR, Mostov KE, Lamm ME, et al. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell. 2000;102:827–37. doi: 10.1016/s0092-8674(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 13.Elm C, Rohde M, Vaerman JP, Chhatwal GS, Hammerschmidt S. Characterization of the interaction of the pneumococcal surface protein SpsA with the human polymeric immunoglobulin receptor (hpIgR) Indian J Med Res. 2004 May;119(Suppl.):61–5. [PubMed] [Google Scholar]
- 14.Lifshitz S, Dagan R, Shani-Sekler M, et al. Age dependent preference in human antibody responses to Streptococcus pneumoniae polypeptide antigens. Clin Exp Immunol. 2002;127:344–53. doi: 10.1046/j.1365-2249.2002.01745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling E, Feldman G, Portnoi M, et al. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol. 2004;138:290–8. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann S, Rohde M, Chhatwal GS, Hammechmidt S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40:1273–87. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 17.Cole JN, Ramirez RD, Currie BJ, Cordwell SJ, Djordjevic SP, Walker MJ. Surface analyses and immune reactivities of major cell wall-associated proteins of group a streptococcus. Infect Immun. 2005;73:3137–46. doi: 10.1128/IAI.73.5.3137-3146.2005. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of the pneumococcal types. J Exp Med. 1944;79:137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson DA, Usher DM. Interruption of capsule production in Streptococcus pneumonia serotype 3 by insertion of transposon Tn916. Infect Immun. 1990;58:3135–8. doi: 10.1128/iai.58.9.3135-3138.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson DA, Musher DM, Verhoef J. Pneumococcal virulence factors and host immune responses to them. Eur J Clin Microbiol Infect Dis. 1995;14:479–90. doi: 10.1007/BF02113425. [DOI] [PubMed] [Google Scholar]
- 21.Briles DE, Nahm M, Schroer K, et al. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J Exp Med. 1981;153:694–705. doi: 10.1084/jem.153.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizrachi Nebenzahl Y, Porat N, Lifshitz S, et al. Virulence of Streptococcus pneumoniae may be determined independently of capsular polysaccharide. FEMS Microbiol Lett. 2004;233:147–52. doi: 10.1016/j.femsle.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Siegel JL, Hurst SF, Liberman ES, Coleman SE, Bleiweis AS. Mutanolysin-induced sphroplasts of Streptococcus mutans are true protoplasts. Infect Immun. 1981;31:808–15. doi: 10.1128/iai.31.2.808-815.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F, Brent R, Kingston R, Moore D. Analysis of protein. In: Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. Canada: Wiley & Sons; 2003. pp. 10.2.1–10.8.18. [Google Scholar]
- 25.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 26.Smith HW, Liggood MA. Further observation on Escherichia coli enterotoxins with particular regard to those produced by atypical piglet strains and calf and lamb strains: the transmissible nature of these enterotoxins and of a K antigen possessed by calf and lamb strains. J Med Microbiol. 1972;5:243–50. doi: 10.1099/00222615-5-2-243. [DOI] [PubMed] [Google Scholar]
- 27.Svanborg C, Bergsten G, Fischer H, et al. Adhesion signal transduction and mucosal inflammation. Adv Mol Cel Microbiol. 2002;1:223–46. [Google Scholar]
- 28.Opitz B, Puschel A, Schmeck B, et al. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279:36426–32. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 29.Svanborg-Eden C, Hansson HA. Escherichia coli pili as possible mediators of attachment to human urinary tract epithelial cells. Infect Immun. 1978;21:229–37. doi: 10.1128/iai.21.1.229-237.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saarela S, Taira S, Nurmiaho-Lassila EL, Makkonen A, Rhen M. The Escherichia coli G-fimbrial lectin protein participates both in fimbrial biogenesis and in recognition of the receptor N-acetyl-D-glucosamine. J Bacteriol. 1995;177:1477–84. doi: 10.1128/jb.177.6.1477-1484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Idanpaan-Heikkila I, Simon PM, Zopf D, et al. Oligosacchride interferes with the establishment and progression of experimental pneumococcal infection. J Infect Dis. 1997;176:704–12. doi: 10.1086/514094. [DOI] [PubMed] [Google Scholar]
- 32.Portnoi M, Ling E, Feldman G, Dagan R, Mizrachi-Nebenzahl Y. The vaccine potential of Streptococcus pneumoniae surface lectin and non-lectin proteins. Vaccine. in press. [DOI] [PubMed]
- 33.Hughes MJ, Moore JC, Lane JD, et al. Identification of major outer surface proteins of Streptococcus agalactiae. Infect Immun. 2002;70:1254–9. doi: 10.1128/IAI.70.3.1254-1259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Costa SS, Romer TG, Boyle MD. Analysis of expression of a cytosolic enzyme on the surface of Streptococcus pyogenes. Biochem Biophys Res Commun. 2000;278:826–32. doi: 10.1006/bbrc.2000.3884. [DOI] [PubMed] [Google Scholar]
- 35.Bergmann S, Rohds M, Hammerschmidt S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun. 2004;72:24416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chhatwal GS. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 2002;10:205–8. doi: 10.1016/s0966-842x(02)02351-x. [DOI] [PubMed] [Google Scholar]
- 37.Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–78. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–97. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro DL, Nardone LL, Rooney SA, Motoyama EK, Munoz JL. Phospholipid biosynthesis and secretion by a cell line (A549) which resembles type II aleveolar epithelial cells. Biochim Biophys Acta. 1978;530:197–207. doi: 10.1016/0005-2760(78)90005-x. [DOI] [PubMed] [Google Scholar]
- 40.Balis JU, Bumgarner SD, Paciga JE, Paterson JF, Shelley SA. Synthesis of lung surfactant-associated glycoproteins by A549 cells: description of an in vitro model for human type II cell dysfunction. Exp Lung Res. 1984;6:197–213. doi: 10.3109/01902148409109248. [DOI] [PubMed] [Google Scholar]
- 41.Asano K, Chee CB, Gaston B, et al. Constitutive and inducible nitric oxide synthase gene expression, regulation, and activity in human lung epithelial cells. Proc Natl Acad Sci USA. 1994;91:10089–93. doi: 10.1073/pnas.91.21.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talbot UM, Paton AW, Paton JC. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect Immun. 1996;64:3772–7. doi: 10.1128/iai.64.9.3772-3777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Muller E, Rohde M. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun. 2005;73:4653–67. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson DA, Kapur V, Musher DM, Jacobson JW, Musser JM. Identification, cloning, and sequencing of DNA essential for encapsulation of Streptococcus pneumoniae. Curr Microbiol. 1995;31:251–9. doi: 10.1007/BF00298383. [DOI] [PubMed] [Google Scholar]
- 45.Neeleman C, Geelen SP, Aerts PC, et al. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by the binding of complement regulatory protein factor H. Infect Immun. 1999;67:4517–24. doi: 10.1128/iai.67.9.4517-4524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDaniel LS, Sheffield JS, Swiatlo E, Yother J, Crain MJ, Briles DE. Molecular localization of variable and conserved regions of pspA and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb Pathog. 1992;13:261–9. doi: 10.1016/0882-4010(92)90036-n. [DOI] [PubMed] [Google Scholar]
- 47.McDaniel LS, Sheffield JS, Delucchi P, Briles DE. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–8. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Briles DE, Ades E, Paton JC, et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun. 2000;68:796–800. doi: 10.1128/iai.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston JW, Myers LE, Ochs MM, Benjamin WH, Jr, Briles DE, Hollingshead SK. Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect Immun. 2004;72:5858–67. doi: 10.1128/IAI.72.10.5858-5867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol Microbiol. 2004;53:889–901. doi: 10.1111/j.1365-2958.2004.04164.x. [DOI] [PubMed] [Google Scholar]
- 51.Luo R, Mann B, Lewis WS, et al. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J. 2005;24:34–43. doi: 10.1038/sj.emboj.7600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musher DM, Phan HM, Baughn RE. Protection against bacteremic pneumococcal infection by antibody to pneumolysin. J Infect Dis. 2001;183:827–30. doi: 10.1086/318833. [DOI] [PubMed] [Google Scholar]
- 53.Hamel J, Charland N, Pineau I, et al. Prevention of pneumococcal disease in mice immunized with conserved surface-accessible proteins. Infect Immun. 2004;72:2659–70. doi: 10.1128/IAI.72.5.2659-2670.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]