Abstract
Summary
Class1 major histocompatibility complex (MHC-I)–antigenic peptide exposed at the target cell surface is crucial for the adaptive immune response exerted in the self/syngeneic context by cytotoxic T lymphocyte (CTL). Such a complex also provides epitopes in the allogeneic context for antibody response directed against the MHC-I polymorphic determinant. In the present report we examined the formation of the MHC-I–peptide complex leading predominantly to the expression of T and/or B cell epitopes in a process of internal versus external antigenic peptide loading onto the binding groove of MHC-I. Analyses using antibodies specific to complex MHC-I–peptide generated in the syngeneic context to mimic T cell receptor (TCR) in comparison with antibodies specific to the MHC-I polymorphic determinant allowed the observation that the external peptide loading to MHC-I, while remaining necessary for inducing the formation of B cell epitopes, was less efficient than the internal one for generating T cell epitopes. Thus, external loading of peptide to the MHC-I appeared to match more closely the allogeneic situation and the humoral immunity in general, while internal peptide loading corresponded with the self/syngeneic context of the cellular CTL response.
Keywords: antigenic peptide loading, MHC-I–antigenic peptide complex, TCR-like antibodies
Introduction
Cytotoxic T lymphocytes of the adaptative immune response operate in the self/syngeneic context [1] via recognition of antigenic peptides presented by the major histocompatibility complex (MHC-I) [2,3]. Antigenic peptides are mainly short sequences of 8–10 amino acids loaded onto the MHC-I peptide binding groove [4]. The way in which protein antigens are processed to antigenic peptides has been well documented [5,6]. Once generated, antigenic peptides are transported and loaded into the MHC-I's groove. The complex MHC-I–peptide is then exposed at the cell surface for immune surveillance via cytotoxic T lymphocyte (CTL) T cell receptors (TCRs). As internal peptides are not the only source of peptides, it was questioned whether the external peptides that circulate in the bloodstream could play a role. On the other hand, in the balance between immunity and tolerance, there could be a possibility for the external peptide to interfere with the presentation and recognition of the internal peptide in the context of the MHC-I. Moreover, it is well known that the humoral and cellular arms of the adaptive immune response follow different pathways for stimulation as well as mode of binding to specific ligands.
In order to examine this feature we used antibodies with a specificity-like TCR, i.e. recognizing a complex MHC-I–antigenic peptide in the syngeneic context. Furthermore, a comparison with antibodies specific to polymorphic determinants of the MHC-I generated in the allogeneic context allowed us to gain insight into the formation of T cell versus B cell epitopes in the case of internal or external antigenic peptides loading onto the MHC-I. We used an experimental model based on mutant cell line RMA-S [7] incubated with ovalbumin (OVA) peptide (SIINFEKL) as representative of external peptide loading [8]. On the other hand, the EL-4 cell line transfected with the gene encoding for OVA, subclone E.G7 [9], was taken as an example of internal peptide loading. Monoclonal antibodies (mAb) isolated in the syngeneic context from C57BL/6 mice and specific to Kb-OVA were used as TCR-like antibodies (anti-T cell epitope) in comparison to those specific to Kb polymorphic determinant (anti-B cell epitope).
Materials and methods
Mice and cell lines
C57BL/6 (H-2b) mice were purchased from IFFA/CREDO (Lyon, France) and maintained in the animal facility according to ECC directives (86/609/CCE).
EL-4 (H-2b) leukaemia cells were from the American Type Culture Collection (ATCC, Rockville, MD, USA); E.G7 cells, a subclone of EL-4 transfected with the OVA gene, were a gift from Dr Bevan (Howard Hughes Medical Institute, Seattle, WA, USA); the X63-Ag8 myeloma cell line was from ATCC. RMA (H-2b) lymphoma and RMA-S mutant cells derived from Rauscher virus-induced murine cell line RB-5 were from Dr K. Kärre’s laboratory (Karolinska Institutet, Stockholm, Sweden). Cell lines were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin (Gibco brl, Cergy Pontoise, France) at 37°C, 5% CO2.
Antibodies, peptides and reagents
Anti-Kb monoclonal antibodies 28·8.6, 34·1.2 and 5F1·3 were from ATCC, fluorescein isothiocyanate (FITC), zLLLal (MG132), goat F(ab′)2 secondary antibody conjugated with FITC was purchased from Sigma (L’Isle d’Abeau-Chesnes, France). OVA peptide257−264 and VSV NP52−59 were purchased from Syntem (Nîmes, France).
Immunization and cell fusion
C57BL/6 mice were immunized subcutaneously with 107 syngeneic splenocytes pulsed with OVA peptide SIINFEKL or with E.G7 cells inactivated by a cycle of − 80 ° C frozen and thawed in phosphate-buffered saline (PBS). Mice were reboosted 4–5 days before killing, spleens were harvested and splenocytes were treated with NH4Cl for red cell lysis. Fusions of splenocytes and myeloma cells X63-Ag8 were performed according to Köhler and Milstein [10].
Flow cytometry analyses
Indirect or direct stainings were performed using standard protocol. When indirect stainings were used, cells were incubated with primary antibodies for 60 min at 4°C, washed with phosphate-buffered saline (PBS), followed by a second incubation with goat F(ab′)2 anti-mouse IgG conjugated with FITC (Sigma Aldrich, L’Isle d’Abeau-Chesnes, France). For control, the first incubation was performed when necessary with the myeloma Ig isotype matched with the tested antibody; in general only FITC–F(ab′)2 anti-mouse Ig secondary antibody was used as there was practically no difference with the myeloma Ig control. In direct stainings all antibodies used were conjugated with FITC. In order to avoid capping when two different antibodies were applied, experiments were performed in the presence of 0·5% of NaN3. Analyses were performed with Becton Dickinson's FACSCalibur and the CellQuest software (Le Pont de Claix, France).
Confocal immunofluorescence
Cells were laid onto microscope glass slides by cytospin, permeabilized with ORTHOPermeafix reagent (Ortho Diagnostic Systems, Roissy, France). Indirect stainings were carried out by incubation of permabilized cells with primary antibodies for 60 min washed with PBS−5% fetal calf serum (FCS). Cell nuclei were stained with propidium iodide. The secondary FITC–goat F(ab′)2 anti-mouse IgG was the same as that used for flow cytometry. Analyses were performed with a confocal Leica microscope and sections < 0·8 µm were scored and treated using the Leica TCS NT software (Wetzlar, Germany).
Enzyme-linked immunosorbent assay (ELISA)
Enzyme-linked immunosorbent assays were performed according to Ternynck and Avrameas [11]; peroxidase conjugated secondary antibodies were purchased from Sigma (Saint-Quentin Fallavier, France) and diaminobenzidine (DAB) was used as substrate.
Western blotting
Cell extracts were prepared in the presence of protease inhibitors [phenylmethylsulphonyl fluoride (PMSF), leupeptin, aprotinin], submitted to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinyl membranes and detected by chemiluminescence (Amersham, Les Illis, France).
Labelling antibodies with FITC
The coupling of antibody Ig 5F1·3, 28·8.6, 20G2·56 and PBE2 (see results) to FITC was performed according to the protocol described by Harlow and Lane [12].
Results
mAbs 20G2·56 and PBE2 derived from syngeneic immunization belonged to the family of antibodies specific to MHC-I-peptide
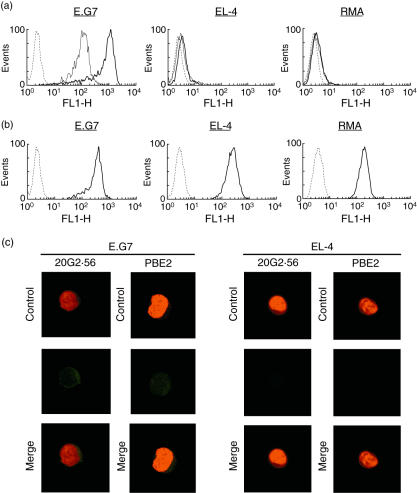
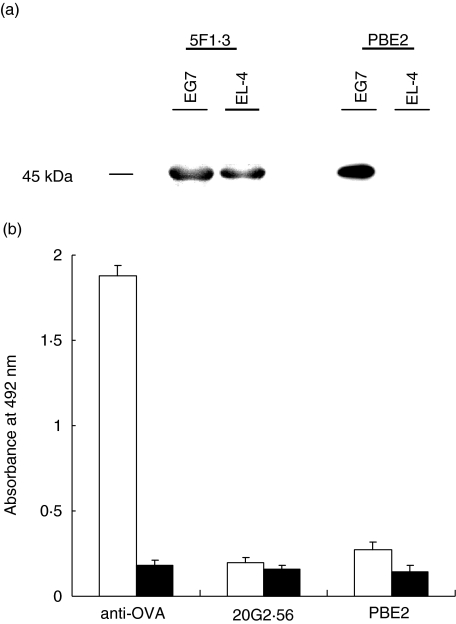
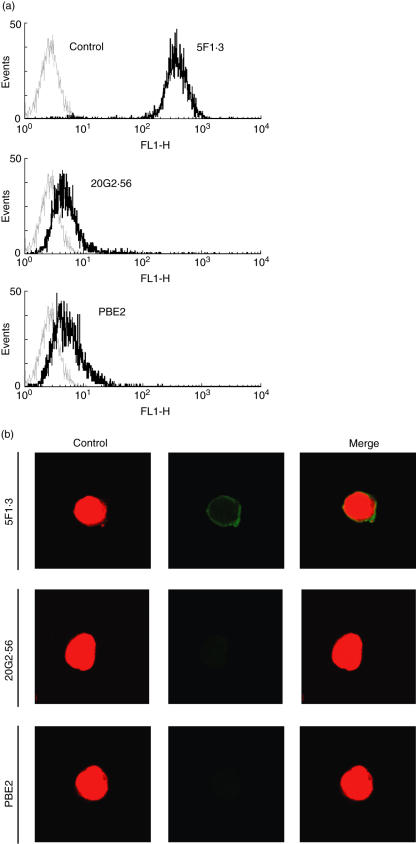
In order to analyse and compare the formation of MHC-I Kb-peptide epitope with that of the MHC-I Kb epitope, we isolated mAbs which recognize complex Kb-OVA. Two such mAbs, namely 20G2·56 and PBE2, were obtained by fusing X63Ag8 myeloma cells with spleen cells from C57BL/6 (H-2b) mice immunized with syngeneic lymphocytes incubated with OVA peptide 257–264 SIINFEKL or EL-4 leukaemia cells (Kb) transfected with OVA gene subclone E.G7, respectively. These two mAbs (IgG1, κ) gave a strong signal in flow cytometry analyses with E.G7 cells but only marginally and inconsistently with untransfected parental cells EL-4 or other Kb cells RMA (Fig. 1a). These three cell lines, E.G7, EL-4 and RMA, expressed Kb which were recognized almost identically by anti-Kb specific mAb antibodies, i.e. 5F1·3, 34·1.2 or 28·8.6. These results are shown in Fig. 1b using mAb 5F1·3. Confocal microscopy confirmed that only E.G7 but not EL-4 was recognized by 20G2·56 and PBE2 (Fig. 1c). Western blotting performed with mAb 5F1·3 allowed us to observe a band corresponding to that of MHC-I with extracts from E.G7 as well as EL-4, while only E.G7 but not EL-4 extract gave this pattern as seen with PBE2 (Fig. 2a). ELISA assays showed that 20G2·56/PBE2 did not recognize OVA or OVA peptide 257–264 alone (Fig. 2b). These results indicated that 20G2·56 and PBE2 recognized essentially the complex Kb-OVA but not the separate elements Kb and OVA; they were members of the family of TCR-like antibodies specific to T cell epitopes.
Fig. 1.
Only ovalbumin (OVA)-transfected E.G7 cells were recognized by 20G2·56 and PBE2. (a) Indirect flow cytometry analyses performed with E.G7, EL-4 or RMA cells in the presence of 20G2·56 (─) and PBE2 (—), control (---). Representative profiles of at least 10 experiments are shown. (b) Indirect flow cytometry analyses with anti-Kb mAb 5F1·3 (─) and E.G7, EL-4, RMA cells, control (---). (c) Confocal microscopy analyses after cell permeation. The indirect staining as above was performed with 20G2·56 and PBE2 as well as with the secondary goat F(ab′)2 anti-mouse Ig–fluorescein isothiocyanate (FITC) together with propidium iodide.
Fig. 2.
Reactivities of 20G2·56/PBE2 as revealed by Western blotting and enzyme-linked immunosorbent assay (ELISA). (a) Western blotting with cell extracts from E.G7 or EL-4 revealed by PBE2 and 5F1·3. (b) ELISA performed with ovalbumin (OVA) (□) and OVA peptide 257–264 (▪) coated to plastic wells (1–5 µg/ml in carbonate buffer pH 9). A mouse immune serum anti-OVA was used as positive control.
Featuring the MHC-I epitopes recognized by 20G2·56/PBE2 and 5F1·3
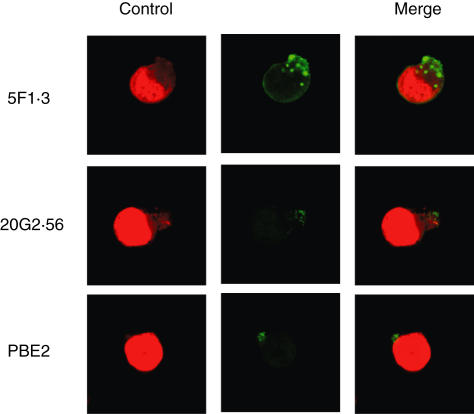
When E.G7 was incubated with mAb 20G2·56/PBE2 at 37°C, capping was occurred. The caps formed were, however, smaller than those induced in the same condition by mAb anti-Kb 5F1·3 (Fig. 3). This treatment decreased recognition of the cells by anti-Kb-peptide antibody, as shown with 20G2·56 conjugated with FITC added in the second step in the presence of PBS−0·5% NaN3. Under such conditions, the recognition of treated E.G7 by 5F1·3 conjugated with FITC was not affected (Fig. 4a). On the other hand, incubation of E.G7 at 37°C with anti-Kb 5F1·3 in the first step decreased its recognition not only by 5F1·3 conjugated with FITC but also 20G2·56 conjugated with FITC added in the second step to treated cells in the presence of PBS−0·5% NaN3. These results implied that epitopes recognized by 20G2·56 were part of Kb epitopes recognized by 5F1·3. Furthermore, when E.G7 was treated with zLLLal, a proteasome inhibitor, the recognition of E.G7 by 20G2·56 was affected, indicating the implication of antigen processing in the formation of anti-Kb peptide-specific epitopes (Fig. 4b).
Fig. 3.
Confocal microscopy. Comparison between the capping of E.G7 induced by 20G2·56/PBE2 and 5F1·3.
Fig. 4.
Involvement of major histocompatibility complex (MHC-I)–Kb and ovalbumin (OVA) antigenic peptide in the formation of 20G2·56/PBE2-specific epitope. (a) Epitopes recognized by 20G2·56/PBE2 were part of MHC-I–Kb recognized by 5F1·3. Incubation of E.G7 with unconjugated 5F1·3 or 20G2·56 at 37°C was followed by that of either 5F1·3–fluorescein isothiocyanate (FITC) (□) or 20G2·56–FITC (▪). (b) Effects of proteasome inhibitor zLLLal on the recognition of E.G7 by 20G2·56, flow cytometry analyses using indirect labelling with secondary antibody goat (Fab′)2 anti-mouse Ig–FITC. Untreated E.G7 (—), zLLLal-treated E.G7 (─), control (---).
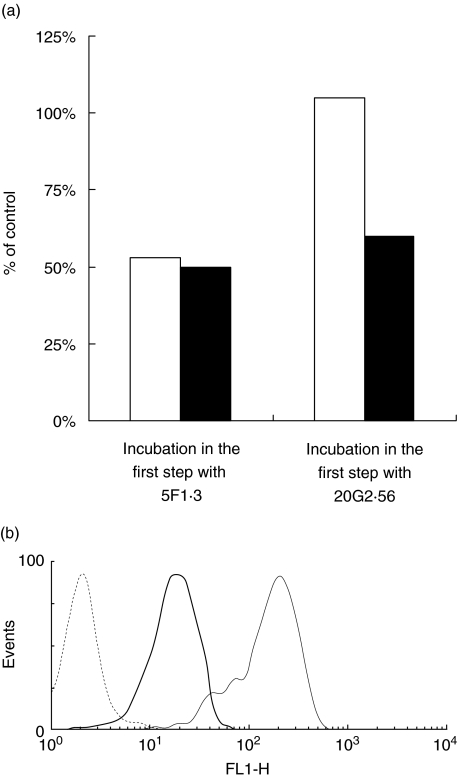
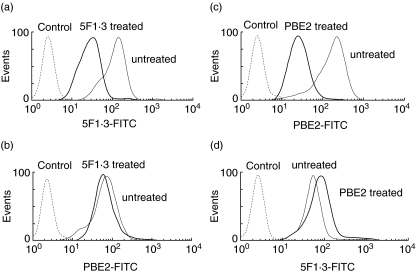
It was further examined whether anti-Kb–peptide antibody (20G2·56 or PBE2) could compete with anti-Kb antibody (5F1·3) for recognizing the epitopes expressed on the E.G7 cell surface. Experiments using two steps of incubation in the presence of PBS−0·5% NaN3 showed that there was no competition between these two types of antibody. Indeed, only unconjugated 5F1·3 reacting to E.G7 in the first step was able, in the second step, to inhibit the binding of 5F1·3 conjugated to FITC, but not PBE2–FITC, to the relevant target (Fig. 5a,b). Conversely, only PBE2 used in the first step to cover the relevant epitope was efficient to decrease the recognition of the treated cell by PBE2 conjugated to FITC but not 5F1·3–FITC added in the second step (Fig. 5c,d).
Fig. 5.
T cell and B cell epitopes were localized at different sites on the major histocompatibility complex (MHC-I). (a) Incubation of E.G7 with unconjugated 5F1·3 in the presence of 0·5% NaN3 and subsequent incubation, after washing, with 5F1·3–fluorescein isothiocyanate (FITC) (—), comparison with untreated E.G7 incubated with 5F1·3–FITC alone (—), control (---). (b) Incubation of E.G7 with unconjugated 5F1·3 in the presence of 0·5% NaN3 and subsequent incubation, after washing, with PBE2–FITC (─), comparison with untreated E.G7 incubated with PBE2–FITC alone (—), control (---). (c). Incubation of E.G7 with unconjugated PBE2 in the presence of 0·5% NaN3 and subsequent incubation, after washing, with PBE2–FITC (─), comparison with untreated E.G7 incubated with PBE2–FITC alone (—), control (---). (d) Incubation of E.G7 with unconjugated PBE2 in the presence of 0·5% NaN3 and subsequent incubation, after washing, with 5F1·3–FITC (─), comparison with untreated E.G7 incubated with 5F1·3–FITC alone (—), control (---).
Implication of endogeneous or exogeneous peptides loading onto MHC-I in the formation of epitopes recognized by 20G2·56/PBE2
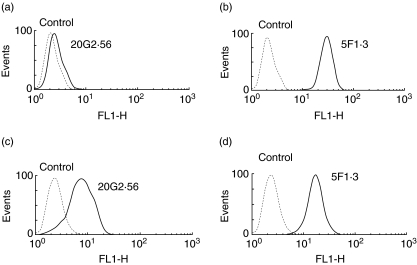
The mutant RMA-S cell line endowed with empty Kb that could be externally loaded with the relevant peptide was used to follow the formation of the epitopes recognized by 20G2·56/PBE2 and 5F1·3. Thus, when RMA-S was incubated with OVA257-264 SIINFEKL (SL8), a strong signal was observed with mAb 5F1·3, as can be seen in flow cytometry analyses. Under such conditions, 20G2·56 and PBE2 gave only a weak signal with a mean fluorescence which rarely exceeded two to three times over control (Fig. 6a). Confocal microscopy confirmed the results observed in flow cytometry showing that only mAb 5F1·3 gave a signal with RMA-S incubated with the peptide, while no signal was observed with 20G2·56/PBE2 (Fig. 6b). When EL-4 (Kb) was used instead of RMA-S and incubated with SL8, OVA257−264 SIINFEKL as previously performed, the target cell, termed EL-4-SL8-inc, was practically not recognized by 20G2·56 (Fig. 7a,b). However, when EL-4 was electroporated with SL8, OVA257−264 SIINFEKL, allowing the peptide to be introduced into the cytosol, the treated cell (EL-4-SL8-elec) was well recognized by 20G2·56 (Fig. 7c,d).
Fig. 6.
External peptide loading to major histocompatibility complex (MHC-I). RMA-S cells incubated with ovalbumin (OVA)257−264 (SIINFEKL) were analysed by indirect immunofluorescence with 5F1·3, 20G2·56, PBE2 and secondary antibody goat (Fab′)2 anti-mouse Ig–fluorescein isothiocyanate (FITC). (a) Flow cytometry. (b) Confocal microscopy (control: propidium iodide, secondary antibody: goat (Fab′)2 anti-mouse Ig–FITC).
Fig. 7.
Comparison between endogeneous and exogeneous peptide loading to major histocompatibility complex (MHC-I) Kb. EL-4 (Kb) only incubated with SIINFEKL (a and b) or electroporated with SIINFEKL (c and d) were examined in flow cytometry with 20G2·56 or 5F1·3 followed by incubation with secondary antibody goat (Fab′)2 anti-mouse Ig–fluorescein isothiocyanate (FITC). Control (---).
Discussion
The MHC-I molecule offers an unique opportunity by expressing two different epitopes, namely the B cell epitopes (i.e. its polymorphic determinants) and the T cell epitopes (i.e. MHC-I antigenic peptide complexes, the specific ligands for CTL TCRs). The presence of these epitopes involved in the humoral and cellular arms of the adaptative immune response made the MHC-I an interesting model to dissect and analyse the modulation of the immune response. Although having a similar structure, antibodies and TCRs are two different molecular entities as soluble and membrane anchored molecules, respectively. For this reason, it was more accurate to perform comparative analyses with homogeneous reagents, either with antibody engineered to be anchored on the cell membrane-like TCR or with TCR-like antibody. We have used the latter strategy and isolated antibodies which reacted similarly to TCRs, i.e. recognizing complex MHC-I–OVA. Several TCR-like antibodies have been described, including those specific to Kb-OVA [13–22]. Our antibodies, 20G2·56 and PBE2, were characterized by the fact that they were obtained in the context of syngeneic immunization, and although derived from two different immunization schemes, mAbs 20G2·56 and PBE2 had almost the same profile of reactivities, i.e. reacting to OVA-transfected Kb cells E.G7, but not or weakly to other Kb cells such as EL-4 or RMA. This aspect was in favour of the same selective process giving rise to the formation of the Fab fragment of 20G2·56 and PBE2. Of particular interest were the reactivities of 20G2·56 to EG.7, although the mAb was derived from the mouse immunized with syngeneic splenocytes incubated with OVA peptide SIINFEKL. Furthermore, E.G7 cells were shown to be able to stimulate the formation of anti-Kb-OVA antibody, as can be seen with the isolation of PBE2. Lastly, no signal was obtained with 20G2·56/PBE2 when the peptide derived from vesicular stomatitis virus nucleocapsid protein (VSV NP52-59) RGYVYQGL was introduced by electroporation into the cytosol of EL-4 cells (results not shown). Overall, like TCR which mainly recognized the target in the self/syngeneic context, mAbs 20G2·56 and PBE2 reacted essentially to the complex MHC-I–peptide. Porgador et al. [16] previously reported data concerning mAbs specific to Kb-OVA peptide which were obtained from Balb/c mice alloimmunized against RMA-S cells loaded with SIINFEKL. The mAbs were then selected for their specificities to RMA-S-SIINFEKL. This approach allowed them to obtain mAbs well-recognizing Kb loaded exogenously with OVA257−264 and probably explained the difference with our antibodies 20G2·56 and PBE2. We also used syngeneic immunization, in which RMA-S cells loaded with SIINFEKL were injected into C57BL/6 mice, but the selection conducted so far did not allow us to isolate a specific antibody. Nevertheless, from the syngeneic combination using either E.G7 or splenocytes incubated with SIINFEKL as immunogens, we have succeeded in selecting PBE2 and 20G2·56 which recognized E.G7 as well as EL-4 electroporated with SIINFEKL. It is worth noting that signals obtained in flow cytometry analyses using EL-4 electroporated with SIINFEKL were less pronounced than those obtained with OVA-transfected E.G7 (see Figs 1a and 7c). The difference observed could be explained by the presence of the OVA-encoded gene on the episome in the case of OVA-transfected cells E.G7, where Kb would be loaded readily and constantly with the peptide. Furthermore, it was shown that EL-4 cells incubated with SIINFEKL were not a good target for 20G2·56/PBE2. Indeed, this aspect was observed in our previous report [13] which showed, in the case of external peptide loading, that radioimmune assays (i.e. 51Cr release or 125I cell surface labelling) were more indicated than immunofluorescence staining to detect these MHC-I–petide complexes. Nevertheless, as reported so far, the few MHC-I–peptide complexes thus formed were sufficient to induce the lytic effects of CTLs [23,24]. The data reported by Krogsgaard et al. [25], showing that the T cell epitope was a heterodimer formed by a MHC-I-endogenous self-peptide linked to a MHC-I-agonist peptide, could explain why Kb-OVA257−264 expressed at the RMA-S cell surface was not well recognized by 2OG2·56/PBE2. Indeed, being contributed mainly by agonist peptide, it was essentially a B cell epitope. The only case where antigenic peptide that was not endogenously generated but could, however, form an appropriate target with the MHC-I for TCR, was reported by Norbury et al. [26] and Neijssen et al. [27]. The former group showed that proteasome substrates rather than peptides were important for antigen transfer in the case of cross-priming a situation observed when antigenic proteins were not synthesized by professional antigen-presenting cells. In the work reported by the second group of authors, the antigenic peptide could be presented by bystander cells to the CTL owing to diffusion through gap junctions, providing the peptide with access into the cytoplasm of bystander cell. Thus, results reported in the present work led to the assumption that the external peptide loading to MHC-I was less efficient than the internal one for inducing a high-level expression of complex MHC-I–peptide (T cell epitope). Peptides from the periphery, therefore, did not represent an effective competitor having the capacity to interfere with the presentation of endogenous peptide.
Finally, the fact that there was no competition between antibody 5F1·3 and 20G2·56/PBE2 might have some application for an eventual synergistic or complementary action resulting from the combined use of these antibodies.
References
- 1.Doherty PC, Zinkernagel RM. T-cell-mediated immunopathology in viral infections. Transplant Rev. 1974;19:89–120. doi: 10.1111/j.1600-065x.1974.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 2.Barber LD, Parham P. Peptide binding to major histocompatibility complex molecules. Annu Rev Cell Biol. 1993;9:163–206. doi: 10.1146/annurev.cb.09.110193.001115. [DOI] [PubMed] [Google Scholar]
- 3.York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–96. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Yewdell J, Anton LC, Bacik I, Schubert U, Snyder HL, Bennink JR. Generating MHC class I ligands from viral gene products. Immunol Rev. 1999;172:97–108. doi: 10.1111/j.1600-065x.1999.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 5.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–50. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky FM, Lem L, Bresnahan PA. Antigen processing and presentation. Tissue Antigens. 1996;47:464–71. doi: 10.1111/j.1399-0039.1996.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 7.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 8.Townsend A, Elliott T, Cerundolo V, Foster L, Barber B, Tse A. Assembly of MHC class I molecules analyzed in vitro. Cell. 1990;62:285–95. doi: 10.1016/0092-8674(90)90366-m. [DOI] [PubMed] [Google Scholar]
- 9.Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–85. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 10.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 11.Ternynck T, Avrameas S. Techniques immuno-enzymatiques. Paris: Les Editions INSERM; 1987. pp. 41–58. [Google Scholar]
- 12.Harlow E, Lane D. Antibodies, a laboratory manual. New York: Cold Spring Harbor Laboratory; 1988. Labeling antibodies with fluorochromes; pp. 353–5. [Google Scholar]
- 13.Duc HT, Rucay P, Righenzi S, Halle-Pannenko O, Kourilsky P. Monoclonal antibodies directed against T cell epitopes presented by class I MHC antigens. Int Immunol. 1993;5:427–31. doi: 10.1093/intimm/5.4.427. [DOI] [PubMed] [Google Scholar]
- 14.Uchanska-Ziegler B, Nossner E, Schenk A, Ziegler A, Schendel DJ. Soluble T cell receptor-like properties of an HLA-B35-specific monoclonal antibody (TU165) Eur J Immunol. 1993;23:734–8. doi: 10.1002/eji.1830230325. [DOI] [PubMed] [Google Scholar]
- 15.Andersen PS, Stryhn A, Hansen BE, Fugger L, Engberg J, Buus S. A recombinant antibody with the antigen-specific, major histocompatibility complex-restricted specificity of T cells. Proc Natl Acad Sci USA. 1996;93:1820–4. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–26. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 17.Polakova K, Plaksin D, Chung DH, Belyakov IM, Berzofsky JA, Margulies DH. Antibodies directed against the MHC-I molecule H-2Dd complexed with an antigenic peptide: similarities to a T cell receptor with the same specificity. J Immunol. 2000;165:5703–12. doi: 10.4049/jimmunol.165.10.5703. [DOI] [PubMed] [Google Scholar]
- 18.Chames P, Hufton SE, Coulie PG, Uchanska-Ziegler B, Hoogenboom HR. Direct selection of a human antibody fragment directed against the tumor T-cell epitope HLA-A1-MAGE-A1 from a nonimmunized phage-Fab library. Proc Natl Acad Sci USA. 2000;97:7969–74. doi: 10.1073/pnas.97.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lev A, Denkberg G, Cohen CJ, et al. Isolation and characterization of human recombinant antibodies endowed with the antigen-specific, major histocompatibility complex-restricted specificity of T cells directed toward the widely expressed tumor T-cell epitopes of the telomerase catalytic subunit. Cancer Res. 2002;62:3184–94. [PubMed] [Google Scholar]
- 20.Denkberg G, Cohen CJ, Lev A, Chames P, Hoogenboom HR, Reiter Y. Direct visualization of distinct T cell epitopes derived from a melanoma tumor-associated antigen by using human recombinant antibodies with MHC-restricted T cell receptor-like specificity. Proc Natl Acad Sci USA. 2002;99:9421–6. doi: 10.1073/pnas.132285699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen CJ, Sarig O, Yamano Y, Tomaru U, Jacobson S, Reiter Y. Direct phenotypic analysis of human MHC class I antigen presentation: visualization, quantitation, and in situ detection of human viral epitopes using peptide-specific, MHC-restricted human recombinant antibodies. J Immunol. 2003;170:4349–61. doi: 10.4049/jimmunol.170.8.4349. [DOI] [PubMed] [Google Scholar]
- 22.Held G, Matsuo M, Epel M, et al. Dissecting cytotoxic T cell responses towards the NY-ESO-1 protein by peptide/MHC-specific antibody fragments. Eur J Immunol. 2004;34:2919–29. doi: 10.1002/eji.200425297. [DOI] [PubMed] [Google Scholar]
- 23.Maryanski JL, Verdini AS, Weber PC, Salemme FR, Corradin G. Competitor analogs for defined T cell antigens: peptides incorporating a putative binding motif and polyproline or polyglycine spacers. Cell. 1990;60:63–72. doi: 10.1016/0092-8674(90)90716-r. [DOI] [PubMed] [Google Scholar]
- 24.Loftus DJ, Chen Y, Covell DG, Engelhard VH, Appella E. Differential contact of disparate class I/peptide complexes as the basis for epitope cross-recognition by a single T cell receptor. J Immunol. 1997;158:3651–8. [PubMed] [Google Scholar]
- 25.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–43. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 26.Norbury CC, Basta S, Donohue, et al. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–21. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 27.Neijssen J, Herberts C, Drijfhout JW, Reits E, Janssen L, Neefjes J. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 2005;434:83–8. doi: 10.1038/nature03290. [DOI] [PubMed] [Google Scholar]