Abstract
Rotavirus infections have been implicated as a possible trigger of type 1 diabetes. We elucidated this connection by comparing peripheral blood T cell responses to rotavirus between children with newly diagnosed type 1 diabetes (n = 43), healthy children with multiple diabetes-associated autoantibodies (n = 36) and control children carrying human leukocyte antigen (HLA)-conferred susceptibility to type 1 diabetes but without autoantibodies (n = 104). Lymphocyte proliferation assays based on stimulation with an antigen were performed using freshly isolated peripheral blood mononuclear cells (PBMC) and IgG and IgA class rotavirus antibodies were measured using plasma samples collected from the children. The expression of interferon (IFN)-γ, interleukin (IL)-4, IL-10 and transforming growth factor (TGF)-β in PBMC was studied with real-time polymerase chain reaction (PCR) in a subgroup of 38 children. No differences were observed in the strength or frequency of positive T cell responses to rotavirus between children with overt diabetes, children with multiple autoantibodies and control children. Children with diabetes-associated autoantibodies had, instead, stronger T cell responses to purified coxsackie B4 virus than control children. Rotavirus-stimulated lymphocytes from autoantibody-positive children produced more IL-4 and phytohaemagglutinin (PHA)-stimulated lymphocytes more IL-4 and IFN-γ than lymphocytes from control children. PHA-stimulated lymphocytes from children with diabetes also produced more IL-4 and purified protein derivative (PPD)-stimulated lymphocytes less TGF-β than lymphocytes from autoantibody-negative control children. In conclusion, our lymphocyte proliferation studies did not provide evidence supporting an association between rotavirus infections and the development of type 1 diabetes or diabetes-associated autoantibodies in young children.
Keywords: cytokine, rotavirus, T cell proliferation, type 1 diabetes
Introduction
Type 1 diabetes (T1D) is caused by immune-mediated destruction of the pancreatic insulin-producing beta cells. Genetic background, largely polymorphism within the class II region of the human leukocyte antigen (HLA) complex, defines the susceptibility to the disease [1]. Environmental factors contribute to the penetrance of the disease, as demonstrated by the conspicuously increased incidence rate of the disease during the last decades [2,3]. Infectious factors including viruses have been implicated as possible triggers of the destructive autoimmune process, and in particular the role of enteroviruses has recently been discussed extensively [1,4].
Honeyman and coworkers [5] have reported that rotavirus infections preceded the appearance of diabetes-associated autoantibodies in an Australian prospective study. They also observed that rotavirus was able to grow in monkey pancreatic islets in vitro [6]. However, we did not find any association between the development of rotavirus antibodies and appearance of diabetes-associated autoantibodies in young children in our prospective study [7].
Rotavirus infections are documented to induce rotavirus-specific T cell and cytokine responses in children [8–11], although in our previous follow-up study T cell responses in young children declined shortly after infection. Persistent responsiveness to rotavirus measured as lymphocyte proliferation was observed in adults [11]. We have now extended our T cell proliferation studies to evaluate further the possible connection between rotavirus and T1D and analysed T cell responsiveness to rotavirus in children with T1D or T1D-associated autoantibodies. Moreover, as a part of cell-mediated immune response to rotavirus, we have evaluated the capability of rotavirus-specific T cells to produce cytokines. As T1D is a T cell-mediated disease [12], we speculated that if a link between rotavirus infections and the disease exists, cellular responsiveness to viral antigens might be altered in children with T1D-associated autoantibodies and/or T1D.
Subjects and methods
Subjects
Altogether 183 children (122 boys) taking part in the Finnish Type 1 Diabetes Prediction and Prevention (DIPP) study [13] at Turku University Hospital, Finland, were included in the study. Forty-three of them had newly diagnosed T1D [median age 8·4 years, interquartile range (IQR) 5·3–11·3 years] and an additional 36 children had developed two or more T1D-associated autoantibodies (median age 6·3 years, IQR 4·5–8·2 years). Altogether, 104 children who had not developed autoantibodies served as control subjects (median age 5·0 years, IQR 3·5–6·9 years). Eleven children with T1D, seven additional children with autoantibodies and 20 autoantibody negative control children were also studied for the expression of interferon (IFN)-γ, interleukin (IL)-4, IL-10 and transforming growth factor (TGF)-β. All children in the study carried the HLA–DR3–DQ2 (DQA1*05–DQB1*02) and/or DR4–DQ8 (DQB1*0302) haplotype, predisposing to T1D, and did not have protective DQ alleles (DQA1*0201–DQB1*02, DQB1*0301 or DQB1*0602). The children were vaccinated according to the standard Finnish vaccination protocol in which, for example, bacillus Calmette–Guérin (BCG) immunization is given to infants within the first few days after birth and diphtheria, pertussis and tetanus (DPT) vaccination at the ages of 3, 4, 5 and 20–24 months. The study was approved by the Ethics Committee of South-West Finland Health Care District and blood was drawn with the consent of all subjects and/or their guardians.
Antigens
The Wa strain (G serotype 1, P serotype 1A) of human rotavirus was propagated in fetal green monkey kidney (MA104) cells in the presence of trypsin. The Nebraska calf diarrhoea (NCD) strain (G serotype 6, P serotype 6) of bovine rotavirus and coxsackie B4 virus were propagated in rhesus monkey kidney epithelial (LLC–MK2) cells. The cells were harvested, as the virus had caused an advanced cytopathic effect. Negative control antigens were prepared identically from uninfected MA104 and LLC cells. Human rotavirus [11] and coxsackie B4 virus [14] were purified with sucrose gradient centrifugation. Purified human rotavirus (PRV) and purified coxsackie B4 virus (PCB), human rotavirus lysate (RV) and uninfected MA104 cell lysate were used at 1 µg/ml concentration and NCDV, coxsackie B4 virus lysate (CBV) and uninfected LLC cell lysate at 10 µg/ml concentration. Tetanus toxoid (TT; 1 µg/ml; National Public Health Institute, Helsinki, Finland) and purified protein derivative (PPD) (2·5 µg/ml; Staatens Seruminstitut, Copenhagen, Denmark) were used as positive control antigens. Phytohaemagglutinin (PHA; 100 µg/ml in T cell proliferation assays and 6·25 µg/ml in cytokine assays; Difco, Detroit, MI, USA) was used as a mitogen control. Cell culture medium served as negative background and control for purified antigens.
T cell proliferation assay
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood samples with Ficoll-Paque gradients (Pharmacia, Uppsala, Sweden). The freshly isolated cells were washed twice with RPMI-1640 medium supplemented with 0·01% gentamycin and resuspended in RPMI-1640 medium containing 7·5% human AB serum (Finnish Red Cross, Helsinki, Finland), 2% HEPES, 0·2% glutamine and 0·01% gentamycin. The cells (5 × 104 PBMC in a final volume of 200 µl of complete medium) were cultured with antigens in quadruplicate in 96-well round-bottomed microtitre plates (Corning Incorporated, Corning, NY, USA). The cells were incubated at 37°C in 5% CO2 for 6 days to be labelled with tritiated thymidine (2 µCi/ml, Amersham, Buckinghamshire, UK) for 18 h. The cells were harvested on glass fibre filters with Tomtec 93 Mach Manual Harvester (Tomtec, Orange, CT, USA) and the incorporated radioactivity was measured with a Micro Beta scintillation counter (Wallac, Turku, Finland). Stimulation indices (SI) were calculated by dividing median experimental counts per minute (cpm) by the median control cpm. Sis ≥ 3 were regarded as positive.
Total RNA extraction and reverse transcription
The cells (1 × 105 PBMC in a final volume of 200 µl of complete medium) were cultured with either PRV, PCB, PPD, PHA or cell culture medium only in duplicate in 96-well round-bottomed microtitre plates (Corning). After 48 h at 37°C in 5% CO2, the cells were collected into 1·5 ml Eppendorf tubes (Sarstedt, Nümbrecht, Germany) and centrifuged for 5 min at 800 g. The supernatant was extracted and the cell pellet was resuspended in 250 µl lysis solution (Applied Biosystems, Foster City, CA, USA) and frozen at −70°C. Total RNA was isolated with the ABI PRISMTM 6100 Nucleic Acid PrepStation (Applied Biosystems). Isolation was performed according to the manufacturer’s instructions, with slight modifications. The loading time in the first step of isolation was extended from 120 s to 180 s and the vacuum was set at 30%. The elution time was also extended from 120 s to 180 s. We used 500 µl, 500 µl and 250 µl wash solution 2 in the three wash steps, respectively. Extracted RNA was suspended in 150 µl elution solution (Applied Biosystems) and stored at −70°C.
The reverse transcription (RT) reaction was performed with 0·04 U pd(T)12−18 primer (Amersham, Piscataway, NJ, USA). The samples were denatured for 5 min at 70°C and cooled for 3 min on ice. RT-enzyme mixture containing 1 × Moloney murine leukemia virus (M-MLV) reverse transcriptase buffer [50 mm Tris-HCl, pH 8·3, 3 mm MgCl2, 75 mm KCl and 10 mm dithiothreitol (DTT)] (Promega, Madison, WI, USA), 0·5 mm deoxyribonucleoside triphosphate (dNTP) (Amersham), 4 U recombinant Rnasin ribonuclease inhibitor (Promega) and 40 U M-MLV reverse transcriptase Rnase H Minus (Promega) was added to the samples. Reaction was performed on a standard polymerase chain reaction (PCR) machine (PerkinElmer Cetus DNA Thermal Cycler, Norwalk, CT, USA) for 60 min at 42°C and the samples were denatured for 5 min at 90°C. The cDNA samples were stored at −70°C.
Primers and probes
The primers and probes are described in Table 1. The probe and reverse primer sequences for the detection of IFN-γ and the primers used for the production of β-actin standard have been published by Stordeur and coworkers [15], the primer sequences for the detection of β-actin and the production of IFN-γ and IL-4 standards were described in our earlier study [16] and the other sequences used were designed for this study with Primer3 software [17] according to the guidelines of Stordeur et al. [15]. The primers and probes were synthesized at Eurogentec (Seraing, Belgium) except for the primers used in the production of TGF-β standards, which were obtained from TIB Molbiol (Berlin, Germany).
Table 1.
Primers and probes.
| Target | Type1,2 | Code3 | Tm4 | Length | Sequence | Exon no. | Product size |
|---|---|---|---|---|---|---|---|
| IFN-γ | St. for | IFN-g1 | 57·7 | 29 | ATGAAATATACAAGTTATATCTTGGCTTT | 1 | 494 |
| St. rev | IFN-g2 | 69·6 | 26 | GATGCTCTTCGACCTCGAAACAGCAT | 4 | 494 | |
| For | F327 | 59·4 | 23 | CTGACTAATTATTCGGTAACTGACTTG | 3/4 | 107 | |
| Rev* | R433 | 60·4 | 19 | GCGACAGTTCAGCCATCAC | 4 | 107 | |
| Probe | P383 | 65·2 | 24 | TCCAACGCAAAGCAATACATGAAC | 4 | ||
| IL-4 | St. for | IL-4/1 | 63·6 | 21 | ATGGGTCTCACCTCCCAACTG | 1 | 614 |
| St. rev | IL-4/2 | 56·7 | 20 | TCAGCTCGAACACTTTGAATAT | 4 | 614 | |
| For | F166 | 60·0 | 20 | ATCTTTGCTGCCTCCAAGAA | 2/3 | 102 | |
| Rev | R267 | 60·3 | 19 | GCAGCGAGTGTCCTTCTCA | 3 | 102 | |
| Probe | P216 | 70·0 | 20 | TGCGACTGTGCTCCGGCAGT | 3 | ||
| TGF-β | St. for | ex6F | 65·8 | 20 | GGCTGGAAGTGGATCCACGA | 6 | 243 |
| St. rev | ex7R | 68·0 | 21 | GCAGGAGCGCACGATCATGTT | 7 | 243 | |
| For | F949 | 60·7 | 20 | GGCTACCATGCCAACTTCTG | 6 | 100 | |
| Rev | R1048 | 60·3 | 20 | CCGGGTTATGCTGGTTGTAC | 7 | 100 | |
| Probe | P987 | 68·8 | 21 | CATTTGGAGCCTGGACACGCA | 6 | ||
| IL-10 | St. for | F109 | 59·2 | 20 | CTGCCTAACATGCTTCGAGA | 1 | 494 |
| St. rev | R602 | 59·5 | 20 | CCAGATCCGATTTTGGAGAC | 5 | 494 | |
| For | F373 | 60·7 | 20 | CGCTGTCATCGATTTCTTCC | 3/4 | 88 | |
| Rev | R460 | 59·6 | 23 | TGCCTTTCTCTTGGAGCTTATTA | 4/5 | 88 | |
| Probe | P405 | 69·9 | 20 | GAGCAAGGCCGTGGAGCAGG | 4 | ||
| β-actin | St. for | F745 | 58·1 | 19 | CCCTGGAGAAGAGCTACGA | 4 | 509 |
| St. rev | R1253 | 58·2 | 20 | TAAAGCCATGCCAATCTCAT | 6 | 509 | |
| For | F926 | 63·9 | 19 | TTGCCGACAGGATGCAGAA | 5 | 76 | |
| Rev | R1001 | 62·3 | 23 | TCAGGAGGAGCAATGATCATTTGAT | 5/6 | 76 | |
| Probe* | P954 | 72·7 | 20 | TGCCCTGGCACCCAGCACAA | 5 |
St., standard; For, forward; Rev, reverse.
A slight modification of the reference sequence is indicated by (*).
Code for primers designed for this study indicates nucleotide position from the start codon (atg).
Tm, primer melting temperature, calculated with Primer 3 software using default parameters. IFN: interferon; IL: interleukin; TGF: transforming growth factor.
Real-time PCR
Either LightCycler (Roche Diagnostics, Mannheim, Germany) or Rotor-Gene 3000 (Corbett Research, Sydney, Australia) was used for analysing the samples. The PCR reaction was carried out in a total volume of 20 µl in glass capillaries (LightCycler) or in 0·1 ml plastic tubes (Rotor-Gene) containing: (1) up to 20 µl H2O; (2) 1 × QuantiTect Probe PCR Master Mix (HotStarTaq DNA polymerase, QuantiTect Probe PCR buffer, dNTP mix and ROX dye) (Qiagen, Hilden, Germany); (3) 6 µm forward and reverse primers, final concentration 300 nm (Table 1); (4) 4 µm probe (final concentration 200 nm, see Table 1); and (5) 2 µl cDNA or standard dilution. The enzyme mixture was pipetted in a separate PCR-clean room in which PCR products are not handled. The PCR protocol consisted of an initial denaturation step at 95°C for 15 min and a two-step temperature cycling consisting of 95°C for 0 s/60°C for 60 s (LightCycler) or 94°C for 5 s/60°C for 60 s (Rotor-Gene). Fluorescence was read at the end of the second step and a total of 50 cycles were performed. A standard curve was constructed from serial dilutions of purified DNA and mRNA amounts were expressed in copy numbers normalized against β-actin mRNA. In later runs with Rotor-Gene, stored parameters were used to calculate the standard curve adjusted with a single standard concentration in duplicate reactions for each run. PBMC were stimulated with PHA and total RNA was isolated for TGF-β, IL-10 and β-actin cDNA standard production. PCR reaction was carried out in a total volume of 20 µl with (1) 1 × QuantiTect SYBR Green PCR master mix (HotStarTaq DNA polymerase, QuantiTect SYBR green PCR buffer, dNTP, SYBR green I and ROX dye) (Qiagen); (2) 6 µm forward and reverse primers, final concentration 300 nM (Table 1); and (3) 5 µl cDNA template. Amplification was performed with an initial denaturation step of 95°C for 15 min followed by 45 cycles of 95°C for 15 s/58°C for 30 s/72°C for 30 s. The PCR products were run on 2% agarose gel with ethidium bromide staining and bands of appropriate size were excised with a scalper. The PCR product was then purified with the GFX DNA and gel band purification kit (Amersham) according to the manufacturer’s recommendations. The calculation of copy numbers was based on spectrophotometry measurements of PCR product concentrations. Standard DNA for IFN-γ and IL-4 were prepared similarly except for the PCR amplification step, which was performed as described in our earlier study [16]. mRNA copy number in each sample was calculated from the standard curve with instrument software and Cycle threshold value, defined as the number of cycles needed for the fluorescence signal to reach the automatically set background threshold. The Ct value was correlated inversely with the number of template nucleic acid present in the reaction.
Rotavirus antibodies
Microtitre plates (Nunc, Roskilde, Denmark) were coated by incubating them with NCD virus (1·0 µg/well) in a carbonate buffer (1·6% Na2CO3, 2·9% NaHCO3, 0·2% NaN3) at room temperature overnight. For the detection of rotavirus IgG-antibodies, plasma samples were diluted 1 : 200 in phosphate-buffered saline (PBS) + 1% bovine serum albumin (BSA) + 0·05% Tween 20 and incubated for 2 h at + 37°C. The plates were washed and peroxidase-conjugated anti-human IgG antibody (Dako, Copenhagen, Denmark) was added in a 1 : 1000 dilution. After a 1-h incubation, O-phenylenediamine and H2O2 in citrate-Na2HPO4 buffer were added. For the detection of rotavirus-specific IgA antibodies, the virus-coated plates were first residual-coated with PBS + 1% BSA. Plasma samples were diluted 1 : 10 in PBS + 0·2% BSA + 0·05% Tween 20 and incubated on the plates for 2 h at + 37°C. The plates were washed and biotinylated anti-human IgA antibody (Vector Laboratories, Burlingame, CA, USA) was added in a 1 : 100 dilution. After a 1·5 h incubation and washes, alkaline phosphatase–streptavidin (Zymed, San Francisco, CA, USA) was used as a secondary conjugate in a 1 : 1000 dilution. The plates were incubated for 1 h, washed and P-nitrophenyl phosphate substrate tablets (Sigma, St Louis, MO, USA) were added in a carbonate buffer (0·05 m NaHCO3, 0·05 m Na2CO3, 0·02% MgCl2, 0·02% NaN3). The colour intensities were measured with spectrophotometer. Each specimen was tested in duplicate. Positive and negative standards were included on each plate in four different concentrations. A ≥threefold absorbance compared to negative control specimen exceeding the cut-off level of seropositivity, 0·150 optical density units, was considered positive.
Autoantibodies
Islet cell antibodies (ICA), GAD antibodies (GADA) and IA-2 antibodies (IA-2 A) were measured as described previously [18,19]. Insulin autoantibodies (IAA) were analysed with a slightly modified method of Williams et al. [20]. The threshold for detection of ICA was 2·5 Juvenile Diabetes Foundation units (JDFU). The sensitivity for our ICA assay was 100% and the specificity 98% in the fourth round of the International Workshop on the Standardization of the ICA assay. The detection limits for positivity for IAA, GADA and IA-2 A were set at the 99th percentile [1·56 relative units (RU) for IAA, 5·36 RU for GADA and 0·43 RU for IA-2 A] in more than 370 non-diabetic Finnish children. The disease sensitivity for our IAA assay was 44%, GADA assay 82% and IA-2 A assay 62% and the disease specificity was 100%, 98% and 100%, respectively, in the 2002 Centers for Disease Control and Prevention-sponsored Diabetes Autoantibody Standardization Program Workshop.
Statistics
Linear regression method was selected to compare T cell proliferation between groups. Age and HLA genotypes (HLA–DR4–DQ8/x or HLA–DR4–DQ8/DR3–DQ2) were included in the analysis as potential confounding factors. Due to skewed distributions, a logarithmic transformation was conducted for stimulation indices (SIs) to TT, PCB and CBV and 1/x transformation for SIs to rotavirus antigens. The Mann–Whitney U-test was used to compare cytokine production between groups. Frequencies of positive T cell responses between two groups were assessed with Fisher’s exact test. Correlations between different parameters were calculated with Spearman’s rank correlation. In correlation analysis, the SIs less than 1 were regarded as 1. Statistical analysis was conducted with the sas system for Windows, release 9·1 (SAS Institute, Cary, NC, USA). The level of significance was set at 0·05.
Results
Lymphocyte proliferation and rotavirus antibodies
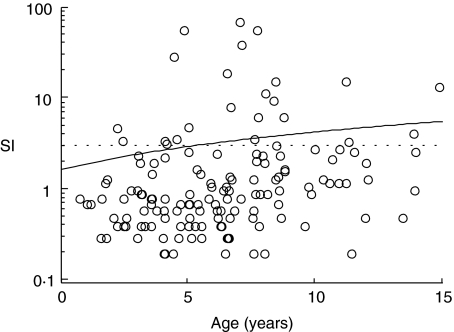
As immunization with BCG and tetanus toxoid (TT) are both included in the routine Finnish vaccination schedule, the lack of an apparent (SI ≥ 3) response to either PPD or TT was considered a probable indicator of a poor condition of the cells and those few samples were excluded from the analysis. The proliferation induced by uninfected MA-104 and LLC cell lysates was similar to background proliferation levels in all groups of children (median SI 1·4 and 1·3 in children with T1D, 1·5 and 1·2 in children with autoantibodies and 1·3 and 1·0 in control children, for MA-104 and LLC, respectively). A significant correlation was observed between age and T cell responses to human RV (r = 0·32 P < 0·0001), bovine NCDV, r = 0·20, P = 0·001), PCB, r = 0·32, P < 0·0001), PPD (r = 0·19, P = 0·01) and a borderline correlation in response to TT (r = 0·15, P = 0·04) (Fig. 1).
Fig. 1.
T cell responses to human rotavirus lysate expressed as stimulation indices (SIs) showed a moderate correlation with age (r = 0·32, P < 0·0001). The dotted line was drawn at SI ≥ 3 as the cut-off limit of a positive response.
Altogether 130 of 180 (72%) children from whom information on rotavirus antibodies was available had IgA and/or IgG antibodies to rotavirus. Eighty-four (65%) of them had both IgA and IgG antibodies, nine (7%) children IgA antibodies only and 37 (28%) children IgG antibodies only. Information on T cell responsiveness to at least two rotavirus antigens was available from 156 children, 54 of whom (35%) had a positive (SI ≥ 3) T cell response to at least one of the rotavirus antigens. In 153 children from whom information on both serological and cellular responsiveness to rotavirus was available, children with rotavirus antibodies had more frequently positive T cell responses to rotavirus (50 of 112, 45%) than children without antibodies to rotavirus (four of 41, 10%) (P < 0·0001). Further, cell responses to all rotavirus antigens were stronger in children with rotavirus antibodies than in children without these antibodies (P = 0·010, P = 0·0031 and P < 0·0001 for PRV, RV and NCDV, respectively) (Table 2). The proportion of children having antibodies to rotavirus was similar in children with T1D (33 of 43, 77%) or T1D-associated autoantibodies (35 of 44, 80%) and control children negative for autoantibodies (68 of 101, 67%) (P = 0·32 and P = 0·17, for children with T1D and T1D-associated autoantibodies and control children, respectively). The frequencies of positive T cell responses to any of the rotavirus or coxsackie B4 virus antigens did not differ between children who had or had not developed T1D-associated autoantibodies (Table 3). Children with clinical T1D had positive responses to coxsackie B4 virus more frequently and they also tended to have responses to TT and rotavirus antigens more frequently than control children. This may be due, however, to age-related changes in cellular responsiveness to these antigens as the children with T1D (median age 8·4 years) were older than the control children (median age 5·0 years).
Table 2.
T cell responsiveness to purified protein derivative (PPD), tetanus toxoid (TT), purified human rotavirus (PRV), human rotavirus lysate (RV) and Nebraska calf diarrhoea virus (NCDV) in children who had or had not IgG and/or IgA antibodies to rotavirus.
| Rotavirus antibody positive children | Rotavirus antibody negative children | ||||||
|---|---|---|---|---|---|---|---|
| Median | n | Interquartile range | Median | n | Interquartile range | P | |
| PPD | 96·9 | 130 | 35·5–195·7 | 82·7 | 50 | 23·3–181·0 | 0·77 |
| TT | 9·1 | 130 | 2·1–31·0 | 9·0 | 50 | 1·7–35·5 | 0·84 |
| PRV | 0·9 | 129 | 0·5–2·0 | 0·7 | 50 | 0·3–1·0 | 0·010 |
| RV | 1·0 | 112 | 0·5–2·6 | 0·7 | 41 | 0·4–0·9 | 0·0031 |
| NCDV | 1·1 | 112 | 0·6–4·4 | 0·6 | 41 | 0·4–0·9 | < 0·0001 |
Table 3.
The proportions of children having a positive T cell response (stimulation index ≥ 3) to at least one of the rotavirus antigens (purified human rotavirus, human rotavirus lysate, bovine Nebraska calf diarrhoea virus), coxsackie B4 virus antigens (purified and lysate coxsackie B4 virus) and tetanus toxoid in children with newly diagnosed type 1 diabetes, children with diabetes-associated autoantibodies and control children.
| Children with diabetes | Control children | P | Children with autoantibodies | Control children | P | |
|---|---|---|---|---|---|---|
| Rotavirus | 18/38 (47%) | 26/84 (31%) | 0·10 | 15/42 (36%) | 26/84 (31%) | 0·69 |
| Coxsackie B4 virus | 27/38 (71%) | 41/84 (49%) | 0·030 | 26/42 (62%) | 41/84 (49%) | 0·18 |
| Tetanus toxoid | 35/43 (81%) | 67/104 (64%) | 0·050 | 34/44 (77%) | 67/104 (64%) | 0·19 |
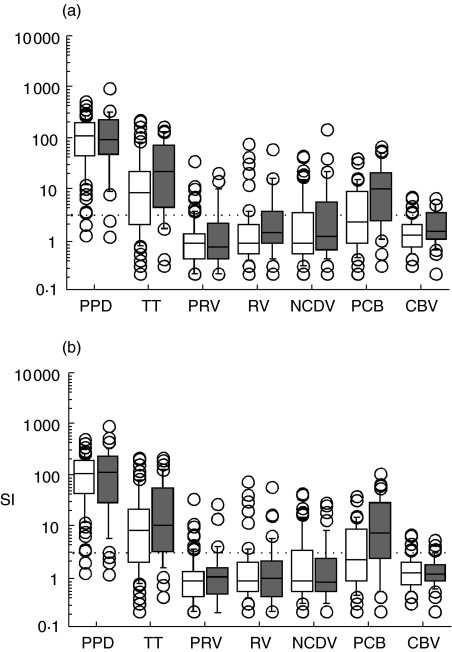
All children carrying either HLA–DR4–DQ8/x (21 of 30 children with T1D, 26 of 44 children with T1D-associated autoantibodies and 43 of 77 control children) or HLA–DR4–DQ8/DR3–DQ2 genotype (nine of 30 children with T1D, 18 of 44 children with T1D-associated autoantibodies and 34 of 77 control children) were included in linear regression analysis. No difference was observed in T cell responses to rotavirus antigens between children who had developed T1D or T1D-associated autoantibodies and autoantibody negative control children (Fig. 2), nor did responsiveness to TT differ between these groups. Responsiveness to PPD was slightly stronger in children with T1D-associated autoantibodies than in control children (P = 0·026), but this was due to differences in the ages of children in these groups (P = 0·015). Children with autoantibodies also had stronger T cell responses to PCB than autoantibody negative control children (P = 0·0003). Autoantibody status (P = 0·033) and age (P = 0·001) were observed to affect cellular responsiveness to PCB. In children with T1D, a similar tendency was observed (P = 0·001; P = 0·058 and P = 0·007 for T1D and age, respectively). Further, the children with T1D had higher responsiveness to CBV lysate antigen (P = 0·013), but this was due to differences in ages and HLA genotypes between the groups (P = 0·028 and P = 0·039, for age and HLA genotype, respectively). Cell-mediated immunity to rotavirus was also analysed separately in those children who, based on the presence of specific antibodies in their sera, had experienced rotavirus infections. This analysis did not either reveal any differences in cellular responsiveness to rotavirus between children with T1D-associated autoantibodies or T1D compared to autoantibody negative control children (data not shown).
Fig. 2.
T cell responses shown as stimulation indices (SIs) to a panel of antigens [purified protein derivative (PPD), tetanus toxoid (TT); purified human rotavirus (PRV); human rotavirus lysate (RV); Nebraska calf diarrhoea virus (NCDV); purified coxsackie B4 virus (PCB); lysate coxsackie B4 virus (CBV)] in children with diabetes-associated autoantibodies (a) and in children with type 1 diabetes (b). T cell responses to PCB were stronger in children with clinical diabetes and autoantibodies (grey boxes, P = 0·0012 and P = 0·0003, respectively) than in control children (white boxes) as well as responses to CBV in children with T1D (P = 0·013) and responses to PPD in children with diabetes-associated autoantibodies (P = 0·026). Median value in each box is shown with a horizontal line in the box. The box plots delineate values between the 25th and 75th percentiles and the whiskers values between the 10th and the 90th percentiles. The values outside this range are indicated with circles.
Cytokine production
We also wanted to assess cytokine responsiveness in association of T cell responses to rotavirus. The production of IL-4, IFN-γ, TGF-β and IL-10 was studied in non-stimulated and PHA-, PPD-, PRV- and PCB-stimulated PBMC from 11 children with T1D, seven additional children with T1D-associated autoantibodies and in 20 control children. In PRV-stimulated cells, a correlation was found between the age of the child and the production of IL-4 and IFN-γ (data not shown).
The production of IFN-γ and IL-4 correlated with the lymphocyte proliferation response to the same antigen in PBMC stimulated with PPD (r = 0·69, P < 0·0001 and r = 0·54, P = 0·0005 for IFN-γ and IL-4, respectively), PRV (r = 0·48, P = 0·003 and r = 0·46, P = 0·004, respectively) and PCB (r = 0·61, P < 0·0001 and r = 0·38, P = 0·019, respectively). The production of TGF-β did not correlate with proliferation responses to any of the studied antigens, but the production of IL-10 in PRV- and PCB-stimulated lymphocytes correlated with T cell responses to these antigens (r = 0·36, P = 0·026 and r = 0·39, P = 0·015, respectively). A trend towards a higher production of IFN-γ was observed in PRV-stimulated PBMC from seropositive children compared to children without rotavirus antibodies (P = 0·084).
In children with T1D-associated autoantibodies, IL-4 expression in PRV-stimulated and IFN-γ and IL-4 expression in PHA-stimulated PBMC were higher than in control children (Table 4). When cytokine production in PRV-stimulated cells was studied in rotavirus seropositive children only, no difference was observed in between children with clinical T1D or T1D-associated autoantibodies and control children, due possibly to the small amount of children in each group. In children with T1D, PHA-stimulated PBMC also expressed more IL-4 and PPD-stimulated cells less TGF-β than in autoantibody negative control subjects (Table 5). No difference was seen in the relative amount of cytokine gene expression in stimulated versus non-stimulated lymphocytes between healthy children with and without autoantibodies. The relative amount of TGF-β in PPD-stimulated versus non-stimulated cultures was lower in children with T1D than in autoantibody negative control children (P = 0·011).
Table 4.
Median cytokine production of peripheral blood mononuclear cells from control children and children with diabetes-associated autoantibodies.
| Children with autoantibodies | Control children | ||||||
|---|---|---|---|---|---|---|---|
| Median | n | Interquartile range | Median | n | Interquartile range | P | |
| IFN-γ | |||||||
| Medium | 6·4 | 10 | 0·0–18·6 | 2·9 | 20 | 0·0–9·0 | 0·44 |
| PHA | 2307·3 | 10 | 588·2–7822·1 | 709·2 | 20 | 177·1–1715·2 | 0·028 |
| PPD | 592·9 | 10 | 452·2–1168·9 | 1027·3 | 20 | 120·2–1743·4 | 0·71 |
| PRV | 126·8 | 10 | 35·0–608·0 | 22·8 | 20 | 7·0–134·0 | 0·18 |
| PCB | 1885·6 | 10 | 64·3–4859·6 | 672·9 | 20 | 48·1–2491·0 | 0·55 |
| IL-4 | |||||||
| Medium | 1·2 | 10 | 0·0–2·2 | 0·0 | 20 | 0·0–1·7 | 0·21 |
| PHA | 38·6 | 10 | 11·5–58·3 | 14·6 | 20 | 3·4–28·6 | 0·039 |
| PPD | 2·2 | 10 | 0·0–6·6 | 6·8 | 20 | 0·0–13·1 | 0·17 |
| PRV | 3·2 | 10 | 0·0–11·0 | 0·0 | 20 | 0·0–1·2 | 0·037 |
| PCB | 1·4 | 10 | 0·0–8·4 | 0·0 | 19 | 0·0–8·6 | 0·82 |
| IL-10 | |||||||
| Medium | 60·6 | 10 | 15·2–167·5 | 15·9 | 20 | 5·9–53·3 | 0·19 |
| PHA | 11·5 | 10 | 4·3–40·0 | 8·5 | 20 | 2·5–23·5 | 0·29 |
| PPD | 14·3 | 10 | 6·0–17·1 | 14·0 | 20 | 7·3–24·6 | 0·79 |
| PRV | 143·0 | 10 | 112·2–201·8 | 101·5 | 20 | 62·8–164·0 | 0·31 |
| PCB | 121·1 | 10 | 29·3–221·3 | 43·1 | 20 | 22·4–177·7 | 0·36 |
| TGF-β | |||||||
| Medium | 29·2 | 10 | 18·8–58·0 | 21·1 | 20 | 16·1–40·6 | 0·43 |
| PHA | 17·3 | 10 | 5·2–23·6 | 11·6 | 20 | 5·2–19·9 | 0·72 |
| PPD | 21·4 | 10 | 13·3–24·7 | 31·2 | 20 | 20·3–44·3 | 0·095 |
| PRV | 36·4 | 10 | 22·0–67·8 | 35·0 | 20 | 30·0–49·3 | 0·57 |
| PCB | 30·4 | 10 | 21·8–47·0 | 21·9 | 20 | 13·5–42·8 | 0·25 |
IFN: interferon; IL: interleukin; TGF: transforming growth factor.
Table 5.
Median cytokine production of peripheral blood mononuclear cells from control children and children with type 1 diabetes.
| Children with diabetes | Control children | ||||||
|---|---|---|---|---|---|---|---|
| Median | n | Interquartile range | Median | n | Interquartile range | P | |
| IFN-γ | |||||||
| Medium | 3·4 | 11 | 0·0–24·9 | 2·9 | 20 | 0·0–9·0 | 0·76 |
| PHA | 588·2 | 11 | 342·2–5165·9 | 709·2 | 20 | 177·1–1715·2 | 0·32 |
| PPD | 717·4 | 10 | 530·0–3495·1 | 1027·3 | 20 | 120·2–1743·4 | 0·84 |
| PRV | 67·2 | 11 | 9·4–123·4 | 22·8 | 20 | 7·0–134·0 | 0·56 |
| PCB | 2081·2 | 11 | 232·7–11285·1 | 672·9 | 20 | 48·1–2491·0 | 0·19 |
| IL-4 | |||||||
| Medium | 0·9 | 11 | 0·0–4·8 | 0·0 | 20 | 0·0–1·7 | 0·24 |
| PHA | 58·3 | 11 | 19·1–97·8 | 14·6 | 20 | 3·4–28·6 | 0·0064 |
| PPD | 5·9 | 10 | 2·7–16·7 | 6·8 | 20 | 0·0–13·1 | 0·96 |
| PRV | 0·0 | 11 | 0·0–11·0 | 0·0 | 20 | 0·0–1·2 | 0·15 |
| PCB | 0·0 | 11 | 0·0–19·8 | 0·0 | 19 | 0·0–8·6 | 0·76 |
| IL-10 | |||||||
| Medium | 48·1 | 11 | 20·2–80·2 | 15·9 | 20 | 5·9–53·3 | 0·099 |
| PHA | 8·7 | 11 | 4·9–22·8 | 8·5 | 20 | 2·5–23·5 | 0·54 |
| PPD | 9·2 | 10 | 6·3–17·1 | 14·0 | 20 | 7·3–24·6 | 0·63 |
| PRV | 180·9 | 11 | 80·4–260·3 | 101·5 | 20 | 62·8–164·0 | 0·12 |
| PCB | 144·6 | 11 | 50·6–278·4 | 43·1 | 20 | 22·4–177·7 | 0·19 |
| TGF-β | |||||||
| Medium | 24·1 | 11 | 18·8–89·3 | 21·1 | 20 | 16·1–40·6 | 0·17 |
| PHA | 29·5 | 11 | 12·9–63·3 | 11·6 | 20 | 5·2–19·9 | 0·060 |
| PPD | 20·3 | 10 | 14·5–26·0 | 31·2 | 20 | 20·3–44·3 | 0·025 |
| PRV | 28·8 | 11 | 24·2–49·4 | 35·0 | 20 | 30·0–49·3 | 0·48 |
| PCB | 21·8 | 11 | 17·9–30·4 | 21·9 | 20 | 13·5–42·8 | 0·59 |
IFN: interferon; IL: interleukin; TGF: transforming growth factor.
Discussion
We did not find any association between rotavirus-specific T cell responses and the presence of T1D-associated autoantibodies or T1D, nor did cytokine responses in rotavirus-stimulated PBMC considerably differ between children with overt T1D, children with autoantibodies or control children. Our results are concordant with those of Jones and Crosby [21], who studied T cell responses to the 69m strain of serotype G8 rotavirus in newly diagnosed T1D patients and control subjects and found strong rotavirus-specific T cell responses in six of 26 diabetics and four of 24 control subjects. The present observations are also consistent with our previous study, in which we did not find any association between serologically diagnosed rotavirus infections and the appearance of T1D-associated autoantibodies in a cohort of 177 children at increased genetic risk for T1D [7]. Twenty-nine children developed multiple T1D-associated autoantibodies during the follow-up, but the appearance of autoantibodies did not occur simultaneously with rotavirus infections and the frequency of infections was similar to that seen in matched control children. Controversially, in a prospective study with 41 Australian children at increased genetic risk to develop T1D, 86% of the increases in IA-2 A, 62% of the increases in IAA and 50% of the increases in GADA were temporally associated with rotavirus antibody seroconversions [5]. The differences between these studies may be due to real differences between populations or predominating virus strains. Also different virus strains used in these studies or methodological differences in interpreting antibody data may affect these results.
Shared epitopes between autoantigens and viruses or a deficient response related to a persistent infection has been suggested to be possible mechanisms for the association between some viral infections and T1D [22]. Honeyman et al. have observed homology between VP7 protein of the P strain of serotype G3 rotavirus and epitope peptides in the T1D-associated autoantigens GAD65 and IA-2 [23]. They reported 75% identity and 92% similarity between GAD65 and VP7 peptide containing amino acids (aa) 17–28 (ILLNYVLKSLTR) and 56% identity and 100% similarity between IA-2 and VP7 peptide containing aa 40–50 (IIVILSPLLNA). In the cases of molecular mimicry between rotavirus and β-cell antigens or an increased number of rotavirus infections, one might expect an increased level of cellular responsiveness to rotavirus. Alternatively, a chronic infection might also be established due to the lack of specific immune responsiveness. Accordingly, we also analysed T cell responses to rotavirus separately in children seropositive for rotavirus IgG and/or IgA antibodies to detect whether they exhibited abnormal responsiveness to rotavirus. However, we did not find any differences in proliferation responses between children with T1D or T1D-associated autoantibodies and control children in these comparisons. In the present study, we have used G1 and G6 serotypes of rotavirus because G1 is the most prevalent serotype in Finland [24] as well as in the whole of Europe, and here serotype G3 represents only a few per cent of rotavirus infections [25]. Of the two peptides in VP7 protein that Honeyman et al. observed to resemble T1D-associated autoantigens, the aa 17–28 sequence of G1 serotype rotavirus contains two different amino acids than G3 serotype. More variation is present in the aa 40–50 sequence, where five amino acids are substituted by different amino acids. The significance of these differences is uncertain, as it has been shown that direct homology between amino acid stretches is not needed in molecular mimicry [26]. Consequently, a few amino acids binding to critical sites in the HLA molecule may be enough to induce a cross-reactive immune response [27].
The ages of children in this study varied from 3·5 to 11·3 years. Naturally, the frequency of positive T cell or antibody responses to viral antigens increases with age. Linear regression models appeared to be the most powerful way to analyse this type of data as age could be included in these analyses as explanatory variables. However, the use of linear regression analysis was not possible in all analysis settings, e.g. when frequencies of positive responses were compared between groups of children. Age differences between the children may thus affect some results, but the effect of age was excluded in the main analyses comparing T cell responsiveness between children with and without T1D and/or T1D-associated autoantibodies.
Coxsackie B4 virus antigens served as a reference in our study as this virus has often been connected to T1D and β cell autoimmunity. In our previous studies, we found increased T cell responsiveness to CBV in children positive for ICA and/or GADA and in T1D patients within 4–72 months from diagnosis, but not in patients with newly diagnosed T1D [28,29]. Jones and Crosby [21], in contrast, found strong T cell responses to a lysate coxsackie B4 virus antigen in newly diagnosed T1D patients. In the present study, we observed stronger cellular responsiveness to PCB but not to CBV in children with T1D-associated autoantibodies compared to autoantibody negative control children. In this study, coxsackie B4 virus lysate antigen was a remarkably poor inducer of any T cell responses. This may reflect technical problems with this particular antigen as cell lysate-type antigens often show inhibitory effects at higher concentrations.
In cytokine expression studies with PRV-stimulated PBMC from children with autoantibodies, the expression of IL-4 was higher than in control children. In PHA-stimulated PBMC, we found higher IL-4 expression in children with clinical T1D and T1D-associated autoantibodies and higher IFN-γ expression in children with autoantibodies than in control children. We also observed lower TGF-β expression in PPD-stimulated PBMC from children with T1D than in control children. We used several stimulators to measure four different cytokines and, as P-values lose their significance if multiplied by the number of comparisons, the differences found may represent chance effects. However, in our earlier study we consistently observed increased mRNA expression of both IL-4 and IFN-γ in PHA-stimulated PBMC in 22 children with newly diagnosed T1D compared to 20 healthy control children [30]. Kallman and coworkers found higher titres of IFN-γ but not IL-4 in PHA-stimulated PBMC from 53 newly diagnosed T1D patients than in 56 control subjects [31]. Kretowski and coworkers reported lower titres of TGF-β1 in PHA-stimulated PBMC from 22 newly diagnosed T1D patients and their 24 first-degree relatives than in 18 control subjects [32]. Controversial observations have been reported by Berman et al. [33] and Lohmann et al. [34], who observed decreased titres of IL-4 and no difference or decreased production of IFN-γ in children with newly diagnosed T1D compared to age-matched control children. These controversial observations may also be related to different methodologies used, as mRNA expression may not be directly comparable with cytokine concentrations measured with enzyme-linked immunosorbent assay (ELISA). Also the length of in vitro stimulation with PHA [35] and the duration of T1D might affect cytokine production pattern.
In conclusion, our cellular immunity studies did not provide any evidence supporting an association between rotavirus infections and T1D or the presence of T1D-associated autoantibodies in young children.
Acknowledgments
We thank Ms Anne Suominen and Mrs Terttu Lauren for their skilful technical assistance and Tero Vahlberg MSc for the help with statistical analysis of the data. We also thank the whole study personnel dedicating their time to the best of the study children and families and, above all, the families participating in the Type 1 Diabetes Prediction and Prevention project. This work was supported by the Juvenile Diabetes Research Foundation (grants 4-1999-731 and 4-2001-435), Sigrid Juselius Foundation, Academy of Finland, Finnish Foundation for Diabetes Research, the Finnish Foundation for Research on Viral Diseases and Jalmari and Rauha Ahokas Foundation. Miia Mäkelä is a student of the National Graduate School of Clinical Investigation (CLIGS).
References
- 1.Gianani R, Eisenbarth GS. The stages of type 1A diabetes: 2005. Immunol Rev. 2005;204:232–49. doi: 10.1111/j.0105-2896.2005.00248.x. [DOI] [PubMed] [Google Scholar]
- 2.Onkamo P, Väänänen S, Karvonen M, Tuomilehto J. Worldwide increase in incidence of Type I diabetes − the analysis of the data on published incidence trends. Diabetologia. 1999;42:1395–403. doi: 10.1007/s001250051309. [DOI] [PubMed] [Google Scholar]
- 3.Hermann R, Knip M, Veijola R, et al. Temporal changes in the frequencies of HLA genotypes in patients with Type 1 diabetes − indication of an increased environmental pressure? Diabetologia. 2003;46:420–5. doi: 10.1007/s00125-003-1045-4. [DOI] [PubMed] [Google Scholar]
- 4.Lammi N, Karvonen M, Tuomilehto J. Do microbes have a causal role in type 1 diabetes? Med Sci Monit. 2005;11:RA63–9. [PubMed] [Google Scholar]
- 5.Honeyman MC, Coulson BS, Stone NL, et al. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes. 2000;49:1319–24. doi: 10.2337/diabetes.49.8.1319. [DOI] [PubMed] [Google Scholar]
- 6.Coulson BS, Witterick PD, Tan Y, et al. Growth of rotaviruses in primary pancreatic cells. J Virol. 2002;76:537–44. doi: 10.1128/JVI.76.18.9537-9544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomqvist M, Juhela S, Erkkilä S, et al. Rotavirus infections and development of diabetes-associated autoantibodies during the first 2 years of life. Clin Exp Immunol. 2002;128:511–15. doi: 10.1046/j.1365-2249.2002.01842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Offit PA, Hoffenberg EJ, Santos N, Gouvea V. Rotavirus-specific humoral and cellular immune response after primary, symptomatic infection. J Infect Dis. 1993;167:1436–40. doi: 10.1093/infdis/167.6.1436. [DOI] [PubMed] [Google Scholar]
- 9.Jaimes MC, Rojas OL, Gonzalez AM, et al. Frequencies of virus-specific CD4(+) and CD8(+) T lymphocytes secreting gamma interferon after acute natural rotavirus infection in children and adults. J Virol. 2002;76:4741–9. doi: 10.1128/JVI.76.10.4741-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas OL, Gonzalez AM, Gonzalez R, et al. Human rotavirus specific T cells: quantification by ELISPOT and expression of homing receptors on CD4+ T cells. Virology. 2003;314:671–9. doi: 10.1016/s0042-6822(03)00507-5. [DOI] [PubMed] [Google Scholar]
- 11.Mäkelä M, Marttila J, Simell O, Ilonen J. Rotavirus-specific T-cell responses in young prospectively followed-up children. Clin Exp Immunol. 2004;137:173–8. doi: 10.1111/j.1365-2249.2004.02509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roep BO. The role of T-cells in the pathogenesis of Type 1 diabetes: from cause to cure. Diabetologia. 2003;46:305–21. doi: 10.1007/s00125-003-1089-5. [DOI] [PubMed] [Google Scholar]
- 13.Kupila A, Muona P, Simell T, et al. Feasibility of genetic and immunological prediction of type I diabetes in a population-based birth cohort. Diabetologia. 2001;44:290–7. doi: 10.1007/s001250051616. [DOI] [PubMed] [Google Scholar]
- 14.Abraham G, Colonno RJ. Many rhinovirus serotypes share the same cellular receptor. J Virol. 1984;51:340–5. doi: 10.1128/jvi.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stordeur P, Poulin LF, Craciun L, et al. Cytokine mRNA quantification by real-time PCR. J Immunol Meth. 2002;259:55–64. doi: 10.1016/s0022-1759(01)00489-6. [DOI] [PubMed] [Google Scholar]
- 16.Halminen M, Sjöroos M, Mäkelä MJ, et al. Simultaneous detection of IFN-gamma and IL-4 mRNAs using RT–PCR and time-resolved fluorometry. Cytokine. 1999;11:87–93. doi: 10.1006/cyto.1998.0392. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Meth Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 18.Savola K, Sabbah E, Kulmala P, Vähäsalo P, Ilonen J, Knip M. Autoantibodies associated with Type I diabetes mellitus persist after diagnosis in children. Diabetologia. 1998;41:1293–7. doi: 10.1007/s001250051067. [DOI] [PubMed] [Google Scholar]
- 19.Savola K, Bonifacio E, Sabbah E, et al. IA-2 antibodies − a sensitive marker of IDDM with clinical onset in childhood and adolescence. Diabetologia. 1998;41:424–9. doi: 10.1007/s001250050925. Childhood Diabetes Finland Study Group. [DOI] [PubMed] [Google Scholar]
- 20.Williams AJ, Bingley P, Bonifacio E, Palmer JP, Gale EA. A novel micro-assay for insulin autoantibodies. J Autoimmun. 1997;10:473–8. doi: 10.1006/jaut.1997.0154. [DOI] [PubMed] [Google Scholar]
- 21.Jones DB, Crosby I. Proliferative lymphocyte responses to virus antigens homologous to GAD65 in IDDM. Diabetologia. 1996;39:1318–24. doi: 10.1007/s001250050576. [DOI] [PubMed] [Google Scholar]
- 22.Hyöty H. Enterovirus infections and type 1 diabetes. Ann Med. 2002;34:138–47. [PubMed] [Google Scholar]
- 23.Honeyman MC, Stone NL, Harrison LC. T-cell epitopes in type 1 diabetes autoantigen tyrosine phosphatase IA-2: potential for mimicry with rotavirus and other environmental agents. Mol Med. 1998;4:231–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Maunula L, von Bonsdorff CH. Frequent reassortments may explain the genetic heterogeneity of rotaviruses: analysis of Finnish rotavirus strains. J Virol. 2002;76:11793–800. doi: 10.1128/JVI.76.23.11793-11800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 26.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maverakis E, van den Elzen P, Sercarz EE. Self-reactive T cells and degeneracy of T cell recognition: evolving concepts-from sequence homology to shape mimicry and TCR flexibility. J Autoimmun. 2001;16:201–9. doi: 10.1006/jaut.2000.0493. [DOI] [PubMed] [Google Scholar]
- 28.Juhela S, Hyöty H, Hinkkanen A, et al. T cell responses to enterovirus antigens and to beta-cell autoantigens in unaffected children positive for IDDM-associated autoantibodies. J Autoimmun. 1999;12:269–78. doi: 10.1006/jaut.1999.0276. [DOI] [PubMed] [Google Scholar]
- 29.Juhela S, Hyöty H, Roivainen M, et al. T-cell responses to enterovirus antigens in children with type 1 diabetes. Diabetes. 2000;49:1308–13. doi: 10.2337/diabetes.49.8.1308. [DOI] [PubMed] [Google Scholar]
- 30.Halminen M, Juhela S, Vaarala O, Simell O, Ilonen J. Induction of interferon-gamma and IL-4 production by mitogen and specific antigens in peripheral blood lymphocytes of Type 1 diabetes patients. Autoimmunity. 2001;34:1–8. doi: 10.3109/08916930108994120. [DOI] [PubMed] [Google Scholar]
- 31.Kallmann BA, Huther M, Tubes M, et al. Systemic bias of cytokine production toward cell-mediated immune regulation in IDDM and toward humoral immunity in Graves’ disease. Diabetes. 1997;46:237–43. doi: 10.2337/diab.46.2.237. [DOI] [PubMed] [Google Scholar]
- 32.Kretowski A, Maciej K, Ida K. The analysis of in vitro transforming growth factor-beta1 (TGF-beta1) production by peripheral blood in overt and pre-clinical type 1 diabetes mellitus. Immunol Lett. 2000;71:85–9. doi: 10.1016/s0165-2478(99)00174-1. [DOI] [PubMed] [Google Scholar]
- 33.Berman MA, Sandborg CI, Wang Z, et al. Decreased IL-4 production in new onset type I insulin-dependent diabetes mellitus. J Immunol. 1996;157:4690–6. [PubMed] [Google Scholar]
- 34.Lohmann T, Laue S, Nietzschmann U, et al. Reduced expression of Th1-associated chemokine receptors on peripheral blood lymphocytes at diagnosis of type 1 diabetes. Diabetes. 2002;51:2474–80. doi: 10.2337/diabetes.51.8.2474. [DOI] [PubMed] [Google Scholar]
- 35.Rapoport MJ, Mor A, Vardi P, et al. Decreased secretion of Th2 cytokines precedes Up-regulated and delayed secretion of Th1 cytokines in activated peripheral blood mononuclear cells from patients with insulin-dependent diabetes mellitus. J Autoimmun. 1998;11:635–42. doi: 10.1006/jaut.1998.0240. [DOI] [PubMed] [Google Scholar]