Abstract
The α-helix is a key structural element in a wide range of peptides and proteins. We report here the design, synthesis, and characterization of a modified peptide in which the helix content can be reversibly photoregulated. The peptide contains two cysteine residues that are cross-linked by an azobenzene derivative in an intramolecular fashion. In accordance with the design, the photoisomerization of the azobenzene cross-linker from the trans to the cis form causes a large increase in the helix content of the peptide, in water.
The α-helix is the classic element of secondary structure in proteins. First described by Pauling and Corey (1), it has been found in proteins performing every sort of biological function. Considerable effort has been expended in understanding the factors that determine α-helix formation and stability (e.g., refs. 2–4 and references therein). Numerous strategies for controlling helix stability have also been developed, including helix-initiator templates and intramolecular peptide cross-links (e.g., refs. 5–9).
The introduction of a photoisomerizable moiety offers the further possibility of reversibly controlling helix stability with light. Photo-control of an α-helix could potentially be used to control the activity of the protein of which it formed part. By analogy with “caged” compounds (10–12), such photo-controlled proteins could be powerful biological tools; they would permit one to probe complex biochemical pathways in situ in a spatially and temporally defined manner. In addition, photo-initiation of helix formation might provide a general way for triggering protein folding. A rapid photochemical trigger could facilitate the study of early events in protein folding reactions (e.g., refs. 13 and 14).
Incorporation of photoisomerizable groups into enzymes has been reported to photomodulate activity in several instances (15). However, only recently has site-specific modification been achieved (16–18), and the conformational consequences of these modifications are in most cases unknown. Chmielewski and coworkers (19) examined the conformational effects of incorporation of a photoisomerizable azobenzene linkage into the backbone of a cyclic β-turn peptide. Moroder and colleagues (20, 21) recently reported a photo-controlled switch between a β-turn conformation and family of less well ordered conformations with another cyclic peptide in DMSO solution. Cerpa et al. (22) reported that a short peptide bearing a single azobenzene side-chain underwent a photo-induced switch from a β-sheet to an α-helical conformation in water. However, the conformational change was not reversible.
In view of the central importance of the α-helix for protein structure and function, we wished to explore the design of a system in which helix formation could be controlled by light. A number of groups have examined the effects of the incorporation of multiple photo-isomerizable groups on the physical properties of helical polymers and polyamino acids (refs. 23–26 and references therein). Conformational changes and shifts in aggregation state have sometimes been observed with these systems with specific solvent mixtures. The effects of photoisomerization have been attributed to changes in polarity of the pendant isomerizable groups that affect the interaction of these groups with solvent or with other parts of the polymer (26, 27). We wished to focus on designs that would operate in water and that were sufficiently simple that the mechanism of the photo-control might be understood in structural terms. The resulting design might then be directly transferable and be of use in controlling a wide variety of proteins.
We report here a simple system that shows a number of desired properties. A short model peptide was designed that is internally cross-linked with a bridge that incorporates a photoisomerizable azobenzene moiety. Circular dichroism spectra demonstrate that the helix content of the peptide can be controlled reversibly by light of appropriate wavelength and intensity.
Materials and Methods
Synthesis of the Azobenzene Cross-Linker.
4,4′-Diaminoazobenzene (0.24 mmol) (Lancaster Synthesis) was dissolved in anhydrous tetrahydrofuran (10 ml) and was stirred under nitrogen for 15 min, protected from light. Three equivalents of triethylamine (0.70 mmol) were added, followed by three equivalents of chloroacetyl chloride (0.70 mmol, dropwise). The reaction mixture was filtered and the solvent evaporated. Sodium iodide (26 mmol) was dissolved in anhydrous acetone (16.25 ml) with anhydrous tetrahydrofuran (5 ml) (degassed) and was added to the reaction flask. This mixture was stirred for 18 h under nitrogen, protected from light. It was then filtered to remove NaCl, and the solvent was removed to give a brownish/yellow solid. The solid was redissolved in tetrahydrofuran (2 ml), and excess NaI was removed by filtration. To the tetrahydrofuran solution, ice-cold water was added in 1-ml increments until precipitate formed (≈3 ml H2O). The slurry was filtered and dried. 1H NMR and high-resolution electron impact MS confirmed the synthesis of the diiodoacetamide azobenzene cross-linker (structure 1) (overall yield 84%). Rf = 0.77 in (C/M: 75/25). 1H NMR (DMSO-d6, 400 MHz): δ 3.86 ppm (s, 4H), δ 7.77 ppm (d, J = 9.52 Hz, 4H), δ 7.86 ppm (d, J = 9.52 Hz, 4H), δ 10.65 ppm (s, 2H). Electron impact MS: (C16H14N4O2I2), observed mass = 547.919897, calculated mass = 547.920630.
Peptide Synthesis and Cross-Linking.
Standard fluorenylmethoxycarbonyl-based solid-phase peptide synthesis methods were used to prepare acetyl-EACARVAibAACEAAARQ-amide (Fig. 1). Peptides were constructed on a Pal-resin (capacity 0.55 mmol/g) (Advanced ChemTech). Coupling used 3 equiv. HATU [O-(7-azobenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate]; 6 equiv. DIPEA (N,N-diisopropylethylamine), 3 equiv. amino acid. Aib-F was synthesized following the procedure of Kaduk et al. (28). Peptides were purified by HPLC [Apex Presil C18 8 μ column (Jones Chromatography, Lakewood, CO) using a linear gradient from 0 to 50% acetonitrile/H2O (+0.1% trifluoroacetic acid) over 40-min elution at 47% acetonitrile]. The peptide primary structure was confirmed by electrospray ionization MS (observed, 1,645.5 Da; calculated, C65H112N24O22S2: 1,645.9) and amino acid analysis, including cysteine analysis via performic acid oxidation (HSC Biotechnology Service Centre, Toronto). The purity by HPLC was >95%.
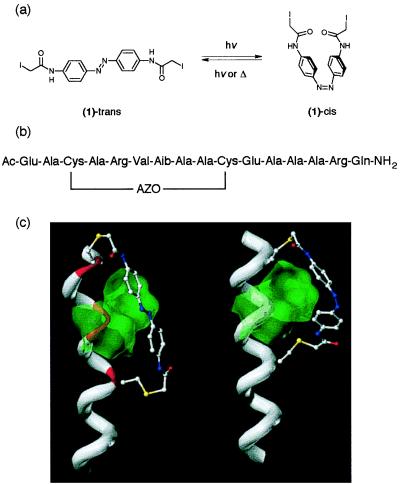
Figure 1.
(a) Chemical structure of the azobenzene cross-linker (1). (b) Primary sequence of the cross-linked peptide. AZO refers to the cross-linker in a after reaction with the two cysteine side-chains. Aib, α-aminoisobutyric acid. (c) Models of the cross-linked peptide with the azobenzene group in the trans (left) and cis (right) conformations. The peptide backbone is shown as a white tube for residues forming normal α-helical hydrogen bonds but in red where the H-bond pattern is disrupted. The ball-and-stick representation is used to display the cross-linker and cysteine side-chain atoms. The Connolly surfaces of residues i + 3 (Val) and i + 4 (Aib) are shown in green (the green overlay makes the red region beneath appear yellow). The trans cross-linker causes a large distortion of the helix by being forced into close contact with the Val and Aib residues whereas the cis cross-linker is fully compatible with the helical structure.
Internal cross-linking of Cys residues in the peptide by compound 1 was performed as follows: In a total volume of 355 μl of 15.5 mM Tris⋅Cl buffer (pH 8), un-cross-linked peptide (1.13 mM) and Tris(carboxyethyl)phosphine (1.13 mM) were combined and incubated for 1 h at room temperature to ensure cysteine residues were in their reduced state. To the aqueous solution, 500 μl DMSO containing 0.56 μmol of 1 was added, giving a total reagent concentration of 0.66 mM. This solution was stirred for 10 min protected from light, then 55.8 μl of a 10 mM solution of 1 was added to the mixture. After another 10 min, a third aliquot was added (55.8 μl of a 10 mM solution of 1) to give a final reagent concentration of 1.93 mM. The reaction mixture was stirred for 10 min protected from light followed by 10 min exposed to light. Unreacted 1 was removed by gel-filtration chromatography on Sephadex-G10, and the modified peptide was purified by HPLC [Zorbax SB-C18 column; 0–80% acetonitrile/H2O (+0.1% trifluoroacetic acid) linear gradient over the course of 45 min; elution at 42% acetonitrile]. The peptide primary structure was confirmed by electrospray ionization MS (observed, 1,938.4 Da; calculated, C81H124N28O24S2 = 1,938.2) and amino acid analysis, including cysteine analysis (which confirmed the absence of Cys).
Molecular Modeling.
Initial models of the photoisomerizable compound in cis and trans conformations were sketched by using the spartan 5.0 package (Wavefunction, Irvine, CA) on an SGI Octane R10000 computer. Cysteine residues were added at each end of the cross-linker (1) then were truncated at the β carbon. This composite of the photoisomerizable cross-linker with cysteine side-chain atoms attached was then subjected to geometry optimization. A comparison between the results of optimizations using the semiempirical representation AM1 (29) and ab initio methods with a minimal basis set (STO-3G) (30) showed only small differences in the structures. A further set of optimizations was undertaken to choose the best interresidue spacing between cysteine residues. A quadratic restraint on the distance between the two “β carbon” atoms at the ends of the composite cross-linker was added to the AM1 potential energy function. The restraint distance was adjusted in small (1 Å or less) increments with geometry optimization after each change.
Polyalanine peptides and the acetyl-EACARVAibAACEAAARQ-amide peptide in an α-helical conformation were built within spartan, and the azobenzene cross-linker was introduced between cysteine residues at an i to i + 7 spacing. The system was now too large for semiempirical calculations to be practicable, so molecular mechanics calculations were conducted by using the spartan implementation of the SYBYL energy function (31). Restraints were introduced to maintain the azobenzene cross-linker near its AM1-minimized conformation because this conformation was found to be fairly rigid in the calculations described above and the azo group has not been fully parameterized in this force field. No restraints were imposed on the χ1, χ2, or χ3 dihedral angles of the cysteine side chains. Work in progress aims to find parameters to represent the cross-linker without imposing restraint terms.
Circular Dichroism Measurements.
Circular dichroism measurements were performed on a Jasco Model J-710 spectropolarimeter. All measurements were made in thermostated quartz cuvettes (0.01–1 cm pathlength). Temperatures were measured by using a microprobe directly in the sample cell. All samples were dissolved in 5 mM phosphate buffer (pH 7). DTT (3 mM) was present in the un-cross-linked peptide sample to ensure that cysteine residues were in the reduced form. Spectra reported are the averages of three or five scans each, with appropriate background spectra subtracted. A scan speed of 10 nm/min, with a 0.5-nm bandwidth and a 4-s response time, was used. A mean residue weight of 102.8 was used for both the un-cross-linked and modified peptide. Peptide concentrations were determined by quantitative amino acid analysis.
UV/Vis Spectra and Photoisomerization.
UV spectra were obtained with a Perkin–Elmer Lambda 2 spectrophotometer using a 1-cm quartz cell in a thermostated holder. To obtain rates of thermal isomerization, absorbance at 360 nm was measured versus time, and the curve was fitted to an exponential decay expression by using the program igor (Wavemetrics). The measuring beam was found to have a negligible effect on the rate of the isomerization process.
Photoisomerization was accomplished by irradiating thermostated peptide solutions with a 450-W xenon arc lamp (Oriel, Stamford, CT) coupled to a high intensity monochromator (2,700 grooves/mm) (Bausch & Lomb) (6-nm slit) or a 380- or 486-nm bandpass filter (Melles Griot, Irvine, CA). Light intensity at the sample was ≈7 mW with the monochromator and 10-fold higher with the bandpass filters. Photoisomerization was complete (as judged by the lack of any further changes in UV spectra) in ≤5 min.
Results
Cross-Linker Design.
The azobenzene chromophore is perhaps the simplest and most robust photoisomerizable unit (32–35). The planar trans conformation is generally more stable than the twisted cis conformation in the dark (Fig. 1a). The barrier for thermal isomerization (i.e., isomerization in the dark) from the cis to the trans conformation is ≈18.5 kcal/mol with the azobenzene derivative considered here (Table 1). Absorption of a photon leads to an excited state in which the barrier for isomerization is substantially smaller. Decay can lead to either a trans or cis ground state (36). Because the trans and cis ground states have different absorption spectra, irradiation of an azobenzene solution leads to a photostationary state that is predominantly trans or predominantly cis, depending on the wavelength. Little photochemical breakdown (bleaching) is observed (34, 35), and the wavelengths required (λ > 330 nm in the present case) are long enough that interaction with intrinsic protein chromophores is avoided.
Table 1.
Temperature dependence of thermal relaxation of the cross-linked peptide
| Temperature, °C* | τ1/2, min† |
| 9 | 115 |
| 18 | 37 |
| 27 | 16 |
| 36 | 6.4 |
| 45 | 2.7 |
Peptide concentration was ≈7 μM in 5 mM NaPO4 buffer (pH 7.0). An activation energy of 18.5 ± 0.5 kcal/mol may be estimated from these data.
*Temperatures are accurate ±1°C.
†Half-times are accurate ±10%.
Because we anticipate the eventual use of such a photo-switching reagent with expressed proteins, a means for site-specific incorporation was desired. The introduction of cysteine residues by site-directed mutagenesis followed by reaction with thiol-specific reagents provides a convenient protocol for site-specific incorporation. A second consideration in the design of the reagent was to minimize its flexibility because we anticipated that excess flexibility would obviate any conformational effects of photoisomerization. The azobenzene derivative (1) (Fig. 1a) incorporates two α-iodoacetamido groups to permit intramolecular cross-linking via reaction with cysteine side-chains and contains a minimum number of single bonds.
Peptide Design.
Our initial idea for altering peptide helix content was to exploit the alteration in length of the cross-linker upon photoisomerization (Fig. 1a). The distance between the β carbon atoms of two residues within an ideal α-helix is fixed by the geometry of the helix. We wished to find which connection points for the cross-linker (i.e., which cysteine residue spacing) would be likely to lead to the largest effects of photoisomerization on helix stability. An analysis was made of the energy cost of deforming the cross-linker (in either cis or trans conformation) as described in Materials and Methods. The largest difference in deformation energy was found for an i to i + 7 spacing (10.4 Å) at over 5 kcal/mol. Whereas relatively little energy is required to deform a cis cross-linker to the ideal i to i + 7 spacing, significant energy would be required to deform a trans cross-linker. The trans cross-linker connected to residues i and i + 7 would thus be expected to destabilize the helix conformation whereas a cis cross-linker would not.
Examination of peptide models with molecular graphics (Fig. 1c) further showed that, in the trans conformation, the azobenzene group was forced to pack tightly against the helix whereas in the cis conformation it was not.‖ We reasoned that, if the helix were made more bulky in the region between the attachment points, steric clashes might destabilize the helix when the cross-linker was in the trans conformation. This could be achieved by making residue i + 3 into a β branched amino acid, such as valine. Additional bulk in the region could also be obtained by altering the hydrogen attached to the α carbon of residue i + 4. One way of doing this would be to alter residue i + 4 to a D rather than L amino acid. However, this is likely to have a destabilizing effect on the helix (38). Instead it was decided to change this residue to the non-standard residue aminoisobutyric acid (Aib), which is known to be helix-promoting (39).
The final stage of modeling was to incorporate the i, i + 3, i + 4, and i + 7 modifications into a peptide known to be soluble, monomeric, and capable of forming α-helices in water. It was also desirable to employ a peptide in which the helix content could be easily manipulated by changes in temperature or by using solvent additives. A helix that was too stable or too unstable might prove difficult to manipulate via photoisomerization of an attached cross-linker. The peptide acetyl-AEAAAREAAAREAAARA-amide, described by Merutka et al. (40), appeared to have the required features.
Energy minimized models of our final designed peptide, acetyl-EACARVAibAACEAAARQ-amide with the cross-linker in cis and trans conformations, are shown in Fig. 1c. As expected for the cis form, all of the hydrogen bonds that stabilize an ideal α-helix are present. In the trans form, significant distortion was observed. In particular, the two hydrogen bonds lying directly under the azobenzene were extended to such an extent that they could be regarded as broken. The molecular mechanics methods used here cannot simulate the unfolding of a helix, as solvent and thermal effects are not explicitly represented. However, if the distorted trans-form helix were solvated, water molecules could hydrogen bond to the exposed amide NH and carbonyl groups and interrupt the intramolecular H-bonding pattern. We can therefore expect the trans form to have a low helix content in water. Molecular dynamics simulations with explicitly represented water molecules are currently underway that are expected to better predict helix stability (4).
Photoisomerization of the Cross-Linker.
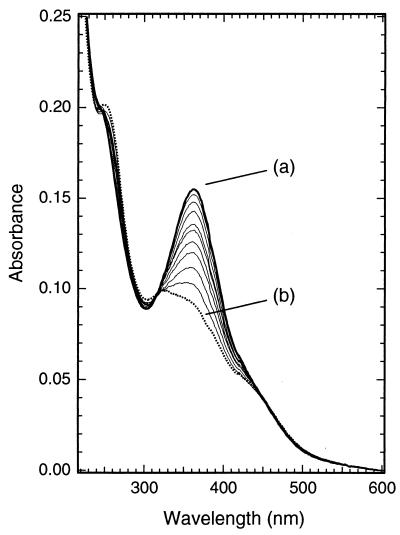
The conformational state (cis or trans) of the azobenzene cross-linker can be monitored by using UV/visible absorption spectroscopy. The absorption spectrum of the dark-adapted form (trans) of the cross-linked peptide in aqueous solution is shown in Fig. 2, curve a. The absorption maximum occurs at 362 nm, typical for an amide-substituted azobenzene π-π* transition (34).
Figure 2.
UV/visible spectra showing photoisomerization of the modified peptide. Curve a (solid heavy line) is the spectrum of the dark adapted peptide [≈5 μM in 5 mM phosphate buffer (pH 7.0), 11.5 ± 0.5°C]. Curve b (dashed line) is the spectrum obtained immediately after exposure of the solution to 380-nm light (7 mW for 5 min). Intermediate curves (thin lines) show spectra obtained at varying times after photoisomerization (20, 40, 60, 80, 100, 140, 190, 240, and 290 min), keeping the sample in the dark at 11. 5 ± 0.5°C.
Irradiation of the peptide solution for 5 min (7 mW) leads to curve b (Fig. 2) in which the peak at 362 nm is markedly reduced. There is a small increase in absorbance at 470 nm (n-π*) and a moderate increase at 250 nm (π-π*, cis form). A comparison with published spectra (20) indicates that curve b (Fig. 2) represents a solution containing ≈75% cis/25% trans conformations of the azobenzene cross-linker. The absorbance change is fully reversible in the dark with a half-life that depends on temperature (Table 1). Irradiation at other wavelengths, e.g., 486 nm, leads rapidly to photostationary states containing varying degrees of cis and trans conformations as expected (spectra not shown).
Effects of Photoisomerization on Peptide Conformation.
The secondary structure of the peptide was investigated by using circular dichroism (CD) spectroscopy. This technique has been used extensively for the determination of helix content in peptides and proteins (41–43). Although the azo group also has transitions in this region, which might in principle complicate analysis of the CD spectrum, Pieroni and colleagues have found no interference of this sort even in polypeptides with up to one azo group for every three peptide bonds (26, 44).
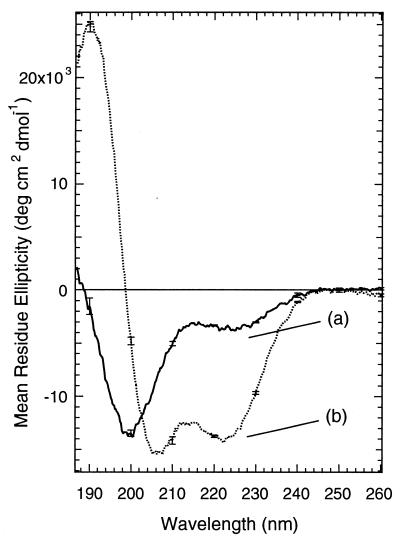
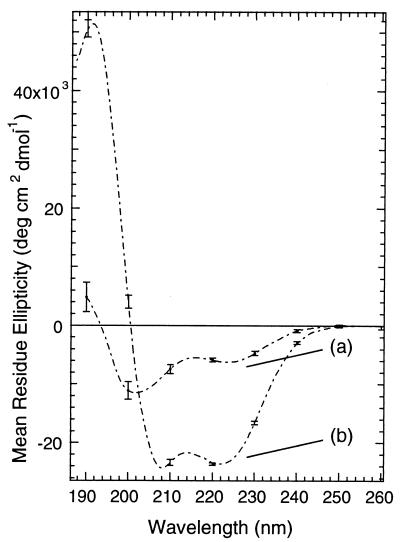
Fig. 3, curve a shows the circular dichroism spectrum of the dark-adapted cross-linked peptide with the azo group in the trans conformation. The spectrum is typical of a largely disordered structure with a small percentage of helix (41).
Figure 3.
CD spectra showing photo-control of helix content in the modified peptide. Curve a (solid line) is the spectrum of the dark adapted peptide [60.5 μM peptide, 5 mM phosphate buffer (pH 7.0), 11 ± 0.5°C]. Curve b (dashed line) is the spectrum obtained immediately after exposure of the solution to 380-nm light (7 mW for 5 min).
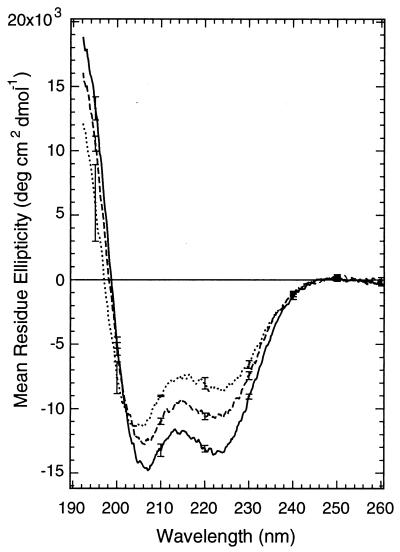
When the cross-linked peptide was irradiated at 380 nm, so that the azobenzene groups were predominantly in the cis conformation, the intensity of the negative CD band at 222 nm increased significantly (Fig. 3, curve b), the minimum at 200 nm shifted to 207 nm, and a strong maximum at 190 nm appeared. The light-induced spectrum was thus characteristic of an α-helix. The magnitude of the change in the CD spectrum of the cross-linked peptide upon photoisomerization depends on temperature. Lower temperatures result in larger changes, primarily because lower temperatures increase the apparent helix content of the irradiated peptide (Fig. 4; Table 2). Lower temperatures have also been found to stabilize the helical conformation of the parent peptide (40). Temperature does not substantially influence the percentage of azo groups that are converted to the cis form upon irradiation.
Figure 4.
Temperature dependence of the CD spectrum of the cross-linked peptide obtained after exposure of the solution to 380-nm light and before substantial thermal reversion to the trans form has occurred. Other conditions were as in Fig. 3. Temperatures: 11°C (—); 18°C (---); 25°C (…).
Table 2.
Peptide helix content under various conditions
| Sample | °C† | θ222 | % helix‡ |
|---|---|---|---|
| Dark adapted cross-linked peptide | 11 | −3,700 ± 200 | 12 |
| 18 | −3,400 ± 200 | 11 | |
| 25 | −3,500 ± 200 | 12 | |
| Irradiated* cross-linked peptide | 11 | −14,400 ± 500 | 48 |
| 18 | −10,700 ± 600 | 36 | |
| 25 | −8,400 ± 400 | 28 | |
| Un-cross-linked peptide | 11 | −7,400 ± 300 | 25 |
| 18 | −6,300 ± 300 | 21 | |
| 25 | −6,100 ± 400 | 20 | |
| Un-cross-linked peptide + 50% TFE | 11 | −23,400 ± 500 | 78 |
| 18 | −22,700 ± 300 | 76 | |
| 25 | −20,700 ± 300 | 69 |
Peptide concentration ≈50 μM in 5 mM NaPO4 (pH 7.0).
*After exposure to 380-nm light for 5 min (7 mW).
†Temperatures are accurate ±1°C.
‡Calculated by using (θ222)/30,000 × 100%.
The CD spectrum of the un-cross-linked peptide under the same conditions is shown in Fig. 5, curve a. The spectrum is consistent with an intermediate degree of helicity. Based on the intensity of the CD band at 222 nm, the un-cross-linked peptide was more helical than the trans form, but much less helical than the irradiated form, of the cross-linked peptide. Light had no effect on the CD spectrum of the un-cross-linked peptide. The effect of adding 50% trifluoroethanol, a strong helix promoting solvent additive (45), to a solution of the un-cross-linked peptide is shown in Fig. 5, curve b. As expected, a spectrum typical of an α-helical peptide is observed.
Figure 5.
(Curve a) CD spectrum of the un-cross-linked peptide (acetyl-EACARVAibAACEAAARQ-amide) under the same conditions as those used with the cross-linked peptide [45.5 μM peptide, 5 mM phosphate buffer (pH 7.0), 11 ± 0.5°C] except that 3 mM DTT was added to prevent oxidation of cysteine residues. (Curve b) Upon the addition of 50% trifluoroethanol.
Although differences in θ222 indicate changes in helix content, assignment of a particular value of θ222 to a particular percentage helix is more difficult (5). By using the simple assumption that θ222 for 100% helix is given by 40,000 × [(n − 4)/n], where n is the number of residues (7), the light-induced transition shown in Fig. 3 is from 12% to 48% helix. For comparison, the addition of 50% trifluoroethanol causes a transition from 25% to 78% helix in the un-cross-linked peptide (Fig. 5). Thus photoisomerization is about as effective (a four-fold increase) at inducing helical secondary structure as the addition of 50% trifluoroethanol, at least in the present case.
Exposure of the cross-linked peptide to wavelengths of light other than 380 nm (that used to cause the isomerization) led to varying degrees of peptide helicity depending on the wavelength chosen. For instance, exposure to 486 nm light gave a photostationary state with ≈25% helix.
Reversible Switching Between Two States.
The helix-coil structural transition observed by CD upon photoisomerization of the cross-linked peptide is fully reversible. If the irradiated form of the peptide is kept in the dark, the CD spectrum reverts to that of the peptide with a trans cross-linker. The time course of the peptide structural change matches, within experimental error, the isomerization of the azo linkage as seen by using UV/visible spectroscopy (Fig. 2). This observation is consistent with isomerization of the azo group being the rate-determining step for the peptide structural change.
The CD spectra of the cross-linked peptide shown in Fig. 3 as well as those obtained by exposing solutions to light of different wavelengths, or obtained during the course of thermal reversion to the trans state, all exhibited an isodichroic point at 203 nm. This observation indicates the existence of a simple two-state equilibrium between helical and disordered forms of the peptide. We found no evidence of peptide self-association. The same CD change and isodichroic point were observed in solutions of varying peptide concentrations (from 5 to 225 μM), and no evidence of light scattering or of β-type secondary structures typical of aggregated peptide (22, 46) were seen.
Discussion
The cross-linked peptide shown in Fig. 1, the product of the design process described, has been found to have properties close to those predicted. Photoisomerization from trans to cis led to a marked increase in helix content in the peptide. Of course, the result might be fortuitous and does not prove the correctness of the design process. Nevertheless, this finding of photoregulation of helix content is significant because the system is simple enough that the factors controlling the process can be systematically assessed and optimized. The initial discovery that certain short peptides could show significant helix content in aqueous solutions (47) led to a systematic analysis of the factors involved and to the synthesis of peptides that are essentially 100% helical in water (40, 48). Systematic structure-activity studies coupled with more detailed, NMR-based, conformational studies are planned for the peptide system at hand.
The introduction of intramolecular cross-links per se can greatly stabilize α-helical secondary structure. For instance, the introduction of a flexible alkanediyl tether between residues i and i + 7 can induce helicity in a peptide that is normally disordered in water (6). This effect is generally attributed to a lowering of the entropy of the disordered form by the cross-link, so that helix formation is promoted (8, 49). Introducing the azobenzene cross-linking reagent in the trans conformation, however, destabilizes the helix relative to the un-cross-linked peptide (Fig. 3, curve a vs. Fig. 5, curve a). The favorable entropic effect of the cross-linker must therefore be counteracted (in the case of the trans form) by factors that destabilize the helix. The modeling suggests that these factors are the mismatch in length of the trans cross-linker, combined with the steric clash between the linker and the residues at position i + 3 and i + 4 . The correctness of this proposal can be checked by altering these residues as well as by increasing the flexibility of the cross-link to relieve the mismatch strain.
These helix-destabilizing factors are removed upon photoisomerization of the cross-linker to the cis form. The length of the cis-cross-linker matches better the ideal i to i + 7 spacing of an α-helix, and the “tent” conformation of the azobenzene group can accommodate bulky residues at positions i + 3 and i + 4. The CD spectra indicate that the conformation of the peptide with the cross-linker in the cis conformation is substantially helical. Because irradiation leads to a photostationary state that is not 100% cis, the secondary structure observed by CD is not expected to be 100% helical. Peptides with cross-linkers in the trans conformation will remain, thus reducing the measured helix content of the sample. The observation that the helix content of the irradiated sample increases as the temperature is lowered (Fig. 4), however, indicates that, at least for temperatures above 11°C, peptides bearing cis cross-linkers are not completely helical. As the temperature is lowered, we expect that the conformation of the peptide with a cis cross-linker will more and more resemble that shown in Fig. 1c; a detailed NMR analysis should establish whether this is true.
If the foregoing analysis proves correct, the system described here may prove a useful model for fundamental studies on the process of peptide (and protein) folding. Simple azobenzene compounds photoisomerize extremely rapidly [<10 ps (36)], so that isomerization of the cross-linker might be used as a trigger in fast time-resolved studies of helix formation (13, 14). The system might also serve as an accessible model for studying the energetics and kinetics of conformational cascades that accompany photoisomerization events in other biological systems (e.g., rhodopsins, phytochromes).
Finally, because protein function is intimately connected with structure, ability to photo-control helix content offers the prospect of photo-control of protein activity. Of course, the ability of the trans cross-linker to unfold a helix may be diminished if the helix is greatly stabilized by its participation in a larger protein structure. Also, it will be important to test whether other residues can substitute for the non-coded residue Aib at position i + 4 if the linker is to function with expressed proteins. However, with careful consideration of the sites of attachment and the conformational consequences of isomerization, photoisomerizable compounds like the cross-linker described here might find wide application for designing light-activated proteins. Such proteins would be very powerful tools for the study of complex biochemical systems.
Acknowledgments
We are grateful to Dr. John Corrie for advice on the conditions for the cross-linking reaction and to Drs. M. Moskovits, R. Kluger, and T. Haslett for the loan of equipment. We thank the Natural Sciences and Engineering Research Council of Canada for support of this work and the Novartis Foundation for a Travel Award (to O.S.S.). J.R.K. is the recipient of an Ontario Graduate Scholarship in Science and Technology graduate fellowship. O.S.S. is supported by the U.K. Medical Research Council.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Initial models also suggested that the right-handed (P) version of the cis-form of the cross-linker was most compatible with the (right-handed) helix. An influence of polypeptide helix sense on the helix or twist sense of appended groups has been noted by Reidy and Green (37).
References
- 1.Pauling L, Corey R B, Branson H R. Proc Natl Acad Sci USA. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohl C A, Fiori W, Baldwin R L. Proc Natl Acad Sci USA. 1999;96:3682–3687. doi: 10.1073/pnas.96.7.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neil K T, DeGrado W F. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 4.Young W S, Brooks C L., 3rd J Mol Biol. 1996;259:560–572. doi: 10.1006/jmbi.1996.0339. [DOI] [PubMed] [Google Scholar]
- 5.Wallimann P, Kennedy R J, Kemp D S. Angew Chem Int Ed Engl. 1999;38:1290–1292. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1290::AID-ANIE1290>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 6.Phelan J C, Skelton N J, Braisted A C, McDowell R S. J Am Chem Soc. 1997;119:455–460. [Google Scholar]
- 7.Jackson D, King D, Chmielewski J, Singh S, Schultz P. J Am Chem Soc. 1991;113:9391–9392. [Google Scholar]
- 8.Ruan F, Chen Y, Hopkins P B. J Am Chem Soc. 1990;112:9403–9404. [Google Scholar]
- 9.Pellegrini M, Royo M, Chorev M, Mierke D F. J Peptide Res. 1997;49:404–414. doi: 10.1111/j.1399-3011.1997.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 10.Adams S R, Tsien R Y. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 11.Mendel D, Ellman J A, Schultz P G. J Am Chem Soc. 1991;113:2758–2760. [Google Scholar]
- 12.Chang C Y, Fernandez T, Panchal R, Bayley H. J Am Chem Soc. 1998;120:7661–7662. [Google Scholar]
- 13.Callender R H, Dyer R B, Gilmanshin R, Woodruff W H. Annu Rev Phys Chem. 1998;49:173–202. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 14.Volk M, Kholodenko Y, Lu H S M, Gooding E A, DeGrado W F, Hochstrasser R M. J Phys Chem B. 1997;101:8607–8616. [Google Scholar]
- 15.Willner I, Rubin I. Angew Chem Int Ed Engl. 1996;35:367–385. [Google Scholar]
- 16.Ueda, T., Murayama, K., Yamamoto, T., Kimura, S. & Imanishi, Y. (1994) J. Chem. Soc. Perkin Trans. 1 225–230.
- 17.Liu D, Karanicolas J, Yu C, Zhang Z, Woolley G A. Bioorg Med Chem Lett. 1997;7:2677–2680. [Google Scholar]
- 18.Hamachi I, Hiraoka T, Yamada Y, Shinkai S. Chem Lett. 1998;6:537–538. doi: 10.1016/s0960-894x(99)00189-4. [DOI] [PubMed] [Google Scholar]
- 19.Ulysse L, Cubillos J, Chmielewski J. J Am Chem Soc. 1995;117:8466–8467. [Google Scholar]
- 20.Behrendt R, Renner C, Schenk M, Wang F, Wachtveitl J, Oesterhelt D, Moroder L. Angew Chem Int Ed Engl. 1999;38:2771–2774. doi: 10.1002/(sici)1521-3773(19990917)38:18<2771::aid-anie2771>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 21.Rudolph-Bohner S, Kruger M, Oesterhelt D, Moroder L, Nagele T, Wachtveitl J. J Photochem Photobiol A Chem. 1997;105:235–248. [Google Scholar]
- 22.Cerpa R, Cohen F E, Kuntz I D. Fold Des. 1996;1:91–101. doi: 10.1016/S1359-0278(96)00018-1. [DOI] [PubMed] [Google Scholar]
- 23.Ueno A, Takahashi K, Anzai J, Osa T. J Am Chem Soc. 1981;103:6410–6415. [Google Scholar]
- 24.Higuchi M, Kinoshita T. J Photochem Photobiol B Biol. 1998;42:143–150. [Google Scholar]
- 25.Cooper T M, Natarajan L V, Crane R L. Trends Polym Sci. 1993;1:400–405. [Google Scholar]
- 26.Pieroni O, Ciardelli F. Trends Polym Sci. 1995;3:282–287. [Google Scholar]
- 27.Pachter R, Cooper T M, Natarajan L V, Obermeier K A, Crane R L, Adams W W. Biopolymers. 1992;32:1129–1140. [Google Scholar]
- 28.Kaduk C, Wenschuh H, Beyermann M, Forner K, Carpino L A, Bienert M. Lett Peptide Sci. 1996;2:285–288. [Google Scholar]
- 29.Dewar M J S, Zoebisch E G, Healy E F, Stewart J J P. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- 30.Hehre W J, Radom L, Schleyer P v R, Pople J A. Ab Initio Molecular Orbital Theory. New York: Wiley; 1986. [Google Scholar]
- 31.Clark M, Cramer R D, III, van Opdensch N. J Comput Chem. 1989;10:982–1012. [Google Scholar]
- 32.Nagele T, Hoche R, Zinth W, Wachtveitl J. Chem Phys Lett. 1997;272:489–495. [Google Scholar]
- 33.Lednev I K, Ye T Q, Hester R E, Moore J N. J Phys Chem. 1996;100:13338–13341. [Google Scholar]
- 34.Rau H. In: Photochromism. Molecules and Systems. Durr H, Bouas-Laurent H, editors. Amsterdam: Elsevier; 1990. pp. 165–192. [Google Scholar]
- 35.Rau H. In: Photochemistry and Photophysics. Rabek J F, editor. II. Boca Raton, FL: CRC; 1990. pp. 119–141. [Google Scholar]
- 36.Wachtveitl J, Nagele T, Puell B, Zinth W, Kruger M, Rudolph-Bohner S, Oesterhelt D, Moroder L. J Photochem Photobiol A Chem. 1997;105:283–288. [Google Scholar]
- 37.Reidy M P, Green M M. Macromolecules. 1990;23:4225–4234. [Google Scholar]
- 38.Fairman R, Anthony-Cahill S J, DeGrado W F. J Am Chem Soc. 1992;114:5458–5459. [Google Scholar]
- 39.Kaul R, Balaram P. Bioorg Med Chem. 1999;7:105–117. doi: 10.1016/s0968-0896(98)00221-1. [DOI] [PubMed] [Google Scholar]
- 40.Merutka G, Shalongo W, Stellwagen E. Biochemistry. 1991;30:4245–4248. doi: 10.1021/bi00231a020. [DOI] [PubMed] [Google Scholar]
- 41.Johnson W C., Jr Proteins. 1990;7:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 42.Greenfield N J. Anal Biochem. 1996;235:1–10. doi: 10.1006/abio.1996.0084. [DOI] [PubMed] [Google Scholar]
- 43.Woody R W. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 44.Fissi A, Pieroni O, Balestreri E, Amato C. Macromolecules. 1996;29:4680–4685. [Google Scholar]
- 45.Cammers Goodwin A, Allen T J, Oslick S L, McClure K F, Lee J H, Kemp D S. J Am Chem Soc. 1996;118:3082–3090. [Google Scholar]
- 46.Pieroni O, Fissi A, Houben J L, Ciardelli F. J Am Chem Soc. 1985;107:2990–2991. [Google Scholar]
- 47.Brown J E, Klee W A. Biochemistry. 1971;10:470–476. doi: 10.1021/bi00779a019. [DOI] [PubMed] [Google Scholar]
- 48.Marqusee S, Robbins V H, Baldwin R L. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skolnick J, Holtzer A. Biochemistry. 1986;25:6192–6202. doi: 10.1021/bi00368a054. [DOI] [PubMed] [Google Scholar]