Abstract
The complete genome sequence of the T4-like, broad-host-range vibriophage KVP40 has been determined. The genome sequence is 244,835 bp, with an overall G+C content of 42.6%. It encodes 386 putative protein-encoding open reading frames (CDSs), 30 tRNAs, 33 T4-like late promoters, and 57 potential rho-independent terminators. Overall, 92.1% of the KVP40 genome is coding, with an average CDS size of 587 bp. While 65% of the CDSs were unique to KVP40 and had no known function, the genome sequence and organization show specific regions of extensive conservation with phage T4. At least 99 KVP40 CDSs have homologs in the T4 genome (Blast alignments of 45 to 68% amino acid similarity). The shared CDSs represent 36% of all T4 CDSs but only 26% of those from KVP40. There is extensive representation of the DNA replication, recombination, and repair enzymes as well as the viral capsid and tail structural genes. KVP40 lacks several T4 enzymes involved in host DNA degradation, appears not to synthesize the modified cytosine (hydroxymethyl glucose) present in T-even phages, and lacks group I introns. KVP40 likely utilizes the T4-type sigma-55 late transcription apparatus, but features of early- or middle-mode transcription were not identified. There are 26 CDSs that have no viral homolog, and many did not necessarily originate from Vibrio spp., suggesting an even broader host range for KVP40. From these latter CDSs, an NAD salvage pathway was inferred that appears to be unique among bacteriophages. Features of the KVP40 genome that distinguish it from T4 are presented, as well as those, such as the replication and virion gene clusters, that are substantially conserved.
Bacteriophage KVP40 and similar Vibrio phages were isolated from polluted seawater off the coast of Japan with a strain of Vibrio parahaemolyticus as the host (41). KVP40 differs from many described vibriophages in having a broad host range; it has been reported to infect eight Vibrio species, including Vibrio cholerae and Vibrio parahaemolyticus, the nonpathogenic species Vibrio natriegens, and Photobacterium leiognathi (41). Additional studies have implicated the Vibrio OmpK outer membrane protein as the phage receptor (23).
Vibriophage KVP40, like the well-studied phage T4 (27, 44), belongs to the Myoviridae family of viruses. This family is characterized by having a double-stranded DNA genome, a prolate icosahedral capsid, and a contractile tail with associated baseplate and extended tail fibers (1). However, there are morphological differences between phage T4 and KVP40. For example, the head of KVP40 is longer (140 nm long and 70 nm wide) than that of T4. Due to the constraints of head size on DNA packaging, this observation suggested that the genome of KVP40 is larger than the 168,903-bp genome of T4.
Beyond morphological similarities, major and minor capsid genes of KVP40 have been sequenced and are related to and functionally conserved with those of T4 (39). However, phylogenetic analysis of Myoviridae capsid genes suggests that the vibriophages, along with T4-like phages infecting species of Aeromonas, are distinct from the closely related T-even group (57). More recently, conservation of gene order and sequence similarity (at roughly 50%) for the major capsid and contractile tail proteins was demonstrated for a T4-like marine cyanophage (15).
Little or nothing is known about biochemical activities carried out by these more distantly related T4-like phages, including the essential functions of DNA replication and metabolism and the overall genome organization. To expand our understanding of the genetic diversity of the classic T-even phage family and to extend functional and comparative analyses of gene products for which, to date, T4 provides the predominant experimental system (27, 44), we determined and described the complete genomic sequence of vibriophage KVP40.
MATERIALS AND METHODS
Strains, media, and purification of bacteriophage DNA.
Vibriophage KVP40 was originally isolated by S. Matsuzaki (41) from Urado Bay, Kochi, Japan, and deposited in the Félix d'Hérelle Reference Center for Bacterial Viruses (Quebec, Canada), where it and the host, V. parahaemolyticus (strain RIMD2210001; here called EB101), were purchased through H.-W. Ackermann. The phage and host were propagated at 30°C in YP-NaCl medium (41), in either broth or soft agar (0.5%) overlays. A KVP40 lysate was prepared by growing strain EB101 in YP-NaCl broth to mid log-phase (3 × 108 cells/ml) and infecting with phage at a multiplicity of infection of 0.01. The phage in 80 ml of lysate (109 PFU/ml) were precipitated twice with 50% polyethylene glycol 8000-0.5 M NaCl, and the two precipitates were pooled in a final volume of 3 ml of saline. The DNA was purified from the particles (1011 PFU) with the proteinase K-CTAB (cetyltrimethylammonium bromide) method essentially as described before (32). Purified KVP40 DNA was confirmed to be sensitive to cleavage with EcoRI and XbaI, as reported previously (41). Agarose gel electrophoresis of the EcoRI- and XbaI-digested genome gave an estimated genome size of circa 200 kbp.
DNA sequencing.
One small insert (2 to 3 kb) plasmid library was generated by mechanical shearing of genomic DNA. Briefly, 50 μg of purified genomic DNA was nebulized for 1 min at 20 lb/in2. The sheared DNA was size fractionated on an agarose gel and purified. The resulting fragments were ligated into pUC-derived sequencing vectors and transformed into Escherichia coli by electroporation. Plasmids were purified from transformed colonies, and the cloned inserts were sequenced with ABI Prism 3700 capillary sequencers. Sequences were trimmed of low-quality bases, and vector sequence was identified and removed. Initially, 2,616 random sequences were determined and assembled with the Institute for Genomic Research assembler. Sequence gaps were closed by primer walking on plasmid clones. Physical gaps were closed by multiplexed (58) and combinatorial PCR, followed by sequencing of the PCR products obtained. The final genome sequence is based on 4,008 total sequences with an average sequence length of 652 bp.
Sequence analysis.
An initial set of open reading frames likely to encode proteins (CDSs) was identified with the program Glimmer (52). CDSs that overlapped were inspected visually and in some cases removed. All CDSs were compared to a nonredundant amino acid database, and search results were inspected visually. Frameshifts were detected and corrected where appropriate (i.e., CDS298 and CDS298-2, and CDS330 and CDS331). CDSs were also analyzed with two sets of hidden Markov models constructed for a number of conserved protein families, pfam version 5.5 (4) and TIGRFAMS 1.0 (14), with the HMMER package (9). TopPredII was used to identify membrane-spanning regions in CDSs (7). tRNAscan-SE was used to search for tRNAs (36). The rho-independent transcription terminators were detected with TransTerm (10). T4-like late promoters were identified with the GCG FindPattern program with various mismatches and base substitutions.
Paralogous gene families were constructed by searching the CDSs against themselves with BlastX, identifying matches with an E of ≤10−5 over 60% of the query search length, and subsequently clustering these matches into multigene families. Multiple alignments were generated with the ClustalW program, and the alignments were examined visually.
Distribution of all 64 trinucleotides (trimers) for each chromosome was determined, and the trimer distribution in 2,000-bp windows that overlapped by half their length (1,000 bp) across the genome was computed. For each window, we computed the χ2 statistic on the difference between its trimer content and that of the whole chromosome. A large χ2 statistic indicates that the trimer composition in this window is different from that of the rest of the chromosome. Probability values for this analysis are based on the assumption that the DNA composition is relatively uniform throughout the genome. Because this assumption may be incorrect, we prefer to interpret high χ2 values merely as indicators of regions on the chromosome that appear unusual and demand further scrutiny.
Comparative genomics.
All the predicted proteins were compared to a data set of proteins from the complete genomes of viruses (phage and eukaryotic viruses), plasmids, and organisms (Bacteria, Archaea, and Eucarya). Comparisons were done with the Fasta3 program (46). Matches were ranked by E value, and only matches with an E value of less than 10−5 were considered significant. Phylogenetic trees were inferred by first aligning all homologs of the gene of interest with ClustalW, followed by manual alignment editing and phylogenetic analysis with the neighbor-joining algorithm of ClustalW or Phylip.
Nucleotide sequence accession number.
The GenBank accession number assigned to the KVP40 genome is B_KVP40 AY283928. CDS refers to the predicted protein coding sequence, with numbers specifying the locus tag in GenBank file AY283928 (i.e., CDS001 is locus KVP40.0001).
RESULTS AND DISCUSSION
Genome overview.
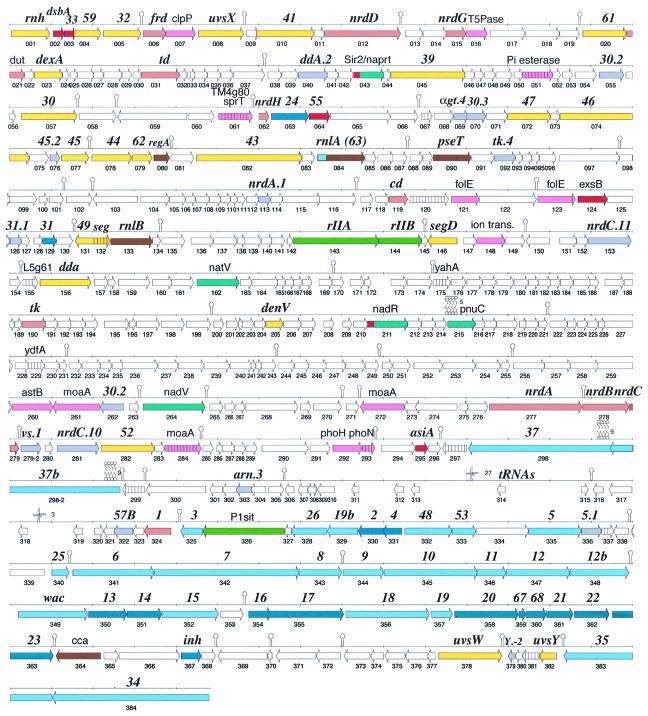
The genome of vibriophage KVP40 consists of one chromosome of 244,853 bp, with an average G+C content of 42.6%. Because assembly of the random library of sequences yielded a closed, circular genome, we conclude that, like phage T4 (44), KVP40 phage particles contain linear, circularly permuted chromosomal DNA. There are a total of 386 predicted protein coding sequences (CDSs), 33 T4-like late promoters, and 57 predicted rho-independent transcription terminators (Fig. 1, Table 1). Of the CDSs that encode proteins with matches to proteins in other complete genomes, most were most similar to proteins from phage (107 in total; 99 most similar to proteins from T4, and 8 similar to proteins from other phages). A smaller fraction was most similar to proteins from Bacteria (23), Eucarya (2), and Archaea (1). The remaining 253 CDSs (65%) had no match and thus were unique to KVP40.
FIG. 1.
KVP40 CDS map. CDSs are labeled with the name of the closest homolog, and thus T4 nomenclature is primarily indicated (bold italics); nonitalics are primarily bacterial orthologs. Numbering below the CDS arrow is that used in the text, tables, and GenBank. Colors correspond to protein functional categories, as follows: orange, DNA metabolism; yellow, DNA replication; red, transcription; brown, translation; light blue, tail and tail fibers; dark blue, head; green, host interactions; turquoise, NAD salvage; pink, bacterial-type genes; gray, hypothetical genes in T4; hatched, conserved hypothetical; white, unique hypothetical. Stem-loops are predicted transcription terminators (Table 5) or other RNA structures; the clover leafs mark the positions of tRNAs, and the circle-squiggle symbols denote predicted transmembrane domains.
TABLE 1.
General features of the KVP40 genome
| Feature | Value |
|---|---|
| Size (bp) | 244,835 |
| Total number of sequences | 4,008 |
| G+C content (%) | 42.6% |
| A, 30.0; C, 20.5; G, 22.1; T, 27.4 | |
| Total CDSs | 386 |
| Average CDS size (bp) | 587 |
| % of genome coding for proteins | 92.1 |
| No. of CDS products similar to known proteins, total | 100 |
| No. of CDS products similar to known T4 proteins | 83 |
| No. of conserved hypothetical proteins, total | 33 |
| No. of CDS products similar to conserved hypothetical T4 proteins | 16 |
| No. of hypothetical proteins | 253 |
| No. of rho-independent terminators | 57 |
| No. of tRNAs | 30 |
| No. of start sites and frequency (%)a | |
| AUG | 321, 83.1% (115, 89.8%) |
| GUG | 30, 7.8% (6, 4.7%) |
| UUG | 35, 9.1% (7, 5.5%) |
Start codons and frequency among all CDSs and, in parentheses, among known CDSs.
A large portion of the KVP40 genome (25% of the genome sequence in 24 separate regions) was not represented in the random clone library and was sequenced by directed walking of PCR products. The largest of these regions was 5,552 bp. There were 145 CDSs contained within these uncloned regions of the genome, consisting of 117 hypothetical proteins, 11 conserved hypothetical proteins, and 17 with a putative function. These regions may be unrepresented due to the presence of genes that code for products toxic to E. coli or to a nonrandom sequencing library. Toxic gene products seem to explain at least part of the unrepresented genomic regions. For example, these regions encode nucleases and DNA repair enzymes (CDS023, dexA; CDS131, denV; CDS131, 49; and CDS132, seg) as well as DNA metabolism and DNA-binding proteins (CDS005, 32; CDS006, frd; CDS015, nrdG; CDS119, cd; and CDS121, folE), which are often toxic to E. coli when cloned (Fig. 1). If such results are common to lytic phage genome sequencing projects, they will introduce added difficulties for completion of such phage genomes by the random shotgun sequencing method but illustrate the importance of closure.
Regions of unusual base composition (codons and di- and trinucleotides) have been seen as evidence of lateral gene transfer. This is because while the overall G+C content can vary greatly within a genome, the pattern of codons and the frequency of di- and trinucleotides remains much more constant (28, 45). Atypical nucleotide composition can also arise in genes with a functional constraint (i.e., rRNA in bacteria). The KVP40 genome was examined for atypical trinucleotide composition in all six frames across the genome. Six areas of the genome with highly different trimer composition were identified (data not shown). These regions correspond to genes also present in the genome of phage T4. We suspect that these genes are native to KVP40 (were not recently acquired) and that their biased trinucleotide compositions are due to functional constraints.
There is distributed synteny between the genomes of KVP40 and T4, with the two largest regions including the DNA replication genes (CDS072 through CDS082) and the virion structural genes (CDS341 through CDS363). Specific gene pairs are at times similarly located in the genomes, but in other instances conserved T4 and KVP40 gene pairs are in different positions relative to each other (e.g., rIIAB, 25, and 48).
Several CDSs appear to have been duplicated in the KVP40 lineage relative to other lineages for which complete genomes are available. To look at this, we searched proteins from KVP40 against a database of proteins from representative complete virus, plasmid, and organismal genomes (including KVP40). Proteins with better matches (based on E value) to other KVP40 proteins were considered candidates for lineage-specific duplications. In total, 16 proteins were identified as likely being duplicated in the KVP40 lineage (Table 2). Most of these are hypothetical proteins.
TABLE 2.
KVP40 CDSs with lineage-specific duplications
| CDS | Most like | Common name |
|---|---|---|
| CDS347 | CDS348 | T4 gp12, short tail fiber protein |
| CDS055 | CDS262 | T4 gp30.2, conserved hypothetical protein |
| CDS058 | CDS115 | Hypothetical proteins |
| CDS058 | CDS116 | Hypothetical proteins |
| CDS061 | CDS195 | Mycobacteriophage TM4 gp80 related |
| CDS238 | CDS239 | Hypothetical proteins |
| CDS252 | CDS270 | Hypothetical proteins |
| CDS260 | CDS272 | Molybdenum cofactor biosynthesis protein A related |
| CDS261 | CDS272 | Molybdenum cofactor biosynthesis protein A related |
In the remaining analysis of the KVP40 genome, we compared the various CDS functions to those of the well-characterized phage T4 (Table 3). Nonetheless, 253 CDSs were unique to KVP40, and it is their identity and biochemical properties that present the greatest challenges emerging from the genomic sequence.
TABLE 3.
KVP40 orthologs to T4 productsa
| Function | CDS or gene (no. of aa)
|
Descriptionb | % Identity % similarity (comments) | |
|---|---|---|---|---|
| KVP40 | T4 | |||
| Transcription | 002 (90) | dsbA (89) | DNA binding | 36/62 |
| 003 (98) | 33 (112) | Late activator | 32/55 | |
| 064 (170) | 55 (185) | Late sigma | 44/64 | |
| 077 (221) | 45 (228) | Late activator | 28/48 | |
| 295 (99) | asiA (90) | Anti-sigma 70 | 32/55 | |
| Translation, RNA | 080 (132) | regA (122) | Repressor | 57/71 |
| 084 (381) | rnlA (374) | RNA ligase 1 | 43/62 | |
| 090 (305) | pseT (301) | Polynucleotide kinase | 33/53 | |
| 133 (335) | mlB (334) | RNA ligase 2 | 33/52 | |
| tRNAs (30) | tRNAs (8) | tRNA | ||
| Nucleotide metabolism | 006 (181) | frd (193) | Dihydrofolate reductase | 32/54 |
| 012 (611) | nrdD (606) | NTP reductase | 53/68 (no intron) | |
| 015 (158) | nrdG (156) | NTP reductase | 50/71 | |
| 031 (300) | td (286) | Thymidylate synthase | 41/57 (no intron) | |
| 062 (79) | nrdH (102) | Glutaredoxin | 25/45 | |
| 119 (150) | cd (193) | dCMP deaminase | 53/67 | |
| 190 (194) | tk (193) | Thymidine kinase | 43/61 | |
| 277 (742) | nrdA (754) | NDP reductase | 58/72 | |
| 278 (374) | nrdB (388) | NDP reductase | 52/75 (no intron) | |
| 279 (99) | nrdC (87) | Thioredoxin | 33/57 | |
| 324 (212) | 1 (241) | dNMP kinase | 28/45 | |
| Replication, recombination, repair | 001 (335) | rnh (305) | RNase H | 46/65 |
| 004 (216) | 59 (217) | Helicase loader | 33/59 | |
| 005 (304) | 32 (301) | SSB | 43/58 | |
| 008 (366) | uvsX (390) | RecA-like protein | 57/74 | |
| 010 (466) | 41 (475) | DNA helicase | 49/69 | |
| 020 (352) | 61 (342) | Primase | 40/58 | |
| 023 (230) | dexA (227) | Exonuclease A | 37/59 | |
| 045 (601) | 39 (516) | Topoisomerase II | 48/66 | |
| 057 (447) | 30 (487) | DNA ligase | 38/52 | |
| 072 (346) | 47 (339) | Recombination nuclease | 40/62 | |
| 074 (745) | 46 (560) | Recombination nuclease | 34/52 (T5 gpD13-like) | |
| 077 (221) | 45 (228) | Clamp | 28/48 | |
| 078 (318) | 44 (319) | Clamp loader | 39/60 | |
| 079 (163) | 62 (187) | Clamp loader | 31/50 | |
| 082 (850) | 43 (898) | DNA polymerase | 44/60 | |
| 131 (151) | 49 (157) | Recombination endonuclease VII | 39/61 | |
| 156 (421) | dda (439) | DNA helicase | 39/55 | |
| 205 (138) | denV (138) | N-Glycosidase | 41/56 | |
| 282 (428) | 52 (442) | Topoisomerase II | 26/43 | |
| 378 (507) | uvsW (587) | RNA-DNA helicase | 55/73 | |
| 382 (144) | uvsY (137) | UvsX assistant; SSB | 36/54 | |
| Head | 063 (298) | 24 (427) | Vertex precursor | 28/45 |
| 330 (198) | 2 (274) | DNA end binding | 46/62 | |
| 331 (151) | 4 (150) | Head completion | 58/71 | |
| 350 (307) | 13 (309) | Head completion | 44/63 | |
| 351 (278) | 14 (256) | Head completion | 45/63 | |
| 354 (219) | 16 (495) | Terminase subunit | 40/58 | |
| 355 (601) | 17 (610) | Terminase subunit | 53/68 | |
| 358 (515) | 20 (524) | Portal vertex protein | 46/64 | |
| 359 (55) | 67 (80) | Prohead core protein | 22/43 | |
| 360 (163) | 68 (141) | Prohead core protein | 38/58 | |
| 361 (213) | 21 (212) | Prohead protease | 49/61 | |
| 362 (283) | 22 (269) | Prohead core protein | 35/57 | |
| 363 (514) | 23 (521) | Head protein | 60/73 | |
| 367 (163) | inh (226) | Head protease inhibitor | (Little similarity) | |
| Tail, tail fiber | 298 (1085) | 37 (1026) | Distal long tail fiber | (Little similarity) |
| 298-2 (1094) | 37 (1026) | Distal long tail fiber | (Duplicated) | |
| 325 (177) | 3 (176) | Tail sheath stabilizer | 33/52 | |
| 328 (282) | 26 (208) | Baseplate hub | 26/46 | |
| 329 (248) | 19 (163) | Tail tube | 20/41 (duplicated) | |
| 332 (379) | 48 (364) | Baseplate-tube cap | 19/42 | |
| 333 (192) | 53 (196) | Baseplate wedge | 27/53 | |
| 335 (421) | 5 (575) | Baseplate hub | 40/58 | |
| 340 (139) | 25 (132) | Baseplate wedge | 39/68 | |
| 341 (646) | 6 (660) | Baseplate wedge | 45/63/PICK> | |
| 342 (1165) | 7 (1032) | Baseplate wedge | 36/55 | |
| 343 (343) | 8 (334) | Baseplate wedge | 42/61 | |
| 344 (327) | 9 (288) | Baseplate; socket | 22/43 | |
| 345 (748) | 10 (602) | Baseplate; pin | 30/45 | |
| 346 (234) | 11 (219) | Baseplate; pin | 24/43 | |
| 347 (512) | 12 (527) | Short tail fiber | 25/46 (duplicated) | |
| 348 (473) | 12 (527) | Short tail fiber | 28/46 (duplicated) | |
| 349 (559) | wac (487) | Whiskers | 24/43 | |
| 352 (450) | 15 (272) | Tail sheath stabilizer | 43/60 | |
| 356 (671) | 18 (659) | Tail sheath | 48/63 | |
| 357 (166) | 19 (163) | Tail tube | 56/74 (duplicated) | |
| 383 (894) | 35 (372) | Fiber hinge | 30/50 | |
| 384 (1290) | 34 (1289) | Proximal long tail fiber | 36/49 | |
| Chaperonins, catalysts | 084 (381) | rnlA (374) | Fiber attachment | 43/62 |
| 129 (112) | 31 (111) | Cochaperonin | 37/61 | |
| Lysis, exclusion | 143 (689) | rIIA (725) | Lysis inhibition | 28/48 |
| 144 (345) | rIIB (312) | Lysis inhibition | 43/67 | |
| 146 (231) | segD (223) | Endonuclease | 39/55 | |
The KVP40 CDS number and the orthologous T4 gene are given, followed by the number of amino acids (aa) in parentheses. Percent similarities and identities were obtained from the WU-BlastP output with the BLOSUM62 matrix.
NTP, nucleoside triphosphate; NDP, nucleoside diphosphate; dNMP, deoxynucleoside monophosphate; SSB, single-strand DNA binding protein.
Transcription proteins, promoters, and terminators.
The temporal control of gene transcription (early, middle, and late) in T-even phages derives specificity from modifications to the host RNA polymerase (which is used throughout infection), phage-encoded transcription factors, and unique promoter sequences (27). Interestingly, KVP40 lacks homologs of the Alt, ModA, and ModB enzymes. In T4, these enzymes ADP-ribosylate many proteins, including the RNA polymerase α subunits (59, 60). Compared to most σ70 promoters, T4 early promoters have an extended −10 region, a −35 region (GTTTAC) that differs from the generalized σ70 −35 (TTGACa), and an A-rich upstream (−42) region (reviewed in reference 44). Pattern searches failed to reveal characteristic T4-like early promoters, although many E. coli-like σ70 promoters were found throughout the genome (not shown). Therefore, it appears that KVP40, which also does not encode the enzymes for cytosine modification of phage DNA (see below), uses an early transcription apparatus that is not distinguished from that of the host.
The transcriptional activator protein MotA, which recognizes a sequence centered at −30 relative to the transcription start site, directs middle-mode transcription in T-even phages (19, 55). However, KVP40 lacks a T4 MotA homolog. Another T4 transcription factor, the RNA polymerase-associated protein RpbA, is also absent. The exact-match MotA binding sites that were identified all occurred in coding regions not associated with other promoter elements. Although a different MotA-like activator with a unique recognition sequence may function in KVP40, the data suggest that the well-characterized middle-mode transcription system of T4 does not function in KVP40. During the course of this work, a similar absence of clear T4-like early and middle promoter sequences was also observed in the genome scan of enterobacteriophage RB49 (8). Alternative strategies for the transcription cycle appear to be used by some of the more distantly related T4-like phages.
In contrast, we can infer that the characteristic T4-style late-mode transcription does function in KVP40. Matsuzaki (38, 40) previously identified T4-like late promoter sites in the major capsid gene region of KVP40; including these, we have identified at least 13 exact-match promoter sequences (5′-TATAAATA-3′) in the KVP40 genome (Table 4). By using minor variations in the sequence, additional late promoters were identified (T4 has 50 late promoters in its 170-kbp genome). The important proteins for late transcription (12, 29, 56), σ55 (phage sigma factor), gp33, DsbA (RNA polymerase-associated proteins), and gp45 (DNA polymerase associated sliding clamp), were all identified in KVP40. It will be interesting to determine whether transcription initiation at KVP40 late promoters is coupled to the activity of the DNA replisome as it is in T4, in light of the reduced RNA polymerase modifications and apparent lack of cytosine modification in the KVP40 DNA template.
TABLE 4.
Representative T4-like late promoters
| Late promoter (strand) | Positiona | 3′ gene start position | 3′ CDS, gene |
|---|---|---|---|
| TATAAATA (+) | 1927 | 2260 | 005, 32 |
| 31657 | 31688 | 058 | |
| 36239 | 36306 | 063, 24 | |
| 89972 | 90005 | 154 | |
| 195983 | 196014 | 340, 25 | |
| 210159 | 210206 | 349, wac | |
| 218048 | 218093 | 356, 18 | |
| 223557 | 223594 | 362, 22 | |
| 224461 | 224507 | 363, 23 | |
| 235271 | 235321 | 378, uvsW | |
| TATAAATA (−) | 85800 | 85768 | 146, segD |
| 161037 | 160989 | 297 | |
| 173437 | 173411 | 311 | |
| aATAAATA (+) | 48436 | 48472 | 080, regA |
| 137598 | 137878 | 263 | |
| 146516 | 146553 | 277, nrdA | |
| 202996 | 203026 | 344, 9 | |
| 229081 | 229119 | 367, inh | |
| aATAAATA (−) | 167683 | 167653 | 298-2, 37b |
| 183476 | 183228 | 323 | |
| TATgAATA (+) | 53849 | 53873 | 086 |
| 58551 | 59687 | 098 | |
| 97052 | 97053 | 168 | |
| 99533 | 99575 | 174 | |
| 191863 | 192498 | 335, 5 | |
| TATgAATA (−) | 184373 | 183876 | 324, 1 |
| TATAAAg/cA (−) | 77123 | 77035 | 131, 49 |
| 85787 | 85768 | 146, segD | |
| 88095 | 87992 | 150 | |
| 178037 | 177990 | tRNA-Leu3 | |
| 178120 | 177990 | tRNA-Leu3 | |
| TTTAAATA (−) | 63226 | 63072 | 103 |
| 168281 | 168239 | 299 |
The position cited is the first base of the promoter sequence. The position of the first gene that would be on the transcript is given, along with its CDS number and name (if known). Lowercase indicates difference from most common sequence (uppercase).
AsiA, the anti-σ70 protein that has been primarily characterized as enhancing MotA-dependent middle-mode transcription in T4, was identified in KVP40 (CDS295). Like T4 AsiA (53), KVP40 AsiA binds σ70 and inhibits transcription by E. coli RNA polymerase (D. Hinton, personal communication). Additional analysis of the sequences and kinetics of promoter recognition (also see reference 29), in light of the apparent differences in phage transcription factors, would be interesting to pursue. KVP40 also encodes transcriptional regulatory proteins (see below) usually associated with the bacterial host, and the possibility exits that these are active in transcribing specific phage genes.
Rho-independent transcription terminators (Fig. 1) (10) were identified and grouped by sequence similarity (80% identity over 80% of the sequence). These 57 rho-independent terminators grouped into 14 singles and 11 sets that contained more than one sequence (Table 5). The major “tetraloop” terminators (e.g., UUCG and GNRA) are present in KVP40. In most instances when RNA polymerase transcribing off one strand would encounter the enzyme transcribing one or more genes off the opposite strand, a terminator is located in the intergenic region. The sequence characteristics of such terminators predict that they function on both strands.
TABLE 5.
Predicted intrinsic transcription terminators
| Familya | Positionb | Sequencec | Tetraloopd | Locuse | Gene |
|---|---|---|---|---|---|
| 1 | 39856 | ATTTATTTGAATAAAAGGGTTGCTTCGGCAACCCTTTTTGCGTATTATAGC | UUCG | 066 | h |
| 1 | 54582 | AAATAATTAAAGAAAAGGGTTGCTTCGGCAGCCCTTTTCTCGTATTATAGC | UUCG | 087 | h |
| 1 | 68356 | TGGATATTAATCAGAAAGGTAGCTTCGGCTACCTTTTTTCGTTTTCAGAGC | UUCG | 116 | h |
| 1 | 109856 | AAATAACCAAAGAAAGGGGTTGCTTCGGCAGCCCTTTTTGCGTATTATAGC | UUCG | 199 | h |
| 1 | 139742 | AAAAATCGTAAATTTAGGGTTGCTTCGGCAGCCCTTTTTTCGTATTATAGC | UUCG | 264 | nadV |
| 1 | 226081# | GAAACAGACAACAAAGGGGACTCTTCGGAGTCCCTTTTTGCGTTTATAGAC | UUCG | 363 | 23 |
| 1 | 233033(−) | TTAACCAACTGTTAAAGGGTGACTTCGGTCGCCCTTTTTTCGTTTACGTCT | UUCG | 373 | h |
| 2 | 9426 | TTAAACACTGTTAAGAAAGCGCCTTCGGGCGCTTTTTTTGTTTTTGTCGGA | UUCG | 012 | nrdD |
| 2 | 72687 | TTTCAAAGAACAAGAATAGCGCCTTCGGGGCGCTCTTTATGAGGTAAACATG | UUCG | 122 | h |
| 2 | 90272 | TAACACAACGAACACAAAGCGCCTACGGGCGCTTTTTTTATGCAAGGAAGG | UACG | 154 | h |
| 2 | 158804 | CGAAACTCAATTAACAAAGCGCCTCCGGGCGCTTTTTTTGTTTAGAACTCC | UCCG | 293 | ch |
| 3 | 21144 | TATTCTTGAAGAAAAGGGTTGACTGCGGTCAGCCCTTTCGCGTATTATAGC | (UGCG) | 037 | h |
| 3 | 59696 | CGAATAATGAAAAAAGGGTTGACAGATGTCAGCCCTTTCGTGTATTCTACG | (AGAU) | 097 | h |
| 3 | 97797 | AAAAGTAATAAAAAAGGGTTGACAGATGTCAGCCCTTTCGCGTATTATACA | (AGAU) | 169 | h |
| 3 | 100191 | CGTAATTGAAGAAAAGGGTTGACGCTTGTCAGCCCTTTCTTGTATTATCTC | (GCUU) | 174 | h |
| 3 | 121374 | AAAACTTTTCGAAAAGGGTTGACTACGGTCAACCCTTTCTTGTATTATAGC | UACG | 231 | h |
| 3 | 148817 | TTTCAATAAAAATAAGGTTGACCATCTTGGTCAACCTCTTTTCTTGGGCGA | 277 | nrdA | |
| 3 | 150224 | ATAATTAAAGAAAAGGGTTGAACAGATGTCAGCCCTTTTTTGTATGCTTCG | (AGAU) | 279 | nrdC |
| 4 | 31635 | GTTCAAGGGTTAATAAAAGGGGCGAAAGCCCCTTTTTTCGTATAAATACTT | GAAA | 057 | 30 |
| 4 | 48888 | ACTGGTTTTAATGAATAAGGGGCATTCGCCCCTTTTATTTGAGGAATACAC | (AUUC) | 080 | regA |
| 4 | 62011(−)# | CTACAGACCATAAAAAAAGGGGCGAATGCCCCTTTTTTTATTCTTTTTCGC | (GAAU) | 103 | h |
| 4 | 154605 | ATTTGGGGTAACGCGGTGGGCACGTAAGTGCCCTTTTATTCCCTTTGAAGT | GUAA | 284 | h |
| 5 | 35862 | CCTGTAATCTCTCATTTAGCCCCGAAAGGGGCTTTTTTAGGTTTGAGCCGG | GAAA | 061 | ch |
| 5 | 184096(−) | GTTAGACACAAGCAATAAGCCCCTATCGGGGCTTTTTTTGTATCTGAATCT | (UAUC) | 325 | 3 |
| 5 | 238328(−) | TACGCAATGCAATAAGAAAGCCCTTCGGGGCTTTTTTTATACGCGAAGCAA | UUCG | 383 | 35 |
| 6 | 32575# | TCGATTGAATAAGAAAAGGGAGCAAATTGCTCCCTTTTTTGATCATGGTGT | 058 | h | |
| 6 | 76557# | AGTATAGTTACACAAAAGGGAGCTAATTGCTCCCTTTTTGCTATTCATCAT | 130 | h | |
| 6 | 87419# | ACGCAGAAAATAAAAAAGGGAGCATTCGCTCCCTTTTTGCTATTTGTTTTT | (AUUC) | 149 | h |
| 6 | 143423# | AATGGTGAAATGAGAAAGGGAGCTTAGTGCTCCCTTTTTTATAACCAATAA | 271 | h | |
| 6 | 236858# | ATAATAGATACGAAAAAGGGAGCAATTTGCTCCCTTTTTTATTAGTCAGAA | 378 | uvsW | |
| 7 | 65690 | ATCACAGACGCAATTTAAGCCGCATTTCGCGGCTTTTTGAGGTTCATATGA | 111 | h | |
| 7 | 229959# | AACGCCATAATAGAAAAGCCCGCATTTAGCGGGCTTTTTTGTGTCTACGAG | 368 | h | |
| 7 | 231296(−) | CCTGATTGATAATTAAAAAGCGCATTTTGCGCTTTTTTTGTATCTATTGGT | 371 | h | |
| 8 | 78629 | GAAGATGAAAACAACAAAGCCCCTTAATTGGGGCTTTTTTATGGGTGAAAG | 134 | h | |
| 8 | 107118 | ACTATTGACTTATCCAGAGCCCCTTAATTGGGGCTTTTTTAGGTCTGAAGA | 194 | h | |
| 8 | 126416 | AGACTGGCGTTAATAAAAGCCCCTTAATTGGGGCTTTTTTAGGTTTGATGA | 243 | h | |
| 8 | 143013 | GACTTGAATCACATAAAAGCCCCTTAATTGGGGCTTTTTTCGTTTTAGCGG | 270 | h | |
| 9 | 3179 | CGATTAATGAATGGAAAGGGGCTTAACAGCCCCTTTGTTCTCTCTATAGGA | UAAC | 005 | 32 |
| 9 | 46933 | CTAGATTTTGTAAGTTACGGGGCGTAACAGCCCCTTTTTTTGAATATTTTA | 077 | 45 | |
| 10 | 5657 | CTAATACCCACGAGACCCGTTGATTTGTCAGCGGGTCTTTTAGCCATTTGA | (UUUG) | 008 | uvsX |
| 10 | 209948 | TCAATGCTAATAAACGGGTGAGATTGATTTCTCACCCTTTTTTGTATGAGC | 348 | 12 | |
| 11 | 61307 | AATAGTTGAATAAAAAGGGTTGCACTGTGCAGCCCTTTTCTATATCATGAG | 101 | h | |
| 11 | 128903 | ATTAATTAAAGAAAAGTGGTTGCTCTATGCAGCCACTTTCTGTATTATAGC | 249 | h | |
| Unrelated | 13732 | CTTCGACTAATTAAAATGCCTCGTTTTCGGGGCTTTTTTATGTTTATCGTA | UUUU | 020 | h |
| 85042(−) | GTCTTTCCAATGTAGAACCCGTCTTCTCGACGGGTTACCACATGAATCTCA | 146 | segD | ||
| 117954 | CCATCGTATGTTTTTTAAGACGCTTCGGCGTCTTTTTTTCGGTTTGAAGAA | UUCG | 221 | h | |
| 132112 | TCCGATGCAGACAACGTCCGTGCCATGTGTATGGACGTTGTTCGTTTATTG | 255 | h | ||
| 138122 | ACAGCTTAATTGAAATGTGCAGATTCGTCTGCACATTTTGTTTAATAACGA | UUCG | 263 | h | |
| 160380# | ACGAATAAAGAAAAGGGAGAGTACAACTGTTACTCTCCCTTTTTTTGGTCA | 296 | |||
| 167720(−) | ATAAGGCAAATCGTAAAGGGCGCAAATAGCGCCCTTTTTTACGGCATAATA | 299 | ch | ||
| 168272(−) | TTTAAATAAGTGATAAGGGCGGCATTGTGCTGCCCTTTTTTGTACAGAGGT | 300 | ch | ||
| 171605# | TAAGCTATAAAAGAAAAGCCCCGTTTTTGGGGCTTTTTTGTTTAATGTGAG | UUUU | 305 | h | |
| 179037(−)# | AAGAAAATCGCAGAAAGTTGAAGTTTTTTTCAATTTTCTGCGAAAATACCG | UUUU | 316 | h | |
| 194945(−) | TTAAATAATCAAAACGGGGCGGCTATGTCGCCCCTTTGTTTCACACGAAAT | 339 | h | ||
| 196440 | ATCGTCTTTAATGAACAGCCCGCCATTGTGCGGGCTTTTTTGTGCATTAAA | 340 | 25 | ||
| 203021 | AAGTAAACAACTCATTAGGCGCGTGTTCGCGCCTTTCTTGGAGATTAAGAT | (UGUU) | 343 | 8 | |
| 215638 | ATGCTTATTTTAACGCTGTGTGCAGCTCGTACGCAGCGTTTGTCTATTTTA | 353 | h |
Terminator family of related sequence (80% identity over 80% of the sequence).
Genome position of the first base of the hairpin. (−), present on the minus strand; #, likely to function on both strands, located between convergently transcribed genes.
Underlined bases are predicted to be in the helix. G:U pairs are allowed.
Terminator-stabilizing tetraloop sequences, when present (those in parentheses are not commonly observed).
Locus refers to the preceding 5′ KVP40 CDS; name refers to the orthologous phage T4 gene aligning with the 5′ locus, except nadV. ch, conserved hypothetical; h, hypothetical.
Nucleotide skew analysis (11, 48) on the KVP40 genome yielded results similar to those for T4 (26); transitions in intrastrand nucleotide biases correspond to a switch in the direction of transcription for gene clusters, such as that between CDS296 and CDS297 (data not shown).
Translation functions and RNAs. (i) Initiation codons, tRNAs, and codon usage.
Among the nonhypothetical KVP40 CDSs, most (89.8%) are initiated with an AUG, while other CDSs initiate with GUG (4.7%), and others use the rare initiator UUG (5.5%). Interestingly, the genome of the KVP40 host V. cholerae (16) is currently annotated with the surprisingly high frequency of 14.4% nonhypothetical genes starting with UUG. It may be that for some of the KVP40 genes starting with UUG, the correct initiation codon is not properly identified, or that KVP40 genes show an adaptation to using a hostlike translation initiation codon not commonly used in E. coli. In contrast, bacteriophage T4 almost exclusively uses AUG as the initiation codon for its 274 genes (44). GUG as an alternative start codon is used for only eight genes (3%) by T4 (40).
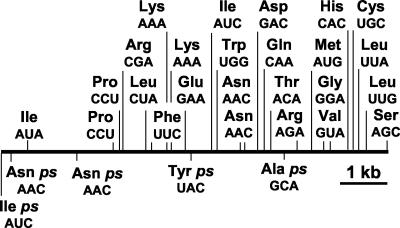
A striking feature of the KVP40 genome is the presence of 25 apparently functional tRNAs and five pseudo-tRNAs in a single region (bp 173138 to 181214; Table 6 and Fig. 2). Such large tRNA clusters are uncommon in sequenced prokaryotic genomes (the Bacillus subtilis 168 chromosome has a single tRNA cluster of 21 tRNAs). Bacteriophage T4 encodes eight tRNAs in one region, which supplement host isoacceptor tRNA species that are present in minor amounts and recognize codons that occur more frequently in the phage genes (33). This situation is not apparent for KVP40. Codon usage for the CDSs of KVP40 and V. cholerae and codons recognized by the KVP40 tRNAs are shown in Table 6. There is biased codon usage for some amino acids for which KVP40 encodes a tRNA (i.e., PheTTC, LeuCTA, ThrACA, AsnAAC, ArgAGA, LysAAA, AspGAC, ValGTA, and GluGGA), but there are also codon usage differences between V. cholerae and KVP40 when there is no corresponding tRNA in KVP40 (i.e., LeuCTT, ValGTT, SerTCA, ThrACT, AlaGCA, TryTAC, and GlyGGT). In other instances, even though KVP40 encodes a tRNA for an alternate codon, there is a trend to greater use of V. cholerae codons (i.e., LeuTTA, LeuTTG, and GlnCAA), or there is no apparent preference (i.e., HisCAC and SerAGC). These observations may reflect adaptation to a broad host range by KVP40 or indicate that further analysis of codon usage in specific gene sets (i.e., early versus late genes; replication versus virion gene sets; and high- versus low-expressed genes) will reveal specialized regulatory processes.
TABLE 6.
Codon usage in phage KVP40 and one of its hosts, V. choleraea
| First nt | 2nd nt T | aa | Usage (%)
|
2nd nt C | aa | Usage (%)
|
2nd nt A | aa | Usage (%)
|
2nd nt G | aa | Usage (%)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V. cholerae | KVP40 | V. cholerae | KVP40 | V. cholerae | KVP40 | V. cholerae | KVP40 | |||||||||
| T | TTT | Phe | 26.80 | 17.9 | TCT | Ser | 11.31 | 12.6 | TAT | Tyr | 15.76 | 18.0 | TGT | Cys | 6.01 | 8.7 |
| TTC | Phe | 13.89 | 24.5 | TCC | Ser | 5.96 | 1.0 | TAC | Tyr | 13.73 | 23.2 | TGC | Cys | 4.51 | 4.0 | |
| TTA | Leu | 19.96 | 11.1 | TCA | Ser | 10.69 | 17.5 | TAA | Stop | 2.02 | 3.4 | TGA | Stop | 0.73 | 1.7 | |
| TTG | Leu | 23.49 | 16.4 | TCG | Ser | 9.34 | 8.7 | TTG | Stop | 0.57 | 0.07 | TGG | Trp | 13.21 | 13.4 | |
| C | CTT | Leu | 12.78 | 18.6 | CCT | Pro | 11.11 | 7.7 | CAT | His | 13.12 | 11.3 | CGT | Arg | 19.70 | 21.1 |
| CTC | Leu | 14.90 | 4.7 | CCC | Pro | 5.99 | 2.1 | CAC | His | 10.76 | 11.7 | CGC | Arg | 17.45 | 12.6 | |
| CTA | Leu | 8.80 | 15.8 | CCA | Pro | 12.20 | 11.6 | CAA | Gln | 33.51 | 23.2 | CGA | Arg | 5.18 | 8.8 | |
| CTG | Leu | 26.75 | 10.3 | CCG | Pro | 10.66 | 9.3 | CAG | Gln | 18.10 | 10.9 | CGG | Arg | 2.76 | 1.1 | |
| A | ATT | Ile | 31.05 | 32.6 | ACT | Thr | 12.87 | 23.4 | AAT | Asn | 19.62 | 22.2 | AGT | Ser | 11.86 | 10.3 |
| ATC | Ile | 24.94 | 28.0 | ACC | Thr | 20.06 | 3.9 | AAC | Asn | 19.33 | 28.0 | AGC | Ser | 10.06 | 9.9 | |
| ATA | Ile | 4.24 | 5.6 | ACA | Thr | 7.79 | 19.4 | AAA | Lys | 35.84 | 42.2 | AGA | Arg | 2.93 | 6.0 | |
| ATG | Met | 26.22 | 28.0 | ACG | Thr | 11.04 | 13.3 | AAG | Lys | 13.33 | 23.8 | AGG | Prg | 11.14 | 0.7 | |
| G | GTT | Vale | 16.00 | 28.3 | GCT | Ala | 20.04 | 22.5 | GAT | Asp | 36.35 | 33.5 | GGT | Gly | 25.84 | 32.4 |
| GTC | Val | 14.43 | 9.8 | GCC | Ala | 21.67 | 4.0 | GAC | Asp | 13.56 | 32.5 | GGC | Gly | 24.18 | 14.4 | |
| GTA | Val | 11.02 | 17.3 | GCA | Ala | 18.84 | 28.2 | GAA | Glu | 37.89 | 52.8 | GGA | Gly | 7.86 | 5.2 | |
| GTG | Val | 29.02 | 13.2 | GCG | Ala | 30.50 | 14.4 | GAG | Glu | 23.78 | 21.6 | GGG | Gly | 8.75 | 5.8 | |
tRNAs found in KVP40 are underlined. AsnAAC, ProCCA, and LysAAA are found twice. V. cholerae usage is shown in the first column, and KVP40 usage is shown in the second. The Met tRNA is the elongator species, and the IleATA specificity is predicted to arise from k2C34 modification (lysidine) in the CAU anticodon. aa, amino acid; nt, nucleotide.
FIG. 2.
KVP40 tRNA gene cluster. tRNAs were identified with tRNAscan-SE (36). ps, possible pseudo-tRNA. The codon recognized is shown below each tRNA. The tRNA cluster extends from nucleotides 173138 to 181214, all encoded on the negative strand.
KVP40 encodes a putative tRNA nucleotidyltransferase (CCA-adding enzyme; CDS364) that could function in the maturation of its tRNAs. The enzyme has 61% similarity over the first 227 residues to the Haemophilus influenzae protein and those from other bacteria. Like other CCA-adding enzymes, it appears to be related to poly(A) polymerases in the N-terminal region. However, the C-terminal 138 residues of CDS364 diverge considerably from those of the other proteins. All of the KVP40 tRNAs are predicted to have a genomically encoded 3′ CCA.
RNA repair processes (see reference 44 for a review) are also encoded by the KVP40 genome, including the T4-like polynucleotide kinase (CDS090, pseT) and two RNA ligases (CDS084, rnlA; and CDS133, rnlB). The abundance of tRNAs encoded by KVP40 suggests that a tRNA repair pathway would be beneficial to the phage in various hosts.
The KVP40 genome appears to be devoid of group I intron RNAs, although at least three have been identified in T-even phages and more yet in Staphylococcus phage Twort (35). Additional RNA pattern-searching tools are needed to more clearly identify and annotate RNA introns.
(ii) RNA-binding proteins.
Three well-characterized T4 translational repressor proteins, gp32 (SSB), gp43 (DNA polymerase), and RegA, are encoded by the KVP40 genome. The first two proteins are essential DNA replication proteins and autoregulate translation initiation rates from their own mRNAs. RegA, although not essential during laboratory growth of T4, binds and translationally regulates mRNAs for several DNA replication enzymes (43). Although the known role (regulation) of RegA appears to be less critical than that of gp32 and gp43, KVP40 regA is at the same location within the replication gene cluster as in T4, and the protein is more similar (70%) to its T4 homolog than are the other two proteins (64 and 59%, respectively). The well-characterized RNA pseudoknot and translational control region of T4 gene 32 (43) is not apparent for KVP40 gene 32, an observation also noted for the gene 32 mRNA of coliphage RB49 (8). The mRNA RNase RegB was not identified in the KVP40 genome, but RNase H (CDS001) was present, and there was a putative RNA-binding protein (CDS154) with a tyrosine-rich RNA-binding motif.
KVP40 DNA metabolism, replication, recombination, and repair. (i) DNA metabolism.
KVP40-directed DNA metabolism is predicted to be highly similar to that of phage T4, with a few possibly important differences. T4 nucleotide metabolism is noteworthy for the phage-encoded pyrimidine biosynthetic enzymes and the presence of the modified base 5-hydroxymethyl-deoxycytosine, which is also extensively glucosylated. The deoxynucleoside triphosphate pool for phage DNA replication arises from phage-encoded enzymes that act on ribosomal nucleoside diphosphates obtained from the host and salvaged from mRNA decay, and on deoxynucleoside monophosphates arising from host DNA degradation (13). Although KVP40 encodes several nucleases and enzymes of nucleotide precursor synthesis, there is no evidence for the existence of T4-like host nucleoid disruption proteins and enzymes for host DNA degradation (34). Homologs to the T4 Alc (alc), Ndd (ndd), endonuclease II (denA), and endonuclease IV (denB) proteins are also absent in KVP40. Therefore, it appears that KVP40 is less efficient than T4 in degrading host DNA.
From a practical perspective, KVP40 may be an effective generalized transducing phage for the bacteria included in its broad host range. If the phage indeed lacks the aforementioned enzymes, it would closely resemble the T4gt mutant strains that do not extensively degrade DNA and therefore transduce large intervals of the host chromosome (64). KVP40 does have exonucleases encoded by the dexA, 46, and 47 genes, which may have multiple functions, including host DNA degradation and phage DNA replication and recombination (Table 3; see below).
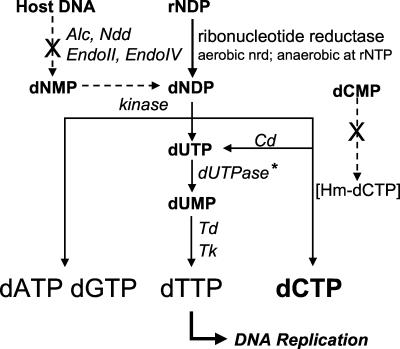
KVP40 encodes most of the characterized enzymes for deoxynucleoside triphosphate conversions and pyrimidine synthesis. Both aerobic (nrdABC) and anaerobic (nrdDGH) ribonucleotide reductase complexes for converting ribonucleotides to deoxyribonucleotides are present. For the more complex pyrimidine pathway, the dUTPase gene (dut) is present, but it is more closely related to bacterial enzymes than to the T4 gene 56-encoded dCTPase/dUTPase enzyme. This agrees with the observation (below) that KVP40 appears to incorporate dCTP and not hydroxymethyl-dCTP into its DNA. The rest of the pyrimidine pathway enzymes are present, including dCMP deaminase (cd), dTMP synthase (td), dihydrofolate reductase (frd), deoxynucleoside monophosphate kinase (gene 1), and thymidine kinase (tk), all of which indicate that KVP40 directs the conversion of ribonucleotides to deoxyribonucleotides and handles conversion of dCMP to dTTP (Table 3 and Fig. 3).
FIG. 3.
Probable KVP40 nucleotide metabolism pathway. KVP40 lacks genes for several enzymes for host DNA breakdown and synthesis of modified cytosine (glucosylated hydroxymethyl [Hm]-deoxycytosine), which are marked by X. The anaerobic ribonucleotide reductase (Nrd) likely acts on ribonucleoside triphosphates (rNTP), like the T4 enzyme. KVP40 encodes a host-like dUTPase (CDS021) rather than the bifunctional T4-like dCTPase/dUTPase. Other abbreviations: Cd, dCMP deaminase; Endo, endonuclease; dNMP, deoxynucleoside monophosphate; dNDP, deoxynucleoside diphosphate; Td, thymidylate synthase; Tk, thymidine kinase. All other nomenclature is as for T4. CDS assignments are listed in Table 3.
Absent among the precursor enzymes are those for cytosine modification, including the dCMP hydroxymethylase which, in T4, plays an important role in protecting the phage DNA from restriction, is thought to be central for assembling the proposed multienzyme deoxynucleoside triphosphate synthetase complex (13), and generates the hydroxymethyl-dCTP that is targeted for glucosylation in the genome. Although the three T-even phages have glucosylated DNA, the α-gt and β-gt genes are absent from KVP40. All of this suggests that there is limited cytosine modification by KVP40. The sensitivity (41) of KVP40 DNA to several restriction endonucleases that do not digest T4 DNA corroborates evidence for the absence of these enzymes in KVP40. The adenosine methylase (dam) that is present in T4 and T2 but absent in T6 is also absent in KVP40.
(ii) Replication, recombination, and repair enzymes.
DNA replication initiates via multiple origins and modes in the T-even phage, so the identification of these elements is not as straightforward as for the Bacteria (31). Replication origins have not yet been identified on the KVP40 genome. However, the T4 replisome is one of the best-characterized DNA replication machines from any organism (2, 5, 25). Many of the T4 DNA replication enzymes are encoded by a single, contiguous gene cluster that is >20 kbp in length. In KVP40, the replication gene cluster is one of two highly conserved regions that are nearly identical to those in the T4 genome. This ca. 10-kbp region of synteny (Fig. 1) includes genes 47, 46, 45, 44, 62, regA, and 43, although some of the smaller CDSs (e.g., 45.2, rpbA, and 43.1) are variable in occurrence and position, as has been observed in the T4-related RB phages (42, 63).
Genes distributed throughout the KVP40 genome encode enzymes directly involved in DNA replication (Table 3) and have been well characterized from T4. Many have overlapping roles in recombination and DNA repair. The helicase (gene 41), helicase loader (gene 59), primase (gene 61), RNaseH (rnh), SSB protein (gene 32), polynucleotide kinase (pseT), DNA ligase (gene 30), DNA helicase (dda), and others are all present in the genome. Similar to phage T2, KVP40 DNA topoisomerase genes 39 and 52 encode the two complete subunits of the enzyme; in T4, the larger gp39 topoisomerase subunit is split (20, 30). The recombination enzyme UvsX as well as UvsY, UvsW, and the heteroduplex resolvase (endonuclease VII; gp49) are all encoded by KVP40. In addition, KVP40 encodes a homolog of the T4 pyrimidine dimer N-glycosidase (endonuclease V; denV) that is used for the repair of UV irradiation-induced DNA damage.
Phage capsid and tail genes.
The KVP40 virion is encoded by the largest single gene cluster, having a gene content and gene order similar to that found in T4 (Fig. 1). The 43.5-kb interval extends from the T4 gene 57B (KVP40 CDS322) through the major capsid protein gene 23 (CDS363). The sequence presented for the gene 16 to gene 23 region agrees with the 10.5-kb sequence previously reported by Matsuzaki and colleagues (38, 40). Although the virion gene order is nearly identical between T4 and KVP40, there are distinctions. First, the proteins that decorate the outside of the capsid, encoded by hoc and soc, and the internal proteins ipI, ipII, and ipIII all appear to be absent from KVP40. In KVP40, gene 12 (the short tail fiber) is tandemly duplicated, and there is a second gene 19-like CDS (the tail tube) 32 kbp removed and adjacent to gene 2. The gene 12 paralogs (CDS347 and CDS348) are 47% similar over their entire length (E value = 1e-48), whereas the two gene 19 proteins are 42% similar over two-thirds the length of the longer protein (E value = 2e-4).
We also note that the T4 ortholog of the GroES chaperonin or assembly catalyst (gp31; CDS129) for the phage head proteins is encoded by KVP40, as it is by the pseudo-T-even coliphage RB49 (3). Although the T4 tail fiber assembly gene 38 is absent, the distal long tail fiber, gp37, is present in two copies (CDS298 and CDS298-2), aligning with the proteins of the other T-even-like phages through the C-terminal amino acids. It appears that the hypervariable properties of gp37 in the T-even phages (reviewed in reference 17) are also exploited by KVP40 for receptor recognition and host range adaptation. Functional studies of the tail fiber and OmpK receptor recognition should now be accessible with KVP40 (23, 24). Overall, duplication of three genes (12, 19, and 37) encoding proteins associated with the phage tail or the tail fiber suggests added flexibility in host range adaptation and the infection process.
Several familiar T4 genes are absent in KVP40.
Genetic and biochemical data on the T4 genome and proteome are so extensive that it is interesting that many familiar T4 genes appear to be absent in KVP40. Many have been cited in the preceding sections. Overall, it is the “nonessential” T4 genes that are frequently absent. Genes involved in immunity, superinfection, membrane interactions, restriction, and lysis regulation (e.g., imm, rI, rIII, rIV, and ac) have not been identified. Many phage DNA modification and endonuclease systems (dam, arn, 42, denA, denB, ndd, and alc; most of the multiple seg and mob genes; and group I intron homing endonucleases) are absent. Phage lysis genes [genes e (lysozyme) and t (holin) in T4] are not yet apparent; no definitive lysis function has been identified in the KVP40 genome (but see P1 Sit homology below).
It will be valuable to determine the transcriptional pattern of KVP40, since most of the proteins and RNA polymerase modification enzymes that determine the classic early and middle transcription modes of T4 are absent. The inhibitor (encoded by T4 pin) of host ATP-dependent protease is absent, but KVP40 does have inh, which encodes the inhibitor of the prohead protease (gene 21). From an RNA perspective, although KVP40 has an impressive number of tRNAs (see above), it lacks the three group I introns found in T4, does not encode the valyl-tRNA synthetase-modifying protein (vs), lacks an apparent stp-dependent tRNA exclusion system, and does not encode the RegB Shine-Dalgarno mRNA RNase. In general, the absence of specific T4 CDSs implies that the computational methods do not allow accurate identification of many phage or viral genes, that KVP40 utilizes gene products or pathways that are not recognized for their ability to carry out these well-studied processes, and/or that the two genomes are evolutionarily distinguished by a history of gene invention, loss, or acquisition of different auxiliary genes that are maintained by different selective pressures.
Conserved hypothetical phage CDSs.
Included among the hypothetical KVP40 CDSs are 16 T4-related CDSs that have no known function (Table 7; also see reference 44). Overall, the pairs of conserved hypothetical proteins from the two phages are of similar length, ranging from 56 to 335 amino acids, with aligned regions of 21 to 47% identity (45 to 68% similarity). The homologous CDSs are distributed throughout the genome. Some are located in the same position relative to adjacent genes in the two genomes (i.e., KVP40 CDS281 and T4 nrdC.10, CDS069 and α-gt.4, and CDS076 and 45.2), while others are not (i.e., CDS092 and tk.4). Indeed, genes 30.2 and 30.3 are adjacent in T4, but their homologs, CDS055 and CDS070, are separated by 14 CDSs and 11,245 bp in KVP40. The presence of these uncharacterized CDSs in both phages suggests that they have important roles in the phage life cycles. Study of the shared hypothetical CDSs, particularly with the tractable T4 system, would be an appropriate starting point for understanding the function of these proteins. The shared T4-KVP40 hypothetical CDSs do not constitute a substantial portion of the hypothetical CDSs of either phage.
TABLE 7.
KVP40 CDSs with homologs to T4 hypothetical, P1, and other phage CDSs
| Phagea | KVP40 CDS (aa)b | Homologous CDS (aa) | No. of residues/total (%)c
|
E value | |
|---|---|---|---|---|---|
| Identical | Similar | ||||
| T4 | 281 (334) | nrdC.10 (325) | 153/323 (47) | 212/323 (65) | 6e-56 |
| 153 (346) | nrdC.11 (336) | 102/346 (29) | 168/346 (48) | 3e-34 | |
| 262 (190) | 30.2 (278) | 54/152 (35) | 85/152 (55) | 4e-19 | |
| 055 (198) | 30.2 (278) | 52/151 (34) | 86/151 (56) | 3e-17 | |
| 092 (170) | tk.4 (155) | 70/166 (42) | 96/166 (57) | 4e-16 | |
| 070 (154) | 30.3 (152) | 58/149 (38) | 81/149 (53) | 9e-16 | |
| 322 (151) | 57B (152) | 56/143 (39) | 81/143 (56) | 9e-15 | |
| 336 (161) | 5.1 (164) | 53/164 (32) | 85/164 (51) | 9e-14 | |
| 069 (116) | α-gt.4 (105) | 30/91 (32) | 51/91 (55) | 8e-06 | |
| 379 (56) | uvsY.-2 (55) | 24/51 (47) | 35/51 (68) | 2e-05 | |
| 113 (94) | nrdA.1 (108) | 34/85 (40) | 50/85 (58) | 3e-05 | |
| 126 (91) | 31.1 (102) | 28/89 (31) | 41/89 (45) | 7e-04 | |
| 076 (88) | 45.2 (62) | 18/52 (34) | 34/52 (64) | 3e-03 | |
| 279-2 (208) | vs.1 (181) | 40/130 (30) | 62/130 (46) | 1e-02 | |
| 040 (235) | dda.2 (248) | 28/131 (21) | 63/131 (47) | 3e-02 | |
| 303 (120) | arn.3 (99) | 23/77 (29) | 40/77 (51) | 1e-01 | |
| P1 | 090 (305) T4 PseT | pap (168) | 44/150 (29) | 73/150 (48) | 1e-08 |
| 010 (427) T4 gp41 | ban (454) | 49/217 (22) | 92/217 (42) | 1e-03 | |
| 341 (646) T4 gp6 | bplA (477) | 50/227 (22) | 96/227 (42) | 6e-03 | |
| 357 (166) T4 gp19 | tubB (203) | 21/76 (27) | 35/76 (46) | 7e-03 | |
| 358 (515) T4 gp20 | proA (569) | 35/147 (23) | 58/147 (39) | 2e-02 | |
| 331 (151) T4 gp4 | pmgV (120) | 19/66 (28) | 34/66 (51) | 2e-02 | |
| 349 (559) T4 wac | 23 (568) | 39/162 (24) | 68/162 (41) | 3e-02 | |
| 344 (253) T4 gp9 | 5 (210) | 14/46 (30) | 23/46 (50) | 7e-01 | |
| 326 (646) | sit (1140) | 114/480 (23) | 193/480 (40) | 1e-04 | |
| 289 (90) | pmfB (140) | 19/73 (26) | 39/73 (53) | 2e-03 | |
| 183 (82) | tciB (54) | 9/42 (21) | 24/42 (57) | 3e-03 | |
| 169 (109) | upfC (94) | 11/36 (30) | 17/40 (42) | 5e-02 | |
| 061 (270) | udrA (203) | 19/66 (28) | 34/66 (51) | 4e-01 | |
| Others | 016 (174) | T5 orf4c (173) | 53/145 (36) | 74/145 (51) | 2e-16 |
| 061 (270) | TM4 80 (192) | 38/105 (36) | 58/105 (55) | 1e-12 | |
| 155 (131) | L5 61 (125) | 42/98 (42) | 55/98 (56) | 8e-11 | |
T4 CDSs are labeled with the GenBank AF158101 nomenclature (44). P1 nomenclature and CDS files were kindly provided by M. Lobocka and M. Yarmolinsky (personal communication).
CDS number and length of encoded protein in amino acids (aa).
BlastP statistics shown are presented as number of identities or similarities and the Expect value obtained with the BLOSUM45 matrix (T4) or WU BlastP 2.0 with BLOSUM62 (others).
Coliphage P1, whose linear double-stranded DNA is maintained as a plasmid-like circle (62), belongs to the Myoviridae morphogenic group (B1 subgroup) (6, 51). All 112 CDSs from the complete 94,800-bp genomic sequence of bacteriophage P1 (M. Lobocka, personal communication) were compared to all KVP40 CDSs with WU-BlastP (BLOSM62). Few CDSs common to both P1 and KVP40 were identified, as is the case between P1 and phage T4. The similar CDSs were often shared between the three phages. The related CDSs that were identified include virion proteins, nucleic acid metabolism enzymes, and a few uncharacterized P1 CDSs (Table 7). Two other similar CDSs should be noted. The product of KVP40 CDS061 (270 amino acids) aligns with both P1 UdrA (203 amino acids) and mycobacterial phage TM4 gp80 (192 amino acids). In P1, udrA may be allelic with gta, a defect affecting generalized transduction (22), and would suggest the capacity for transduction by KVP40 as well. P1 Sit (1,140 amino acids) aligns with KVP40 CDS326 (646 amino acids). Sit is similar to bacterial lytic transglycosylases and to the phage T7 internal virion protein D (1318 amino acids), suggesting that it could be the KVP40 lysis function that replaces the T4-like lysozyme.
Two CDSs in the KVP40 genome align with proteins from the lysogenic mycobacterial phages L5 and D29. CDS155 (131 amino acids) aligns over almost the entire length with gp61 (125 residues) of L5 and D29. CDS016 (174 amino acids) aligns at the N-terminal region with gp66 of L5 and D29 and is an apparent phosphoesterase; a similar CDS (orf4c) occurs in phage T5 (Table 7), and related proteins are predicted from the Archaea Methanococcus jannaschii and Clostridium acetobutylicum. The ClpP endoprotease homolog (KVP40 CDS007; see below) is primarily found in Bacteria and Eukarya, but also occurs in bacteriophage infecting lactic acid bacteria (i.e., phage adh). Overall, relatively few KVP40 proteins resemble proteins encoded by other phage, except those noted for T4.
CDSs of cellular genomes: a pyridine nucleotide salvage pathway.
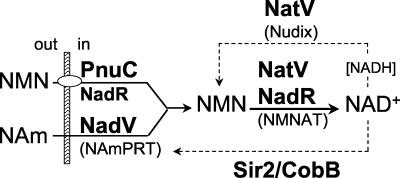
Several CDSs in the KVP40 genome have homologs that have been identified primarily from genomes of cellular organisms. These are summarized in Table 8. Of particular note is the apparent pyridine nucleotide (NAD+) salvage pathway that can be deduced from the KVP40 CDSs, a pathway that has not been described previously for phages (Fig. 4). A membrane-associated nicotinamide mononucleotide (NMN) transporter (PnuC, CDS215; with the associated multifunctional [50] NadR protein, CDS211), together with nicotinamide phosphoribosyltransferase (NadV; CDS264) and NMN adenylyltransferase activities (NadR and the bifunctional Nudix; CDS162), may serve for precursor transport and synthesis of NAD+ (47). nadV was identified on a plasmid as conferring NAD independence on Haemophilus ducreyi and other members of the Pasteurellaceae and was identified in the genomes of other bacteria, including the marine cyanobacterial species Synechocystis (37). A Nudix hydrolase (nudE) was recently characterized from T4 (61), but the larger KVP40 enzyme aligns with a bifunctional enzyme also described from a Synechocystis sp. that has both Nudix and NMN adenylyl transferase activities (49). Interestingly, KVP40 also encodes activities that would cycle NADH back to precursors, including the Nudix hydrolase (which we designate natV for Nudix and adenylyltransferase of vibriophage KVP40). The role of this pathway (Fig. 4) in phage metabolism and its relevance to the host redox or energy status are of interest.
TABLE 8.
KVP40 CDSs with homologs primarily in cellular organisms
| KVP40 CDS (aa)a | Homolog (aa)b | Descriptionc | Organismd | No. of residues/total (%)e
|
E value | |
|---|---|---|---|---|---|---|
| Identical | Similar | |||||
| 264 (497) | NadV (495) | Nicotinamide phosphoribosyl transferase | Haemophilus; B E | 163/500 (32) | 246/500 (48) | 2e-53 |
| 162 (341) | NADM_SYNY3 (339) | Nudix/NMN adenylyltransferase | Synechocystis; B, A | 115/343 (33) | 177/343 (51) | 3e-44 |
| 043 (240) | Sir2 (234) | NAD hydrolysis | Helicobacter; B, A, E | 71/182 (39) | 104/182 (57) | 1e-28 |
| 215 (221) | PnuC (241) | NMN transport | Yersinia; B | 70/224 (31) | 106/224 (47) | 2e-19 |
| 211 (326) | NadR (323) | NMN-transport, adenylyltransferase transcriptional regulator | Mycobacterium; B | 82/315 (26) | 142/315 (45) | 1e-13 |
| 284 (293) | MoaA (251) | Fe-S protein | Helicobacter; B, A | 76/295 (25) | 135/295 (45) | 5e-10 |
| 272 (350) | MoaA/NirJ (394) | Fe-S protein | Clostridium; B, A | 48/170 (28) | 79/170 (46) | 2e-06 |
| 261 (374) | MoaA (298) | Fe-S protein | Methanococcus; B, A | 44/155 (28) | 75/155 (48) | 3e-04 |
| 260 (329) | AstB (323) | Fe-S protein | Thermotoga; B, A | 36/139 (25) | 62/139 (44) | 1e-03 |
| 123 (302) | NP_518569 (277) | GTP cyclohydrolase | Ralstonia; B | 103/266 (38) | 150/266 (56) | 3e-40 |
| 121 (222) | FolE (230) | GTP cyclohydrolase | Vibrio; B, E | 112/207 (54) | 145/207 (69) | 9e-40 |
| 292 (235) | PhoH (307) | Pi starvation; ATP | Caulobacter; B, A | 71/181 (39) | 106/181 (58) | 2e-25 |
| 293 (102) | NP_103983 (231) | Acid phosphatase | Mesorhizobium | 14/61 (22) | 28/61 (44) | 2.8 |
| 364 (365) | CCA (416) | tRNA transferase | Haemophilus; B, E | 103/223 (46) | 137/223 (61) | 6e-44 |
| 124 (274) | ExsB (221) | PP-loop regulator | Oceanobacillus; B, A | 77/225 (34) | 121/225 (53) | 1e-29 |
| 021 (170) | Dut | dUTPase | Desulfitobacterium; B, E | 54/138 (39) | 79/138 (57) | 1e-20 |
| 051 (242) | ZP00049534 (254) | Metallo-Pi esterase | Magnetospirillum; B, A | 78/249 (31) | 114/249 (45) | 3e-19 |
| 120 (308) | NP_717779 (291) | PTP synthase domain | Shewanella; B | 87/318 (27) | 142/318 (44) | 5e-14 |
| 007 (239) | ClpP (241) | ATP-dependent protease | Arabidopsis; B, E | 44/153 (28) | 75/153 (48) | 5e-07 |
| 148 (228) | CAC01596 (288) | K+, ion transport | Streptomyces; E | 28/123 (22) | 59/123 (47) | 2e-06 |
| 040 (235) | NP_283776 (275) | Amino acid permease | Neisseria; B, E | 32/125 (25) | 56/125 (44) | 0.62 |
| 300 (478) | NP_199153 (655) | Unknown | Arabidopsis | 122/423 (28) | 202/423 (47) | 2e-36 |
| 299 (176) | NP_213077 (146) | Unknown | Aquifex; B | 63/144 (43) | 83/144 (56) | 6e-17 |
| 381 (137) | NP_520338.1 (149) | Unknown | Ralstonia; B | 41/120 (34) | 62/120 (51) | 8e-06 |
| 229 (129) | YdfA (144) | Unknown | Plasmid ColIb-P9 | 40/144 (27) | 64/144 (44) | 6e-05 |
| 175 (139) | YahA (107) | Unknown | E. coli | 33/72 (45) | 43/72 (58) | 7e-03 |
CDS number and length of encoded protein in amino acids (aa).
Proteins are listed by name or GenBank locus, with the length given.
Descriptions, when available, apply to the cited protein. NMN, nicotinamide mononucleotide; PP, pyrophosphate; PTP, 6-pyruvoyl-tetrahydropterin.
The organism with the longest, best Blast alignment is cited, with presence of the protein as noted in Bacteria (B), Archaea (A), or Eukarya (E).
FIG. 4.
Inferred pyridine nucleotide salvage cycle encoded by KVP40. The components of the NAD salvage pathway (solid lines) are most related to enzymes of bacteria, none of which have been previously identified in phages. PnuC (CDS215) and NadR (CDS211) are shown as a complex for active transport of nicotinamide mononucleotide (NMN) across the bacterial membrane (hatched bar). Nicotinamide (NAm) is assimilated, but an active transport system has not been described. NadV (CDS264) is a nicotinamide phosphoribosyltransferase (NAmPRT) resembling the enzyme from Haemophilus ducreyi that catalyzes the formation of nicotinamide mononucleotide from nicotinamide. NadR is bifunctional, having an apparent nicotinamide mononucleotide adenylyltransferase domain similar to that of bacterial enzymes. The two-domain Nudix hydrolase (here designated NatV; CDS162) resembles one found in the marine cyanobacterial species Synechocystis, which also has a nicotinamide mononucleotide adenylyltransferase domain. Hydrolysis of NADH (dashed lines) can occur by the NatV Nudix hydrolase and by a KVP40 enzyme (CDS043) that is a Sir2/CobB-like enzyme prevalent in eukaryotes and bacteria (also not previously seen in a phage). See references 37, 47, 49, 50, and 61 for details on the pathway and related enzymes.
KVP40 encodes four Fe-S center-containing proteins (CDS260, -261, -272, and -284) whose precise function is not clear. Another gene pair (CDS292 and CDS293) aligns with the phosphate starvation-inducible PhoH protein and an acid phosphatase (although the latter protein displays a relatively poor Blast E value). Three potential transcriptional regulatory proteins of cellular origin were identified (CDS043, -124, and -211), but only the NadR protein (CDS211) has a clear target gene (PnuC; see above) known to be regulated (47). A permease (CDS040), an ion channel protein (CDS148), and an uncharacterized membrane protein (CDS300) are also encoded. KVP40 could direct protein degradation through a ClpP homolog (CDS007) and could affect folate or GTP levels through GTP cyclohydrolase activity (CDS121 and CDS123). Expounding further on the roles of these CDSs in the KVP40 developmental cycle would be speculative, yet their putative identities suggest biochemical or growth experiments to directly evaluate their functions.
Many of the genes in KVP40 without a phage homolog are not closely related to V. cholerae genes. This may be a result of the broad host range of KVP40 and acquisition of genes from other host genomes, suggesting that the phage has an even greater host range than the Vibrio and Photobacterium genera noted previously (41).
Hypothetical CDSs unique to KVP40.
More than 60% of the 386 KVP40 CDSs are of unidentified function and are without characterized protein homologs. Many of these, like the auxiliary genes of T4, are likely to be proteins required for propagation in specific hosts or under specific growth conditions. The presence of these numerous uncharacterized genes, together with the absence of several well-understood T4 genes, suggests a host range and gene pool that are distinct and isolated for the two phages. Specific adaptation of T4 to coliform bacteria versus a KVP40 adaptation to predominantly marine vibrios would have contributed to the evolution of the distinct gene sets. The fundamental structure of the virion and replisome is conserved and likely originated before the divergence of the two phages.
There is an alternative possibility for the origin of the large number of unique genes in KVP40. With the T4-like “headful” DNA packaging system, the extended, prolate head of KVP40 relative to T4 (140 nm versus 110 nm, yielding a larger head volume) may have selected for additional, potentially “junk,” DNA for stability of the virion in the marine environment. Consequently, the hypothetical CDSs may be degenerate relics of redundant phage genes or may be genes that were “hijacked” from the genomes of marine bacteria, which are presently underrepresented in the GenBank database. Evaluating the function and significance of these unique CDSs presents a significant challenge. Genetic systems utilizing the broad host range of KVP40 should facilitate this analysis.
Conclusions.
The genomic sequence of phage KVP40 reveals that only a small portion of its 386 CDSs (99 CDSs, 26%) are clearly related to T4. Many of the remaining CDSs (circa 65%) are of unidentified function. It appears that KVP40 and T4 share what would constitute the minimal genome unit of the widely distributed T-even-like phages (1). The shared genes encode three major “molecular machines” central to the existence of the phage: the DNA replisome, the late transcriptional apparatus, and the virion structural proteins. Trinucleotide analysis suggests that these genes were not acquired by horizontal gene transfer but were retained from the ancestral phage. KVP40 encodes deoxyriboendonucleases, the RecA-like UvsX recombination protein, and an ortholog of the T4 SegD endonuclease (54), the last being related to group I intron homing endonucleases. Components for horizontal gene transfer, from host or phage genomes during mixed infections, are therefore present in the KVP40 genome.
By their absence, the KVP40 sequence lends support to the nonessential role ascribed to many of the T4 proteins. Conversely, the presence in KVP40 of other characterized “nonessential” T4 genes (e.g., the illustrious rII, regA, rnh, and uvsW genes and others) implies that both phages encounter growth conditions for which these products are beneficial.
The complete KVP40 phage genome sequence provides an important resource for structure-function studies of the conserved proteins and an opportunity to examine a phage transcriptional pattern that appears to lack the well-characterized T4 early and middle elements and presents a large number of hypothetical CDSs of unknown function. Because of its broad host range and apparent reduced capacity for host DNA degradation, we also suggest that KVP40 will be useful for gene transfer studies (i.e., transduction) of Vibrio spp. conducted in the laboratory and in the environment. Finally, this perspective on a complete T4-like phage genome that infects a bacterium other than E. coli complements other efforts in phage genomics (see http://phage.bioc.tulane.edu/), expanding what is still a limited view of phage genomes.
Acknowledgments
H. Ackermann and M. Matsuzaki provided bacterial and bacteriophage strains to initiate this work. We are grateful to Gisela Mosig for comments on the manuscript and to many other individuals in the phage research community who provided thoughtful insights. We thank M. Heany, S. Lo, V. Sapiro, B. Lee, R. Karamchedu, and M. Holmes for database and software support and J. Peterson, L. Umayam, I. Holt, and D. Haft for informatics support. M. Bessman, J. Grose, and J. Roth provided stimulating discussion on NAD metabolism.
This work was supported by discretionary funds of the Institute for Genomic Research. E.S.M. was supported by the North Carolina Agricultural Research Service.
The first two authors contributed equally to the manuscript.
REFERENCES
- 1.Ackermann, H.-W., and H. M. Krisch. 1997. A catalogue of T4-type bacteriophages. Arch. Virol. 142:2329-2345. [DOI] [PubMed] [Google Scholar]
- 2.Alberts, B., and R. Miake-Lye. 1992. Unscrambling the puzzle of biological machines: the importance of the details. Cell 68:415-420. [DOI] [PubMed] [Google Scholar]
- 3.Ang, D., A. Richardson, M. P. Mayer, F. Keppel, H. Krisch, and C. Georgopoulos. 2001. Pseudo-T-even bacteriophage RB49 encodes CocO, a cochaperonin for GroEL, which can substitute for Escherichia coli's GroES and bacteriophage T4's Gp31. J. Biol. Chem. 276:8720-8726. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. Sonnhammer. 1999. Pfam 3.1: 1313 multiple alignments and profile hidden Markov models match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhagwat, M., and N. G. Nossal. 2001. Bacteriophage T4 RNase H removes both RNA primers and adjacent DNA from the 5′ end of lagging-strand fragments. J. Biol. Chem. 276:28516-28524. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, D. E. 1967. Ultrastructure of bacteriophage and bacteriocins. Bacteriol. Rev. 31:230-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686.7704669 [Google Scholar]
- 8.Desplats, C., C. Dez, F. Tetart, H. Eleaume, and H. M. Krisch. 2002. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. J. Bacteriol. 184:2789-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy, S. R. 1995. Multiple alignment with hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:114-120. [PubMed] [Google Scholar]
- 10.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 11.Frank, A. C., and J. R. Lobry. 2000. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 16:560-561. [DOI] [PubMed] [Google Scholar]
- 12.Fu, T. J., E. P. Geiduschek, and G. A. Kassavetis. 1998. Abortive initiation of transcription at a hybrid promoter. An analysis of the sliding clamp activator of bacteriophage T4 late transcription, and a comparison of the σ70 and T4 gp55 promoter recognition proteins. J. Biol. Chem. 273:34042-34048. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg, R. G., P. He, J. Hilfinger, and M.-J. Tseng. 1994. Deoxynucleoside triphosphate synthesis and phage T4 DNA replication, p. 14-27. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 14.Haft, D. H., B. J. Loftus, D. L. Richardson, F. Yang, J. A. Eisen, I. T. Paulsen, and O. White. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henning, U., and S. Hashemolhosseini. 1994. Receptor recognition by T-even-type coliphages, p. 291-298. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 18.Herr, A. J., N. M. Wills, C. C. Nelson, R. F. Gesteland, and J. F. Atkins. 2001. Drop-off during ribosome hopping. J. Mol. Biol. 311:445-452. [DOI] [PubMed] [Google Scholar]
- 19.Hinton, D. M., and S. Vuthoori. 2000. Efficient inhibition of Escherichia coli RNA polymerase by the bacteriophage T4 AsiA protein requires that AsiA binds first to free sigma70. J. Mol. Biol. 304:731-739. [DOI] [PubMed] [Google Scholar]
- 20.Huang, W. M. 1986. Nucleotide sequence of a type II DNA topoisomerase gene, bacteriophage T4 gene 39. Nucleic Acids Res. 14:7751-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, W. M., S. Z. Ao, S. Casjens, R. Orlandi, R. Zeikus, R. Weiss, D. Winge, and M. Fang. 1988. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science 239:1005-1012. [DOI] [PubMed] [Google Scholar]
- 22.Iida, S., R. Hiestand-Nauer, H. Sandmeier, H. Lehnherr, and W. Arber. 1998. Accessory genes in the darA operon of bacteriophage P1 affect antirestriction function, generalized transduction, head morphogenesis, and host cell lysis. Virology 251:49-58. [DOI] [PubMed] [Google Scholar]
- 23.Inoue, T., S. Matsuzaki, and S. Tanaka. 1995. A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 125:101-105. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, T., S. Matsuzaki, and S. Tanaka. 1995. Cloning and sequence analysis of Vibrio parahaemolyticus ompK gene encoding a 26-kDa outer membrane protein, OmpK, that serves as receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 134:245-249. [DOI] [PubMed] [Google Scholar]
- 25.Jones, C. E., T. C. Mueser, K. C. Dudas, K. N. Kreuzer, and N. G. Nossal. 2001. Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination. Proc. Natl. Acad. Sci. USA 98:8312-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kano-Sueoka, T., J. R. Lobry, and N. Sueoka. 1999. Intra-strand biases in bacteriophage T4 genome. Gene 238:59-64. [DOI] [PubMed] [Google Scholar]
- 27.Karam, J. D., J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.). 1994. Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 28.Karlin, S., A. M. Campbell, and J. Mrazek. 1998. Comparative DNA analysis across diverse genomes. Annu. Rev. Genet. 32:185-225. [DOI] [PubMed] [Google Scholar]
- 29.Kolesky, S., M. Ouhammouch, E. N. Brody, and E. P. Geiduschek. 1999. Sigma competition: the contest between bacteriophage T4 middle and late transcription. J. Mol. Biol. 291:267-281. [DOI] [PubMed] [Google Scholar]
- 30.Kreuzer, K. N. 1998. Bacteriophage T4, a model system for understanding the mechanism of type II topoisomerase inhibitors. Biochim. Biophys. Acta 1400:339-347. [DOI] [PubMed] [Google Scholar]
- 31.Kreuzer, K. N., and S. W. Morrical. 1994. Initiation of DNA replication, p. 28-42. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 32.Kricker, M., and K. Carlson. 1994. Isolation of T4 phage DNA, p. 455-456. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular Biology of Bacteriophage T4. ASM Press, Washington, D.C.
- 33.Kunisawa, T. 1992. Synonymous codon preferences in bacteriophage T4: a distinctive use of transfer RNAs from T4 and from its host Escherichia coli. J. Theor. Biol. 159:287-298. [DOI] [PubMed] [Google Scholar]
- 34.Kutter, E., T. White, M. Kashlev, M. Uzan, J. McKinney, and B. Guttman. 1994. Effects on host genome structure and expression, p. 357-368. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 35.Landthaler, M., and D. A. Shub. 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. USA 96:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuzaki, S., T. Inoue, and S. Tanaka. 1998. A vibriophage, KVP40, with major capsid protein homologous to gp23* of coliphage T4. Virology 242:314-318. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzaki, S., M. Kuroda, S. Kimura, and S. Tanaka. 1999. Major capsid proteins of certain Vibrio and Aeromonas phages are homologous to the equivalent protein, gp23*, of coliphage T4. Arch. Virol. 144:1647-1651. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki, S., M. Kuroda, S. Kimura, and S. Tanaka. 1999. Vibriophage KVP40 and coliphage T4 genomes share a homologous 7-kbp region immediately upstream of the gene encoding the major capsid protein. Arch. Virol. 144:2007-2012. [DOI] [PubMed] [Google Scholar]
- 41.Matsuzaki, S., S. Tanaka, T. Koga, and T. Kawata. 1992. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol. Immunol. 36:93-97. [DOI] [PubMed] [Google Scholar]
- 42.Miller, E. S., and C. E. Jozwik. 1990. Sequence analysis of conserved regA and variable orf43.1 genes in T4-like bacteriophages. J. Bacteriol. 172:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, E. S., J. D. Karam, and E. K. Spicer. 1994. Control of translation initiation: mRNA structure and protein repressors, p. 193-205. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 44.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, W. R. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132:185-219. [DOI] [PubMed] [Google Scholar]
- 47.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-730. In F. C. Niedhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 48.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 2000. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 10:1594-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raffaelli, N., T. Lorenzi, A. Amici, M. Emanuelli, S. Ruggieri, and G. Magni. 1999. Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltrransferase and “Nudix” hydrolase activities. FEBS Lett. 444:222-226. [DOI] [PubMed] [Google Scholar]
- 50.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reanney, D.C., and H. W. Ackermann. 1982. Comparative biology and evolution of bacteriophages. Adv. Virus Res. 27:205-280. [DOI] [PubMed] [Google Scholar]
- 52.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification with interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Severinova, E., K. Severinov, and S. A. Darst. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 279:9-18. [DOI] [PubMed] [Google Scholar]
- 54.Sharma, M., R. L. Ellis, and D. M. Hinton. 1992. Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc. Natl. Acad. Sci. USA 89:6658-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma, M., P. Marshall, and D. M. Hinton. 1999. Binding of the bacteriophage T4 transcriptional activator, MotA, to T4 middle promoter DNA: evidence for both major and minor groove contacts. J. Mol. Biol. 290:905-915. [DOI] [PubMed] [Google Scholar]
- 56.Sieber, P., A. Lindemann, M. Boehm, G. Seidel, U. Herzing, P. van der Heusen, R. Muller, W. Ruger, R. Jaenicke, and P. Rosch. 1998. Overexpression and structural characterization of the phage T4 protein DsbA. Biol. Chem. 379:51-58. [DOI] [PubMed] [Google Scholar]
- 57.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 59.Tiemann, B., R. Depping, and W. Ruger. 1999. Overexpression, purification, and partial characterization of ADP-ribosyltransferases ModA and ModB of bacteriophage T4. Gene Expr. 8:187-196. [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkens, K., B. Tiemann, F. Bazan, and W. Ruger. 1997. ADP-ribosylation and early transcription regulation by bacteriophage T4. Adv. Exp. Med. Biol. 419:71-82. [DOI] [PubMed] [Google Scholar]
- 61.Xu, W., P. Gauss, J. Shen, C. A. Dunn, and M. J. Bessman. 2002. The gene e.1 (nudE.1) of T4 bacteriophage designates a new member of the Nudix hydrolase superfamily active on flavin adenine dinucleotide, adenosine 5′-triphospho-5′-adenosine, and ADP-ribose. J. Biol. Chem. 277:23181-23185. [DOI] [PubMed] [Google Scholar]
- 62.Yarmolinsky, M. B., and N. Sternberg. 1988. Bacteriophage P1, p. 291-438. In R. Calendar (ed.), The bacteriophages. Plenum Press, New York, N.Y.
- 63.Yeh, L. S., T. Hsu, and J. D. Karam. 1998. Divergence of a DNA replication gene cluster in the T4-related bacteriophage RB69. J. Bacteriol. 180:2005-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, K. K., G. J. Edlin, and G. G. Wilson. 1982. Genetic analysis of bacteriophage T4 transducing bacteriophages. J. Virol. 41:345-347. [DOI] [PMC free article] [PubMed] [Google Scholar]