Abstract
In bacteria, the biosynthetic pathway for the hydroxymethyl pyrimidine moiety of thiamine shares metabolic intermediates with purine biosynthesis. The two pathways branch after the compound aminoimidazole ribotide. Past work has shown that the first common metabolite, phosphoribosyl amine (PRA), can be generated in the absence of the first enzyme in purine biosynthesis, PurF. PurF-independent PRA synthesis is dependent on both strain background and growth conditions. Standard genetic approaches have not identified a gene product singly responsible for PurF-independent PRA formation. This result has led to the hypothesis that multiple enzymes contribute to PRA synthesis, possibly as the result of side products from their dedicated reaction. A mutation that was able to restore PRA synthesis in a purF gnd mutant strain was identified and found to map in the gene coding for the TrpD subunit of the anthranilate synthase (AS)-phosphoribosyl transferase (PRT) complex. Genetic analyses indicated that wild-type AS-PRT was able to generate PRA in vivo and that the P362L mutant of TrpD facilitated this synthesis. In vitro activity assays showed that the mutant AS was able to generate PRA from ammonia and phosphoribosyl pyrophosphate. This work identifies a new reaction catalyzed by AS-PRT and considers it in the context of cellular thiamine synthesis and metabolic flexibility.
Metabolic processes in a living cell are connected through the integration of biochemical pathways. This metabolic integration allows bacteria to survive under diverse conditions and is essential for their adaptation to changing environments. The structure and function of networks created by the integration and overlap of metabolic processes are largely uncharacterized. The thiamine biosynthetic pathway in Salmonella enterica has direct biochemical connections with other metabolic pathways, including purine and isoprenoid biosynthesis (5, 33, 40), and indirect connections with a number of other cellular processes (1, 16, 19, 25, 36). Thus, this biosynthetic pathway serves as a focal point for studies to identify and characterize metabolic interactions, redundancy, and regulation that are essential in a robust metabolism.
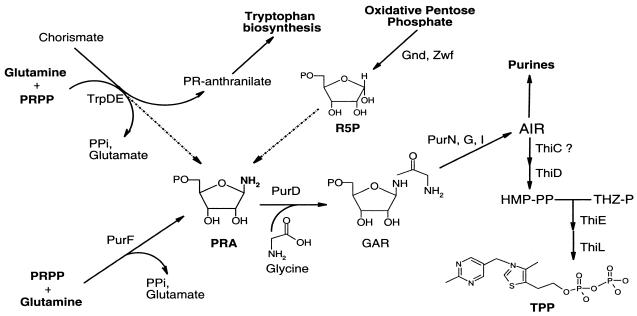
Thiamine consists of a 4-amino-5-hydroxymethyl-2-methyl pyrimidine (HMP) pyrophosphate moiety and a 4-methyl-5-(β-hydroxyethyl)-thiazole phosphate moiety, which are synthesized independently prior to their condensation (6) (Fig. 1). HMP is generated from an intermediate in the de novo purine biosynthetic pathway, aminoimidazole ribotide, the last intermediate common to purine and thiamine biosynthesis (18, 33, 34). Mutants lacking the first step in purine synthesis, catalyzed by the PurF enzyme (phosphoribosyl pyrophosphate [PRPP] amidotransferase), lack synthesis of purines but retain HMP synthesis under a number of growth conditions and genetic backgrounds (14, 15, 36). PurF is the only purine biosynthetic enzyme that can be bypassed in thiamine synthesis, indicating that phosphoribosyl amine (PRA) is the compound generated in a PurF-independent mechanism (15, 36) (Fig. 1). Mutations in gnd (gluconate-6-phosphate dehydrogenase) or zwf (glucose-6-phosphate dehydrogenase) prevented thiamine-independent growth of a purF mutant under otherwise permissive conditions (16, 36). These results demonstrated a role for the oxidative pentose phosphate pathway in this process. The requirement for the oxidative pentose phosphate pathway in PRA formation can be overcome by exogenous ribose (16, 36) or a null mutation in yjgF (17). Genetic attempts to define an activity responsible for PurF-independent PRA synthesis have been unsuccessful, leading to a model in which multiple enzymes contribute to PRA formation in vivo.
FIG. 1.
Inputs to the synthesis of PRA in S. enterica. Schematically represented are the purine, thiamine, and tryptophan biosynthetic pathways. When present, gene names are indicated by the biosynthetic step catalyzed by their product. The dotted lines represent the pathways proposed to contribute to PRA synthesis in the absence of the PurF enzyme. PR-anthranilate, phosphoribosyl anthranilate; PPi, pyrophosphate; R5P, ribose-5-phosphate; AIR, aminoimidazole ribotide; THZ-P, 4-methyl-5-(β-hydroxyethyl)-thiazole phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methyl pyrimidine pyrophosphate; TPP, thiamine pyrophosphate.
Here we report the isolation and characterization of a mutant derivative of a purF gnd strain that is capable of thiamine-independent growth. The causative mutation in this strain was in the trpD gene, encoding anthranilate synthase (AS) component II. The catalytic similarities between AS-phosphoribosyl transferase (AS-PRT) and PurF, and the results obtained in this study, support the conclusion that the AS-PRT complex can generate PRA in vivo. We suggest that the increased synthesis of PRA by the mutant complex is sufficient to allow thiamine synthesis in the absence of PurF.
MATERIALS AND METHODS
Bacterial strains.
All strains used in this study are derivatives of S. enterica serovar Typhimurium strain LT2 and are listed with their genotypes in Table 1. Tn10d(Tc) refers to the transposition-defective mini-Tn10 (Tn10Δ16Δ17) (44). MudJ is used to refer to the Mud1734 transposon, described previously (10).
TABLE 1.
Bacterial strains
| Strain | Genotype |
|---|---|
| DM1 | Wild type (LT2) |
| DM728 | purF2085 gnd-181 |
| DM1231 | purF2085 gnd-174::MudJa |
| DM1936 | purF2085 |
| DM6417 | purF2085 gnd-181 zdd-9147::Tn10d(Tc)btrpD3611 |
| DM6418 | purF2085 gnd-181 zdd-9147::Tn10d(Tc) |
| DM6808 | purF2085 gnd-181(pSU19) |
| DM6809 | purF2085 gnd-181(pIR-EDW) |
| DM6810 | purF2085 gnd-181(pIR-EDMS) |
| DM6811 | purF2085 gnd-181(pIR-DW) |
| DM6812 | purF2085 gnd-181(pIR-DMS) |
| DM6813 | purF2085 gnd-181(pIR-S40F) |
| DM6942 | purF2085 gnd-181 yjgF3::MudJ(pIR-S40F) |
| DM7091 | trpR3612::Tn10d(Tc) |
| DM7092 | purF2085 trpR3612::Tn10d(Tc) |
Culture media and chemicals.
No-carbon E medium of Vogel and Bonner (12, 43) supplemented with MgSO4 (1 mM) and a carbon source (11 mM) was used as minimal medium. When present in the culture media, the following compounds were used at the indicated concentrations: adenine, 0.4 mM; thiamine, 0.5 μM; and tryptophan, 0.5 mM. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) and Luria-Bertani broth were used as rich media. Difco BiTek agar was added (15 g/liter) for solid medium. Antibiotics were added as needed to the following concentrations in rich and minimal media, respectively: tetracycline, 20 and 10 μg/ml; kanamycin, 50 and 125 μg/ml; and chloramphenicol, 20 and 4 μg/ml.
Genetic methods. (i) Transduction methods.
All transductional crosses were performed by using the high-frequency general transducing mutant of bacteriophage P22 (HT105/1 int-201) (39) as described previously (13). Transductants were purified by streaking on nonselective green indicator plates, and putative phage-free clones, identified by their light green color (11), were verified to be phage-free by cross-streaking with phage P22.
(ii) Isolation of the suppressor mutant.
A pool of >70,000 cells containing random Tn10d(Tc) insertions was generated as described elsewhere (27, 29) and mutagenized with nitrosoguanidine (12, 26). A P22 lysate grown on this pool of cells was used to transduce strain DM1231 (purF2085 gnd-174::MudJ) to tetracycline resistance (Tcr) on nutrient agar plates. The Tcr transductants that remained Knr (i.e., retained the gnd-174::MudJ insertion) and were able to grow on glucose and gluconate minimal media supplemented with adenine were tested for growth in the presence of 0.5 mM serine. To eliminate yjgF mutants (17), only those transductants that were able to grow in the presence of serine were investigated further. All putative mutants were reconstructed by transduction into the parental strain. The presence of the two selected phenotypes among transductants was interpreted to mean that the selected Tn10d insertion was linked to the suppressor mutation. The chromosomal location of the relevant insertion was determined by sequencing with a PCR-based protocol (7). A DNA product was amplified with degenerate primers and primers derived from the Tn10d(Tc) insertion sequences as described previously (45) and was sequenced by the University of Wisconsin Biotechnology Center-Nucleic Acid and Protein Facility.
(iii) Isolation of trpR mutation.
A phage P22 lysate grown on a pool of Tn10 insertion mutants was used to transduce a thr mutant strain to Tetr. The Tetr transductants were screened for Thr+ and resistance to 5-methyltryptophan (0.45 mM) to be consistent with map locations and phenotypes described for trpR mutants (2, 41). The resulting trpR3612::Tn10d(Tc) mutation was 80% linked to the threonine operon and 100% linked to 5-methyltryptophan resistance (41).
(iv) Phenotypic analysis.
Nutritional requirements were assessed both on solid and agar overlays and in liquid growth media as described below.
(a) Liquid growth.
Strains to be analyzed were grown to full density in nutrient medium at 37°C. After overnight incubation, cells were pelleted and resuspended in an equal volume of saline (85 mM). A 0.2-ml sample of this suspension was used to inoculate 5 ml of the appropriate medium (2% [vol/vol] inoculum). Culture tubes were incubated at 37°C with shaking, and growth was monitored as optical density at 650 nm on a Bausch and Lomb Spectronic 20D spectrometer. Alternatively, 2 μl of the cell suspension was used to inoculate 200 μl of the appropriate medium contained in each well of a 96-well microtiter plate. Growth at 37°C was monitored with a Spectra-Max Plus microplate spectrophotometer. The specific growth rate was determined as μ = ln(X/X0)/T, where X is optical density at 650 nm during the linear portion of the growth curve and T is time.
(b) Solid media.
Nutritional requirements were measured in soft agar overlays as follows. A 0.2-ml aliquot of saline cell suspension (prepared as described above) was added to 4 ml of molten 0.7% agar and spread on an appropriate plate. Compounds to be tested were spotted at the indicated volume and concentration after the agar had solidified. Growth was scored after 24 and 48 h.
Molecular biology techniques.
Open reading frames were amplified by PCR with cloned Pfu DNA polymerase and the appropriate primers listed in Table 2. The resulting PCR products were purified and blunt-end ligated into SmaI-cut pSU19 (3). Plasmids were transformed into Escherichia coli strain DH5α and screened for vectors containing inserts. Analysis of restriction digest patterns for the resulting plasmids was performed, and the identities and orientations of the plasmid inserts were confirmed by sequencing.
TABLE 2.
Plasmids
| Plasmida | Insert | Primers used to amplify insert |
|---|---|---|
| pIR-EDW | trpEDb | 5′-GAGAATAACCATGCAAACAC-3′ |
| 5′-TCAGAAGGTCTCCTGTGCAT-3′ | ||
| pIR-EDMS | trpEDc | 5′-GAGAATAACCATGCAAACAC-3′ |
| 5′-TCAGAAGGTCTCCTGTGCAT-3′ | ||
| pIR-DW | trpDb | 5′-CAGGAGACCTTCTGATGGCT-3′ |
| 5′-TCAGAAGGTCTCCTGTGCAT-3′ | ||
| pIR-DMS | trpDc | 5′-CAGGAGACCTTCTGATGGCT-3′ |
| 5′-TCAGAAGGTCTCCTGTGCAT-3′ | ||
| pIR-S40F | trpEDd | 5′-GAGAATAACCATGCAAACAC-3′ |
| 5′-TCAGAAGGTCTCCTGTGCAT-3′ |
For all plasmids, the parent plasmid was pSU19 (Cmr).
The insert was PCR amplified from LT2 chromosomal DNA.
The Insert was PCR amplified from DM6417 chromosomal DNA.
The insert was PCR amplified from plasmid pSTG92 containing a feedback-resistant allele of trpE (9).
Preparation of cell extracts.
Cells were grown for 16 h at 37°C with agitation in the minimal medium of Vogel and Bonner (12, 43) supplemented with the following: acid-hydrolyzed casein, 0.005% (21, 22); MgSO4, 1 mM; glucose, 11 mM; adenine, 0.4 mM; and thiamine, 0.5 μM. Cells were harvested by centrifugation at 4°C, washed twice with 0.05 M potassium phosphate buffer (pH 7.5) containing 1 mM EDTA and 1 mM dithiothreitol, and resuspended in the same buffer. The cells were disrupted by using a French pressure cell at 104 kPa. Cell debris was removed by centrifugation (30 min at 20,000 × g and 4°C). The clear supernatant constituted the crude extract.
Enzyme assays.
AS activity was determined as described previously (20, 42, 49) by measuring the amount of anthranilate produced from chorismate and glutamine or from chorismate and NH4Cl. The initial rate of anthranilate formation was determined at room temperature with a SpectraMax GeminiEM spectrofluorometer. The reaction mixture contained 0.5 mM chorismate, 5 mM glutamine, 5 mM MgCl2, 50 mM potassium phosphate buffer (pH 7.5), and crude cell extract in a final volume of 200 μl. When ammonium-dependent anthranilate synthesis was assayed, the reaction mix contained 0.5 mM chorismate, 50 mM NH4Cl, 5 mM MgCl2, 50 mM Tris-HCl buffer (pH 8.7), and crude cell extract in a final volume of 200 μl. One unit of activity was defined as the appearance of 1 nmol of anthranilate in 1 min per mg of protein.
The PRT activity of AS component II was also assayed fluorometrically (20, 24) by measuring the rate of disappearance of anthranilate at room temperature. The reaction mixture contained 15 μM anthranilic acid, 0.3 mM PRPP, 10 mM MgCl2, 100 mM Tricine buffer (pH 7.6), and crude cell extract in a final volume of 200 μl. One unit of activity was defined as the disappearance of 1 nmol of anthranilate in 1 min per mg of protein. In all assays, between 0.5 and 0.7 μg of protein was added per reaction. To assay allosteric inhibition, tryptophan was added to a final concentration of 0.5 mM.
PRA-forming activity was determined by using a modified assay initially described for PurF (38), where synthesis of PRA from PRPP and glutamine is determined as a function of [14C]glycinamide ribonucleotide (GAR) produced in a coupled reaction catalyzed by GAR synthetase. A molecule of provided [14C]glycine is incorporated into the PRA structure. The reactions were performed in 50 mM potassium phosphate buffer (pH 8.0) in the presence of 6 mM Mg acetate, 2.5 mM ATP, 25 mM [14C]glycine (26 nCi), 35 mM NH4Cl (or 6 mM glutamine), 2 μg of GAR synthetase, cell extract (0.25 to 0.35 μg of protein), and 0.35 mM tryptophan (when added). Reactions were started by the addition of PRPP (6 mM final concentration), and reaction mixtures were incubated at 37°C for 4 h. Labeled GAR and glycine were separated by thin-layer chromatography on polyethyleneimine-cellulose with a methanol-pyridine-water system. The positions of radioactive spots were detected with a Cyclone Storage Phosphor system (Packard Instrument Company), and their identities were confirmed with known standards.
RESULTS
Isolation of a mutation that allows PurF-independent PRA synthesis.
In continuing efforts to identify cellular processes (other than PurF) involved in in vivo PRA generation, Thi+ derivatives of a purF gnd mutant strain were isolated. A pool of Tn10d(Tc) insertions was mutagenized with nitrosoguanidine, and a P22 phage lysate grown on these cells was used to transduce DM1231(purF gnd) to Tcr. From a screen of approximately 16,000 Tcr transductants that retained the gnd-174::MudJ from the recipient, 20 grew on glucose minimal medium supplemented with adenine. Since lesions in yjgF can result in this phenotype (17), the 20 mutants were tested for the serine sensitivity associated with yjgF mutations. Eleven of the mutants were able to grow in the presence of 0.5 mM serine. Genetic reconstruction experiments determined that the causative mutation in one of these strains was linked (30%) to the selected Tn10d(Tc) insertion. This strain was further characterized.
A mutation in the trpD gene allows PRA synthesis.
To define the site of the lesion responsible for allowing PRA synthesis in a purF gnd background, the location of the linked Tn10d(Tc) insertion [zdd-9147::Tn10d(Tc)] was determined by arbitrarily primed PCR (7). Sequence analysis determined that the insertion was in the acnA-cysB intergenic region at ∼28.7 min on the S. enterica chromosome. Further linkage analysis and data from three-factor cross experiments determined that the relevant mutation was located clockwise of the cobA gene on the circular chromosomal map. When an insertion in the nearby trp operon was used as a selected marker in three-factor crosses, PRA formation could not be recovered, suggesting that one or more of the tryptophan biosynthetic genes were involved in this phenomenon.
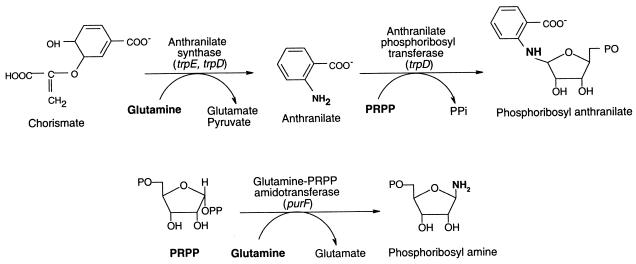
When the catalytic reactions of the tryptophan biosynthetic enzymes and glutamine-PRPP amidotransferase (PurF) are compared, a similarity between AS-PRT (encoded by trpED) and PurF is obvious (Fig. 2). The causative mutation was hypothesized to be an allele of trpD or -E that resulted in a mutant AS-PRT that is able to generate PRA in vivo. To address this possibility, trpE and -D were amplified from both the wild-type (DM1) and mutant (DM6417)strains and cloned into the mid-copy-number vector pSU19 (3), generating plasmids pIR-EDW and pIR-EDMS, respectively. Sequence analysis identified a single nucleotide difference between the genes amplified from the mutant and the wild type. The insert derived from strain DM6417 carried a C-T transversion at nucleotide 1084 of trpD, causing proline 362 of the peptide to be replaced by leucine (trpD3611). Subsequent experiments confirmed the assignment of this change as the causative lesion (see below). Pro362 resides in the PRT subdomain of AS component II, which consists of residues 202 to 531 of TrpD (28, 46). A comparison of multiple ASs from a variety of organisms shows that this residue is highly conserved. The crystal structure of the dimeric anthranilate PRT of Sulfolobus solfataricus has been published (30). While Pro362 is not conserved in S. solfataricus, this amino acid is located next to a conserved residue in the proposed anthranilate binding site, R364.
FIG. 2.
TrpDE- and PurF-catalyzed reactions show biochemical similarity. Biochemical reactions catalyzed by the AS-PRT complex (TrpDE) and glutamine phosphoribosyl amidotransferase (PurF) are illustrated to emphasize similarities. Each enzyme has a glutaminase activity and transfers a phosphoribosyl group to an amidated substrate.
Tryptophan prevents PRA formation by trpD3611.
The trpD3611 mutation was selected to allow thiamine-independent growth of a purF gnd mutant on solid glucose medium. This phenotype was quantified by liquid growth analyses. The specific growth rates (μ) for isogenic strains DM6418 (purF gnd) and DM6417 (purF gnd trpD3611) grown on glucose-adenine medium with the indicated supplements are shown in Table 3. These quantitative growth analyses confirmed that the trpD3611-containing strain (DM6417) grew in absence of thiamine, while the parental strain (DM6418) did not. Addition of thiamine restored growth to the parental strain and had no stimulatory effect on the growth of DM6417. A similar effect of the trpD3611 allele was observed in a purF mutant grown with glucose and when gluconate was provided as the sole carbon source for the purF gnd mutant strains (data not shown).
TABLE 3.
The trpD3611 allele restores thiamine-independent growth of a purF gnd mutant
| Strain | Relevant genotype | Specific growth rate (μ) in minimal glucose medium supplemented witha:
|
|||
|---|---|---|---|---|---|
| Ade | Ade + Thi | Ade + Trp | Ade + Trp + Thi | ||
| DM6417 | purF gnd trpD3611 | 0.413 | 0.362 | NGb | 0.428 |
| DM6418 | purF gnd | NG | 0.274 | NG | 0.285 |
Strains were grown in minimal glucose medium with the indicated supplements at 37°C as described in Materials and Methods. Ade, adenine; Trp, tryptophan; Thi, thiamine. The specific growth rate (μ) was determined by the equation ln(X/X0)/T, where X is A650, X0 is A650 at time zero, and T is time (in hours).
NG, specific growth rate was below detection (≤0.02).
During the genetic mapping experiments described above, it was found that the addition of exogenous tryptophan prevented the thiamine-independent growth of DM6417 (purF2085 gnd trpD3611). As shown in Table 3, the growth inhibition caused by tryptophan (0.5 mM) was completely overcome by the addition of thiamine to the medium, indicating that this amino acid was simply negating the effect of the trpD3611 mutation. Nutritional analyses confirmed that the requirement for thiamine caused by the presence of tryptophan could be satisfied by providing the HMP moiety of thiamine but not by providing the 4-methyl-5-(β-hydroxyethyl)-thiazole moiety. This result was consistent with the conclusion that tryptophan had simply restored the thiamine requirement of the parental strain. Since AS is negatively regulated at the levels of both transcription and activity by tryptophan (8, 37), it was reasonable that tryptophan was having a regulatory effect.
Catalytic activity of AS-PRT is required to generate PRA in the absence of PurF.
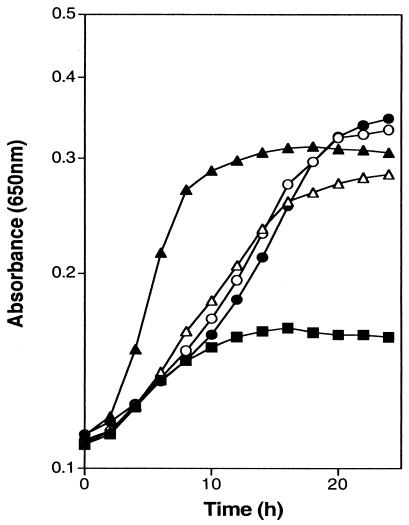
The trpD3611 allele could result in a form of AS-PRT that is able to generate PRA by stimulating an existing activity or generating a new one. Plasmids pIR-EDW and pIR-EDMS were used to address whether the trpD3611 allele had generated a new activity. These plasmids were introduced by electroporation into strain DM728 (purF gnd), and the resulting strains were tested for growth in the absence of thiamine. As shown in Fig. 3, both plasmids allowed the purF gnd mutant strain to grow in the absence of thiamine. However, pIR-EDMS, carrying the mutant trpD allele, resulted in an approximately twofold-higher growth rate. From this result it was concluded that the wild-type AS-PRT was able to generate PRA in vivo and that the trp3611 mutation had stimulated this activity.
FIG.3.
TrpDE, provided in trans, allows thiamine-independent growth. Growth analyses were preformed at 37°C as described in Materials and Methods. Growth of DM6808 [purF2085 gnd-181(pSU19)] (▪), DM6809 [purF2085 gnd-181(pIR-EDW)] (▵), DM6810 [purF2085 gnd-181(pIR-EDMS)] (▴), DM6811 [purF2085 gnd-181(pIR-DW)] (○), and DM6812 [purF2085 gnd-181(pIR-DMS)] (•) in glucose-adenine medium is shown.
As with the strain carrying trpD3611 in the chromosome (DM6417), suppression of the thiamine requirement of purF gnd, by plasmids pIR-EDW and pIR-EDMS, was prevented by tryptophan (data not shown). Significantly, in these plasmid constructs, the native trp promoter was not present. Rather, transcription depended on the plasmid-encoded lac promoter, suggesting that tryptophan was mediating its effect via allosteric regulation of AS-PRT activity. To confirm this interpretation, a plasmid (pSTG92) that carried a feedback-resistant allele of AS-PRT (trpES40FD) was obtained (9). From this plasmid a DNA fragment corresponding to the insert in plasmid pIR-EDW was cloned into pSU19. As expected, the resulting plasmid (pIR-S40E) restored growth to a purF gnd mutant on adenine medium, but, significantly, thiamine-independent growth of this strain was not sensitive to the presence of tryptophan (data not shown).
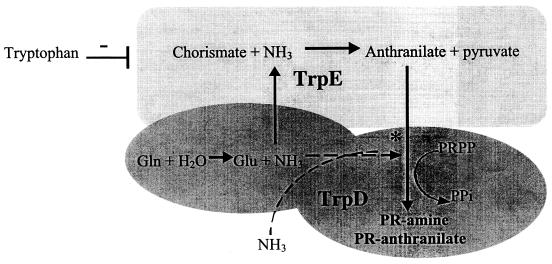
The AS-PRT complex is a dimeric enzyme with complex intersubunit interactions (4, 23, 31, 35) (see Fig. 5). To assess the subunits independently, wild-type and mutant trpD genes were amplified and cloned into pSU19 to generate plasmids pIR-DW and pIR-DMS, respectively. When these plasmids were introduced into DM728 (purF gnd), both allowed growth in glucose-adenine medium, as shown in Fig. 3. Significantly, growth allowed by the wild-type and mutant trpD plasmids was indistinguishable in both rate and yield. Interestingly, the rate of thiamine-independent thiamine synthesis allowed by the trpD genes alone was similar to that allowed by pIR-EDW, the plasmid carrying wild-type trpDE. Further experiments showed that a plasmid carrying only the trpE gene did not restore thiamine-independent growth of the purF gnd mutant strain (data not shown). These results indicate that at least in multicopy, TrpD was sufficient to allow PRA synthesis in vivo, but the trpD3611 allele failed to stimulate that synthesis. Unexpectedly, the thiamine-independent growth allowed by pIR-DW and pIR-DMS was prevented by tryptophan, possibly reflecting inhibition of the PRT activity previously reported (20, 22, 35). Because of common problems with assessing complementation and growth phenotypes with plasmids present, data from the experiments described above were considered only as a qualitative assessment of function and regulation; continued experiments utilized the chromosomal mutation(s).
FIG. 5.
Schematic representation of wild-type and mutant AS-PRT complexes. AS-PRT catalyzes the first two steps in tryptophan biosynthesis, which involve amidotransferase and PRT activities of component II (TrpD). The site for allosteric inhibition by tryptophan is on TrpE, and its approximate location is indicated. Solid lines represent the defined reaction path. Dotted lines reflect the proposed reaction catalyzed by the AS complex leading to PRA synthesis. An asterisk indicates the position of the trpD3611 mutation in TrpD.
trpR mutations allow PRA synthesis in vivo.
The results from the plasmid studies showed that overproduction of TrpDE, even when the mutant allele is absent, allowed PRA synthesis sufficient for thiamine synthesis in vivo. A simple extrapolation of this conclusion suggested that regulatory mutations derepressed for the trp operon (i.e., trpR) would allow thiamine-independent growth of purF mutants. A trpR3612::Tn10d(Tc) mutation was transduced into strain DM1936 (purF), and the resulting strain was assessed for thiamine-independent growth. When assessed on solid media, the trpR mutation allowed thiamine-independent growth, although it was somewhat weaker than that allowed with the trpD3611 allele (data not shown).
Can AS catalyze PRA synthesis in vitro.
Crude cell extracts were generated from stains DM6418 (wild type) and DM6417 (trpD3611) grown under conditions previously determined to induce the trp operon (21, 22). Three biochemical activities previously attributed to the AS-PRT complex were assayed. These assays measured glutamine-dependent AS, ammonium-dependent AS, and PRT activities, and the results are presented in Table 4. As anticipated, the addition of tryptophan inhibited each of these reactions. Further, the data showed that the trpD3611 allele decreased the PRT activity by ∼40%, consistent with the location of the mutant residue in the enzyme. While the PRT activity was reduced, the in vivo data suggested that this decrease was not sufficient to negatively affect the tryptophan biosynthetic capacity of the cell. The other two activities of the AS-PRT complex were not altered by the mutation.
TABLE 4.
The trpD3611 allele alters catalytic properties of the AS-PRT complex
| Strain | Relevant genotype | Activitya
|
|||||
|---|---|---|---|---|---|---|---|
| PRT
|
AS(Gln)b
|
AS(NH4)c
|
|||||
| Without Trp | With Trp | Without Trp | With Trp | Without Trp | With Trp | ||
| DM6418 | purF gnd | 10.2 ± 1.3 | 3.87 ± 0.92 | 10.2 ± 1.8 | 2.6d | 1.4 ± 0.1 | NDe |
| DM6417 | purF gnd trpD3611 | 6.58 ± 1.1 | 2.50 ± 0.28 | 10.5 ± 1.7 | 1.9d | 1.5 ± 0.2 | ND |
Strains were grown in minimal glucose medium supplemented with acid-hydrolyzed casein, adenine, and thiamine, as indicated in Material and Methods. Except as indicated, the data represent the averages and standard deviations from three independent experiments performed in triplicate. Trp, tryptophan. For activity units, see Materials and Methods.
As(Gln), glutamine-dependent AS activity.
AS(NH4), NH4-dependent AS activity.
Assays were performed only once in presence of tryptophan.
ND, not detectable.
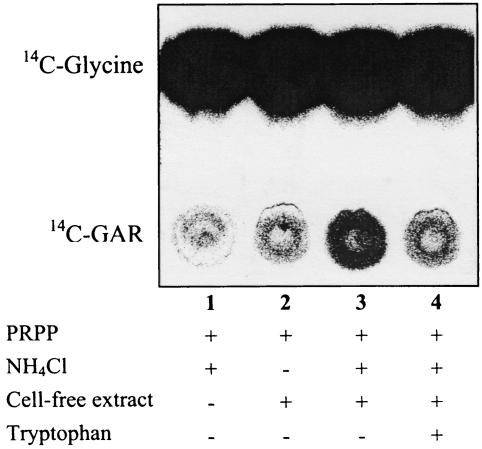
In addition to the activities described above, the proposed PRA synthetase activity in these cell extracts was assayed. These assays utilized PRPP in combination with either ammonia or glutamine. When glutamine was present, no PRA formation was detected (data not shown). However, when NH4Cl was provided, the cell extracts of DM6417, but not those of DM6418, generated detectable PRA, as shown in Fig. 4. Significantly, the PRA-forming activity was inhibited by tryptophan, as predicted by the in vivo results. The amount of PRA detected was small, a result not unexpected given the cellular requirement for thiamine and the extremely short half-life of PRA in aqueous solution (38). However, this was the first demonstration of enzymatic generation of PRA in a strain lacking the PurF enzyme, and it served to explain the growth data presented throughout this paper.
FIG. 4.
Synthesis of PRA from PRPP and NH4Cl. PRA-forming activity in a cell extract from strain DM6417 was determined by using a coupled assay with GAR synthetase. In the coupled reaction a molecule of provided [14C]glycine is incorporated into the PRA structure. Synthesis of PRA from PRPP and NH4Cl was determined as a function of [14C]GAR synthesis. Labeled GAR and glycine were separated on polyethyleneimine-cellulose with a methanol-pyridine-water (20:1:5) solvent system. Quantification determined that lane 3 contained twofold more GAR than the control lane without NH4Cl. No increase in GAR formation was seen with cell extracts from DM6418.
DISCUSSION
This study describes the isolation of a mutation allowing thiamine synthesis to be independent of both PurF and the oxidative pentose phosphate pathway. Characterization of this mutation determined that it caused a proline-to-leucine change at residue 362 of the trpD gene product. Together, trpD and trpE encode the AS-PRT complex, which catalyzes the first two steps in the tryptophan biosynthetic pathway (Fig. 5). Since previous work demonstrated that PurF-independent thiamine synthesis is synonymous with PRA formation (15), the simplest interpretation of the thiamine-independent growth of the purF gnd trpD3611mutant was that PRA was being generated by the mutant AS-PRT complex. This hypothesis is supported by the biochemical and genetic data presented here.
AS-PRT is a multifunctional enzyme with complex regulation and multiple intersubunit and interdomain interactions (9, 23, 24, 32). Although the different activities associated with this enzyme complex appear to be functionally independent and can be physically separated (4, 28, 46, 48), interactions between the AS-II glutamine domain, AS-II PRT domain, and AS-I have been reported (9, 20, 22, 32, 35). The primary PRA-forming enzyme in the cell, glutamine amidophosphoribosyl transferase (PurF), shares several features with AS-PRT. Both enzymes have NH3- and glutamine-dependent activities (47, 48) that ultimately result in the condensation of a phosphoribosyl molecule and an amino group (Fig. 2). Significantly, P362 is located in the PRT subdomain of TrpD, consistent with the proposed effect of this substitution on PRA formation. Compared to the S. solfataricus sequence, P362 is positioned in the anthranilate binding site near the conserved R364 residue. In the recently solved crystal structure, R364 is proposed to provide a ligand to the pyrimidine ring of anthranilate (30).
The position of P362 in a critical binding site suggested that altered catalytic properties of the AS-PRT complex might be detected in vitro. Analysis of catalytic activities of AS-PRT in crude cell extract showed that the trpD3611 mutation decreased the PRT activity of the enzyme while leaving the AS activities unchanged (Table 4). Based on growth of the mutant strains, any reduction in phosphoribosyl anthranilate caused by the trpD3611 mutation was not sufficient to disrupt the normal role of this enzyme in tryptophan biosynthesis. Significantly, the mutant extract was able to generate detectable PRA from PRPP and ammonia. As expected, all catalytic activities detected were inhibited by the addition of tryptophan. To our knowledge, this is the first case of enzymatic PRA formation being detected in the absence of a functional PurF, and it justifies the interpretation of the in vivo growth results presented here.
Together, the biochemical results are consistent with the location of the P362L substitution within the anthranilate binding site in the PRT domain of the enzyme. One scenario for the flow of metabolites in the AS-PRT enzyme that could result in PRA formation from known substrates is schematically illustrated in Fig. 5. In the analyzed structure of AS-PRT, the binding sites for PRPP and anthranilate are too distant to undergo catalysis; therefore, domain rearrangements have been proposed to occur during substrate binding (30). We hypothesize that the P362L substitution could decrease the efficiency of anthranilate binding, affecting the proper conformational changes required for phosphoribosyl anthranilate synthesis, and favor PRA production by allowing condensation between PRPP and ammonia. Kinetic analysis of purified enzymes will be required to test this prediction.
While the trpD3611 mutation restored full thiamine-independent growth to a purF gnd mutant strain, the wild-type AS-PRT enzyme or the TrpD subunit alone was able to partially satisfy the PRA requirement of thiamine synthesis if provided in multicopy. This result suggested that in the wild-type enzyme either glutamine or the NH3 released on cleavage of the glutamine has the potential to react with PRPP in the AS-PRT complex to generate PRA. These results suggested that derepression of the trp operon might similarly generate PRA for thiamine synthesis. Although not obtained in numerous mutant screens for PRA synthesis, when trpR mutations were introduced into strains lacking purF, thiamine-independent growth was observed. This finding confirmed that under some conditions, wild-type AS-PRT is able to contribute to PRA synthesis in vivo.
While PRA formation by the wild-type AS-PRT complex was not detected in the purF mutant extract, it was possible that this enzyme contributed to thiamine synthesis under some conditions in vivo. The finding that tryptophan inhibited PurF-independent thiamine synthesis of a purF gnd yjgF mutant strain on glucose medium (data not shown) suggested that PRA formation in this situation involved AS-PRT activity. Multiple enzymes are expected to participate in PRA synthesis, making it difficult to define the contributors in vivo. In addition, the low levels of PRA needed to satisfy the cellular thiamine requirement, combined with the instability of PRA (38), make the biochemical approach equally laborious. The work described here has taken advantage of a mutant allele to identify the AS-PRT complex as a potential contributor to the formation of PRA in vivo. In addition, a new catalytic activity for a well-studied enzyme has been demonstrated.
Acknowledgments
We thank Ronald Bauerle (University of Virginia) for providing plasmid pSTG92, which carries the trpES40F allele.
This work was supported by competitive grant GM47296 from the NIH. Funds were also provided from a 21st Century Scientist Scholars Award from the J. S. McDonnell Foundation. I. Ramos was supported by a predoctoral fellowship from CONACyT (Mexico).
REFERENCES
- 1.Allen, S., J. L. Zilles, and D. M. Downs. 2002. Metabolic flux in both the purine mononucleotide and histidine biosynthetic pathways can influence synthesis of the hydroxymethyl pyrimidine moiety of thiamine in Salmonella enterica. J. Bacteriol. 184:6130-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balbinder, E., R. Callahan, P. P. McCann, J. C. Cordaro, A. R. Weber, A. M. Smith, and F. Angelosanto. 1970. Regulatory mutants of the tryptophan operon of Salmonella typhimurium. Genetics 66:31-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 4.Bauerle, R., J. Hess, and S. French. 1987. Anthranilate synthase-anthranilate phosphoribosyltransferase complex and subunits of Salmonella typhimurium. Methods Enzymol. 142:366-386. [DOI] [PubMed] [Google Scholar]
- 5.Begley, T. P. 1996. The biosynthesis and degradation of thiamin (vitamin B1). Nat. Prod. Rep. 13:177-185. [DOI] [PubMed] [Google Scholar]
- 6.Begley, T. P., D. M. Downs, S. E. Ealick, F. W. McLafferty, A. P. Van Loon, S. Taylor, N. Campobasso, H. J. Chiu, C. Kinsland, J. J. Reddick, and J. Xi. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171:293-300. [DOI] [PubMed] [Google Scholar]
- 7.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 8.Caligiuri, M. G., and R. Bauerle. 1991. Identification of amino acid residues involved in feedback regulation of the anthranilate synthase complex from Salmonella typhimurium. Evidence for an amino-terminal regulatory site. J. Biol. Chem. 266:8328-8335. [PubMed] [Google Scholar]
- 9.Caligiuri, M. G., and R. Bauerle. 1991. Subunit communication in the anthranilate synthase complex from Salmonella typhimurium. Science 252:1845-1848. [DOI] [PubMed] [Google Scholar]
- 10.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Downs, D. M., and L. Petersen. 1994. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 176:4858-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downs, D. M., and J. R. Roth. 1991. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J. Bacteriol. 173:6597-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enos-Berlage, J. L., and D. M. Downs. 1999. Biosynthesis of the pyrimidine moiety of thiamine independent of the PurF enzyme (phosphoribosylpyrophosphate amidotransferase) in Salmonella typhimurium: incorporation of stable isotope-labeled glycine and formate. J. Bacteriol. 181:841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enos-Berlage, J. L., and D. M. Downs. 1996. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 178:1476-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enos-Berlage, J. L., M. J. Langendorf, and D. M. Downs. 1998. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 180:6519-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estramareix, B., and S. David. 1986. Biosynthesis of thiamine: origin of the methyl carbon atom of the pyrimidine moiety in Salmonella typhimurium. Biochem. Biophys. Res. Commun. 134:1136-1141. [DOI] [PubMed] [Google Scholar]
- 19.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding gamma-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grieshaber, M., and R. Bauerle. 1974. Monomeric and dimeric forms of component II of the anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase complex of Salmonella typhimurium. Implications concerning the mode of assembly of the complex. Biochemistry 13:373-383. [DOI] [PubMed] [Google Scholar]
- 21.Grove, T. H., and H. R. Levy. 1975. Anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyl-transferase from Salmonella typhimurium. Inactivation of glutamine-dependent anthranilate synthetase by agarose-bound anthranilate. Biochim. Biophys. Acta 397:80-93. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, E. J., H. Nagano, H. Zalkin, and L. H. Hwang. 1970. The anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase aggregate. Purification of the aggregate and regulatory properties of anthranilate synthetase. J. Biol. Chem. 245:1416-1423. [PubMed] [Google Scholar]
- 23.Henderson, E. J., and H. Zalkin. 1971. On the composition of anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase from Salmonella typhimurium. J. Biol. Chem. 246:6891-6898. [PubMed] [Google Scholar]
- 24.Henderson, E. J., H. Zalkin, and L. H. Hwang. 1970. The anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase aggregate. Catalytic and regulatory properties of aggregated and unaggregated forms of anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase. J. Biol. Chem. 245:1424-1431. [PubMed] [Google Scholar]
- 25.Hoffmeyer, J., and J. Neuhard. 1971. Metabolism of exogenous purine bases and nucleosides by Salmonella typhimurium. J. Bacteriol. 106:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, J. S., B. N. Ames, and N. Hong. 1971. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc. Natl. Acad. Sci. USA 68:3158-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes, K. T., and J. R. Roth. 1985. Directed formation of deletions and duplications using Mud(Ap, lac). Genetics 109:263-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, E. N., and C. Yanofsky. 1974. Localization of two functions of the phosphoribosyl anthranilate transferase of Escherichia coli to distinct regions of the polypeptide chain. J. Bacteriol. 117:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleckner, N., J. Roth, and D. Botstein. 1977. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J. Mol. Biol. 116:125-159. [DOI] [PubMed] [Google Scholar]
- 30.Mayans, O., A. Ivens, L. J. Nissen, K. Kirschner, and M. Wilmanns. 2002. Structural analysis of two enzymes catalyzing reverse metabolic reactions implies common ancestry. EMBO J. 21:3245-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morollo, A. A., and R. Bauerle. 1993. Characterization of composite aminodeoxyisochorismate synthase and aminodeoxyisochorismate lyase activities of anthranilate synthase. Proc. Natl. Acad. Sci. USA 90:9983-9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagano, H., H. Zalkin, and E. J. Henderson. 1970. The anthranilate synthetase-anthranilate-5-phosphorribosylpyrophosphate phosphoribosyltransferase aggregate. On the reaction mechanism of anthranilate synthetase from Salmonella typhimurium. J. Biol. Chem. 245:3810-3820. [PubMed] [Google Scholar]
- 33.Newell, P. C., and R. G. Tucker. 1968. Biosynthesis of the pyrimidine moiety of thiamine. A new route of pyrimidine biosynthesis involving purine intermediates. Biochem. J. 106:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newell, P. C., and R. G. Tucker. 1968. Precursors of the pyrimidine moiety of thiamine. Biochem. J. 106:271-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pabst, M. J., J. C. Kuhn, and R. L. Somerville. 1973. Feedback regulation in the anthranilate aggregate from wild type and mutant strains of Escherichia coli. J. Biol. Chem. 248:901-914. [PubMed] [Google Scholar]
- 36.Petersen, L., J. Enos-Berlage, and D. M. Downs. 1996. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics 143:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pittard, J. 1996. The various strategies within the TyrR regulation of Escherichia coli to modulate gene expression. Genes Cells. 1:717-725. [DOI] [PubMed] [Google Scholar]
- 38.Schendel, F. J., Y. S. Cheng, J. D. Otvos, S. Wehrli, and J. Stubbe. 1988. Characterization and chemical properties of phosphoribosylamine, an unstable intermediate in the de novo purine biosynthetic pathway. Biochemistry 27:2614-2623. [DOI] [PubMed] [Google Scholar]
- 39.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 40.Sprenger, G. A., U. Schorken, T. Wiegert, S. Grolle, A. A. de Graaf, S. V. Taylor, T. P. Begley, S. Bringer-Meyer, and H. Sahm. 1997. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 94:12857-12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuttard, C. 1972. Location of trpR mutations in the serB-thr region of Salmonella typhimurium. J. Bacteriol. 111:368-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamir, H., and P. R. Srinivasan. 1969. Purification and properties of anthranilate synthase from Salmonella typhimurium. J. Biol. Chem. 244:6507-6513. [PubMed] [Google Scholar]
- 43.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 44.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 45.Webb, E., F. Febres, and D. M. Downs. 1996. Thiamine pyrophosphate negatively regulates transcription of some thi genes of Salmonella typhimurium. J. Bacteriol. 178:2533-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanofsky, C., V. Horn, M. Bonner, and S. Stasiowski. 1971. Polarity and enzyme functions in mutants of the first three genes of the tryptophan operon of Escherichia coli. Genetics 69:409-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zalkin, H. 1993. The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 66:203-309. [DOI] [PubMed] [Google Scholar]
- 48.Zalkin, H. 1983. Structure, function, and regulation of amidophosphoribosyltransferase from prokaryotes. Adv. Enzyme Regulat. 21:225-237. [DOI] [PubMed] [Google Scholar]
- 49.Zalkin, H., and D. Kling. 1968. Anthranilate synthetase. Purification and properties of component I from Salmonella typhimurium. Biochemistry 7:3566-3573. [DOI] [PubMed] [Google Scholar]