Abstract
The cyclic AMP receptor protein (CRP) activates transcription of the Escherichia coli acs gene, which encodes an acetate-scavenging enzyme required for fitness during periods of carbon starvation. Two promoters direct transcription of acs, the distal acsP1 and the proximal acsP2. In this study, we demonstrated that acsP2 can function as the major promoter and showed by in vitro studies that CRP facilitates transcription by “focusing” RNA polymerase to acsP2. We proposed that CRP activates transcription from acsP2 by a synergistic class III mechanism. Consistent with this proposal, we showed that CRP binds two sites, CRP I and CRP II. Induction of acs expression absolutely required CRP I, while optimal expression required both CRP I and CRP II. The locations of these DNA sites for CRP (centered at positions −69.5 and −122.5, respectively) suggest that CRP interacts with RNA polymerase through class I interactions. In support of this hypothesis, we demonstrated that acs transcription requires the surfaces of CRP and the C-terminal domain of the α subunit of RNA polymerase holoenzyme (α-CTD), which is known to participate in class I interactions: activating region 1 of CRP and the 287, 265, and 261 determinants of the α-CTD. Other surface-exposed residues in the α-CTD contributed to acs transcription, suggesting that the α-CTD may interact with at least one protein other than CRP.
The Escherichia coli cyclic AMP (cAMP) receptor protein (CRP; also known as the catabolite activator protein) activates transcription from more than 100 promoters. When bound to its allosteric effector, cAMP, the CRP homodimer binds specific DNA sites near target promoters, enhancing the binding of RNA polymerase holoenzyme (RNAP) and facilitating the initiation of transcription. Simple CRP activation operates through two related mechanisms, designated class I and class II. Both mechanisms depend upon specific interactions between CRP and RNAP. At class I promoters, a CRP dimer binds to DNA at a site centered near position −61.5, −71.5, −82.5, or −92.5. CRP bound at any of these positions uses a defined surface (activating region 1 [AR1]) in the downstream subunit of CRP to contact a specific surface determinant, 287, of the C-terminal domain of the α subunit of RNA polymerase (α-CTD). Two additional α-CTD determinants contribute to class I activation: the 261 determinant, proposed to interact with the σ subunit of RNAP, and the 265 determinant, known to interact with DNA, especially A+T-rich sequences, most notably the UP element.
Class I interactions appear to recruit RNAP to promoter DNA by increasing the binding constant for closed-complex formation. At class II promoters, a CRP dimer binds to DNA at a site centered near position −41.5. When bound at this position, CRP uses AR1 of the upstream subunit of CRP to contact the 287 determinant of the α-CTD and a second surface (AR2) to contact the 162 to 165 determinant of the N-terminal domain of α. The α-CTD 265 determinant also contributes by interacting with DNA. Class II interactions appear to increase closed-complex formation and the rate of isomerization to the open complex. A variation of CRP-dependent activation, designated class III, utilizes two or more CRP dimers or a combination of CRP and other activators to achieve maximal transcription activation (reviewed in references 5 and 10).
Acetyl-coenzyme A synthetase (Acs; acetate:coenzyme A ligase [AMP forming]; EC 6.2.1.1) irreversibly catalyzes the conversion of acetate to acetyl-coenzyme A through an enzyme-bound acetyladenylate intermediate (2). This activity permits cells to use acetate, a two-carbon compound, as a source of both energy (via the tricarboxylic acid cycle) and biosynthetic subunits (via the glyoxylate shunt) (3). Escherichia coli cells require this high-affinity acetate-scavenging enzyme (3, 15) to compete successfully during periods of carbon starvation (A. J. Wolfe, unpublished data). Thus, the induction of Acs represents a survival response for cells entering a nutrient-poor environment.
Previously, we reported that cells control Acs activity primarily at the level of transcription initiation, inducing transcription during exponential phase, achieving maximal transcription during the transition to stationary phase, and partially repressing that transcription following entry into stationary phase. Transcription occurs from two σ70-dependent promoters: the distal promoter acsP1 (P1) and the proximal promoter acsP2 (P2) (Fig. 1) (4, 13, 14). Transcription depends upon CRP, the anaerobically triggered transcription activator FNR, the glyoxylate shunt repressor IclR and its activator FadR, and several enzymes involved in acetate metabolism. While multiple factors influence transcription, CRP appears to function directly as the critical transcription factor (4, 13).
FIG. 1.
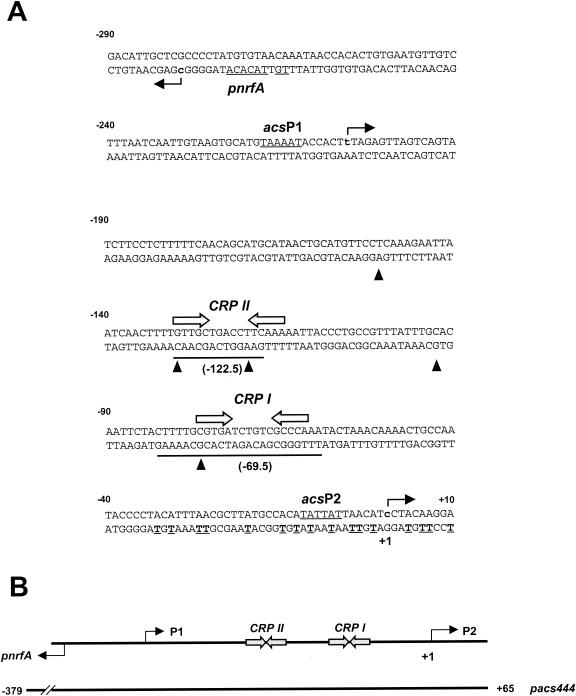
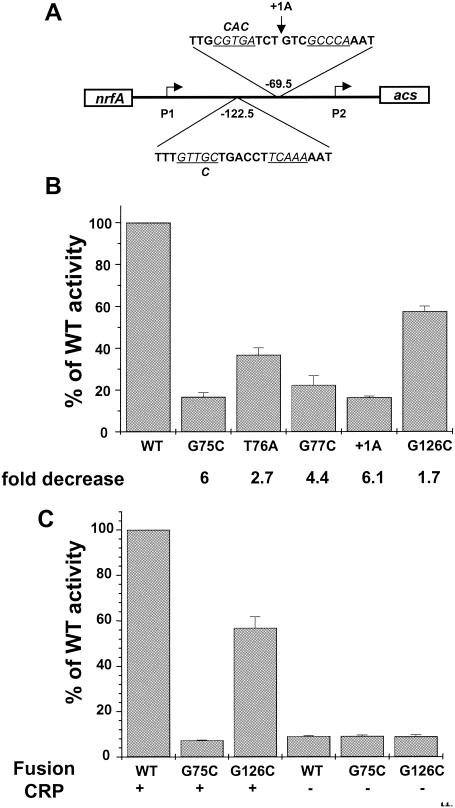
Organization of the acs promoter region. (A) The sequence of the acs promoter region from positions −290 to +10 relative to the acsP2 transcription start site (+1). The acsP1, acsP2, and pnrfA −10 elements are underlined, and the predicted start points of transcription for acsP1, acsP2, and pnrfA are designated in bold lowercase letters, with the direction of transcription indicated by a horizontal arrow. Inverted arrows specify the location of DNA sites for CRP binding (CRP I and CRP II), while underlining shows the extent of protection afforded by CRP in DNase I footprint experiments. Triangles indicate hypersensitive sites within the relevant strand. The cleavage sites produced by potassium permanganate footprint analysis are in bold and underlined. (B) Schematic of the region, showing the locations of the +1 sites associated with each promoter, each binding site, and the extent of the pacs444 fragment used for DNase I footprint experiments (Fig. 4).
This study demonstrated that P2 can function as the primary acs promoter and that its activation in vitro and in vivo requires only CRP, which binds tandem DNA sites (centered at positions −69.5 and −122.5). acs activation required the proximal DNA site for the CRP site, involved the distal DNA binding site for CRP, required AR1 of CRP, and involved the 287, 261, and 265 determinants of the α-CTD. On the basis of these observations, we propose that P2 is a synergistic class III promoter that consists of tandem DNA sites for CRP, each located at a class I position. To our knowledge, this represents the first report of a native promoter with this particular architecture (23). Intriguingly, several α-CTD residues not within the 287, 265, and 261 determinants also affected acs transcription, suggesting that the α-CTD may interact with at least one factor other than CRP.
MATERIALS AND METHODS
Chemicals and biological reagents.
Chemical reagents were purchased from Fisher Scientific (Pittsburgh, Pa.) and Sigma Chemical Company (St. Louis, Mo.). β-Galactosidase assay reagents were purchased from Pierce Biochemicals (Rockford, Ill.). Restriction endonucleases and modifying enzymes were purchased from Promega Corp. (Madison, Wis.), New England Biolabs (Beverly, Mass.), or Gibco-BRL (Gaithersburg, Md.). Purified Escherichia coli RNAP was purchased from Epicentre (Madison, Wis.). Primers were purchased from Integrated DNA Technologies (Ames, Iowa). Medium components were made by Difco.
Bacterial strains, plasmids, and bacteriophage.
All strains used in this study were derivatives of E. coli K-12 and are listed in Table 1.
TABLE 1.
Strains, promoters, plasmids, and phages used in this study
| Strain, promoter, plasmid, or phage | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| AJW678 | thi-1 thr-1(Am) leuB6 metF159(Am) rpsL1136 ΔlacX74 | 13 |
| AJW1941 | AJW678 λCB12 | This study |
| AJW1942 | AJW678 λCB13 | This study |
| AJW1945 | AJW678 λCB16 | This study |
| AJW1969 | AJW678 λCB20 | This study |
| AJW1970 | AJW678 λCB21 | This study |
| AJW2162 | AJW678 λCB21a | This study |
| AJW2163 | AJW1941 crp::Km | This study |
| AJW2175 | AJW678 λCB36 | This study |
| AJW2181 | AJW1969 crp::Km | This study |
| AJW2182 | AJW2175 crp::Km | This study |
| CB369 | MG1655 crp::Km | C. Bausch |
| DH5α | lacZΔM15 recA | Bethesda Research Laboratories |
| P90C | ara Δ(pro-lac) thi | 22 |
| Promoters | ||
| pacs444 | Fragment carrying acs sequences from −379 to +65 | This study |
| Plasmids | ||
| pRS415 | bla lacZ+ (transcriptional fusion vector) | 22 |
| pCB33 | pRS415 pacs444 | This study |
| pCB34 | pCB33 derivative A214G/A215G | This study |
| pCB35 | pCB33 derivative C16G/T10C | This study |
| pCB37 | pCB33 derivative C16G/A11C | This study |
| pCB45 | pCB33 derivative T76A | This study |
| pCB46 | pCB33 derivative G77C | This study |
| pCB47 | pCB33 derivative G76C | This study |
| pCB50 | pCB33 derivative +A | This study |
| pDCRP | pBR322 derivative carrying crp gene | 24 |
| pDCRP/H159L | pDCRP derivative carrying crpH159L allele | 24 |
| pDCRP/K101E | pDCRP derivative carrying crpK101E allele | 24 |
| pDCRP/H159L, K52N | pDCRP derivative carrying crpH159L,K52N allele | 24 |
| pDU9 | pDCRP derivative with crp deleted | 24 |
| pREII (and derivatives) | Plasmid carrying rpoA gene (and alanine substitution derivatives) | 8 |
| pGEM t | pUC19-derived TA cloning vector | Promega |
| pCB26 (and derivatives) | pGEM-t carrying −379 acs through the end of the acs open reading frame; template for all site-directed mutants | This study |
| pSR | pBR322 derivative containing transcription terminator | 12 |
| pSRacs444 | pSR pacs444 | This study |
| Phages | ||
| λCB12 | pCB33-based phage | This study |
| λCB13 | λCB12 derivative A214G/A215G | This study |
| λCB14 | λCB12 derivative C16G/T10C | This study |
| λCB15 | λCB12 derivative T76A | This study |
| λCB16 | λCB12 derivative C16G/A11C | This study |
| λCB20 | λCB12 derivative G77C | This study |
| λCB21 | λCB12 derivative G76C | This study |
| λCB21a | λCB12 derivative +A | This study |
| λCB28 | λCB12 derivative T31C | This study |
| λCB36 | λCB12 derivative G126C | This study |
The plasmids and bacteriophage used in this study are also listed in Table 1. For cloning and transformation, plasmids were prepared with the Wizard miniprep kit (Promega), following the protocol provided. Large-scale DNA preparations for electrophoretic mobility shift assays and footprint analyses were purified over cesium chloride gradients, as described before (18). All restriction digestions and ligations were performed according to standard methods (18).
To construct the β-galactosidase fusion plasmids pCB33 and its mutant derivatives, 444-bp fragments were PCR amplified from pCB26 and its mutant derivatives with primers 2927 and 2926. These PCR products were subcloned into pGEM-T and verified by sequencing. Promoter fragments were excised from the resultant plasmids with EcoRI and BamHI restriction enzymes and subcloned into pRS415. To construct strains that carry single-copy acs::lacZ fusions, β-galactosidase fusion plasmids were converted to hybrid λ phage, as described (22). pSR derivatives were generated by PCR amplification from pCB26 with 2926 and 2927HindIII (AAGCTTCTTGTCATAGGGGCTTC). This PCR product was digested with EcoR1 and HindIII and subcloned into pSR. The Δcrp::Km allele was transferred from strain CB369 by generalized transduction with P1kc (21).
PCRs.
All PCRs were performed in a 50-μl reaction volume. The reactions contained 1× PCR buffer (Promega), 3 mm MgCl2, 20 nM each primer, 0.2 mM deoxynucleoside triphosphate mix, and 1 U of Taq polymerase. Reactions were subjected to one cycle of 95°C for 5 min and then 30 cycles 95°C for denaturing, 68°C for annealing, and 72°C for extension. Site-directed mutations were generated either by megaprimer PCR or the Gene Editor site-directed mutagenesis kit (Promega), following the protocol provided. Megaprimers were produced by standard PCRs with the 5′ primer 2926 (GGATCCGTTGGGTCTGCGATGTTG) and a 3′ mutagenic primer (either ttgcgtCatctgtcgccca, ttgcgAgatctgtcgcca, ttgcCtgatctgtcgcca, or ttgcgtgatctAgtcgccca). Megaprimers were gel purified and quantified by spectrophotometry. A second PCR was carried out with the megaprimer and the 3′ primer 2927 (GAATTCCTTGTCATAGGGGCTTC). The resultant PCR products were subcloned into pGEM-t, and the successful incorporation of mutations was verified by sequencing.
Medium and growth conditions.
For strain constructions, cells were grown in Luria broth (LB; 1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 0.5% [wt/vol] sodium chloride). For β-galactosidase assays, cells were grown at 37°C in tryptone broth (TB; 1% [wt/vol] tryptone and 0.5% [wt/vol] sodium chloride). The medium was supplemented with antibiotics or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), as needed.
Promoter activity assays.
β-Galactosidase activity was determined quantitatively with the Y-PER β-galactosidase assay kit (Pierce Biochemical). The values expressed are the mean and standard error of the mean of three independent measurements. Each experiment was repeated two to five times.
Protein purification.
CRP was purified as described previously (13).
DNase I and potassium permanganate footprint experiments.
All footprint experiments were performed on α-32P-end-labeled AatII-HindIII fragments containing the acs-nrfA intergenic region, as described previously (19). For DNase I footprinting experiments, each reaction (20 μl) contained a final concentration of 4 nM template DNA. The buffer composition was 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM dithiothreitol, 500 μg of bovine serum albumin per ml, and 25 μg of herring sperm DNA per ml. When CRP was present in the reactions, 0.2 mM cAMP also was included. For potassium permanganate footprint experiments, herring sperm DNA was omitted and E. coli RNA polymerase (Epicentre Technologies) was used at a final concentration of 50 nM. All samples were analyzed by denaturing gel electrophoresis. Gels were calibrated with Maxam-Gilbert G+A sequencing reactions of the labeled fragment and quantified with a Typhoon 8600 variable-mode imager and ImageQuant software (Molecular Dynamics).
In vitro transcription assays.
In vitro transcription reactions (20 μl) were performed in a buffer containing 100 mM KCl, 40 mM Tris-acetate (pH 7.9), 10 mM MgCl2, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 200 μM ATP, 200 μM CTP, 200 μM GTP, 10 μM UTP, and 2 μCi of [α-32 P]UTP (Perkin Elmer). When CRP was present in the reactions, 0.2 mM cAMP was also included. CsCl-ethidium bromide gradient-purified supercoiled template DNA (pSRacs444) was added at a final concentration of 10 nM. Reactions were initiated by the addition of RNA polymerase (Epicentre Technologies) to a final concentration 50 nM and incubated at 30°C for 10 min. Samples were analyzed by denaturing gel electrophoresis and quantified with a Typhoon 8600 variable-mode imager and ImageQuant software (Molecular Dynamics).
RESULTS
P2 can function as the major acs promoter.
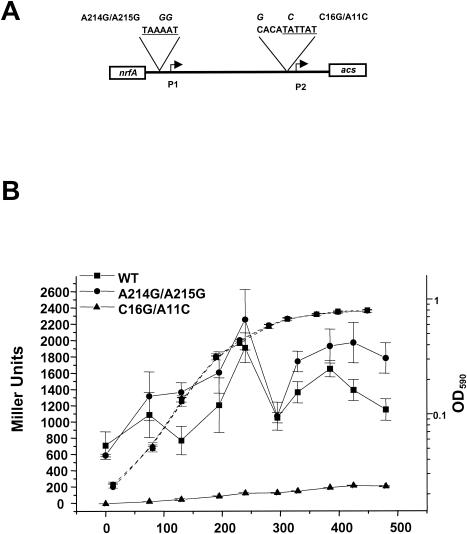
On the basis of reverse transcription-PCR (14), in vivo reporter assays (5), in vitro transcription, and potassium permanganate footprint analyses (data not shown), we previously proposed the existence of two acs promoters, the distal P1 and a proximal P2. To determine the relative contribution of P1 and P2 to acs transcription, we generated point mutations in the predicted −10 element of each promoter. For P1, we generated A214G/A215G; for P2, we generated C16G/A11C (Fig. 2A) and C16G/T10C (data not shown). We incorporated each mutant allele into an acs::lacZ transcriptional fusion (λCB12, Table 1) that encompasses sequences from position −379 to position +65 of acs (Fig. 1B). We then constructed a set of lysogens in a wild-type strain (AJW678, Table 1), with each lysogen carrying a single copy of either λCB12 (AJW1941) or one of the mutant derivatives (Table 1). We grew the resultant lysogens in tryptone broth (TB), harvested cells at several intervals throughout growth, and determined the β-galactosidase activity.
The C16G/A11C allele resulted in a complete loss of activity, while the A214G/A215G allele exhibited no loss of activity (Fig. 2B). To generate a restriction site, the C16G/A11C allele combines a mutation (A11C) at a highly conserved position within the −10 element with a mutation (C16G) outside the −10 element. To determine whether the C16G mutation exerts an effect on transcription, we constructed a second mutant allele (C16G/T10C) that included a mutation (T10C) at a less conserved position within the −10 element and was thus predicted to cause less severe consequences to promoter activity. The C16G/T10C allele behaved like its wild-type parent (data not shown), supporting the conclusion that the T11C mutation was responsible for the decrease in transcription. A 3′ deletion from the middle of the −10 element resulted in activity similar to that exhibited by the C16G/A11C allele, whereas a 5′ deletion that removed the P1 −10 region exhibited transcription comparable to that of the wild type (data not shown). Taken together, these results support the hypothesis that P2 functions as the major acs promoter, at least under the conditions tested.
FIG. 2.
Evidence that P2 is the major acs promoter. (A) Schematic of the acs promoter region, showing the mutant alleles. The sequences of the P1 and P2 −10 elements are underlined. The italic letters above the sequences indicate site-directed mutations. (B) Optical density at 590 nm (OD590, dashed lines) and β-galactosidase activity (solid lines) of acs::lacZ transcriptional fusions, either wild type (WT) or mutant for P1 (A214G/A215G) or P2 (C16G/A11C). Wild-type strain AJW678 was lysogenized with a hybrid λ that carried either the wild-type acs::lacZ fusion (λCB12) or one of its mutant derivatives (λCB13 or λCB16). The resultant lysogens were grown in TB, samples were harvested during a growth curve, the β-galactosidase activity was determined, and the activity was expressed as a Miller units. Each value represents the mean ± standard error of the mean (SEM) of at least three independent measurements.
CRP activates P2 in vitro.
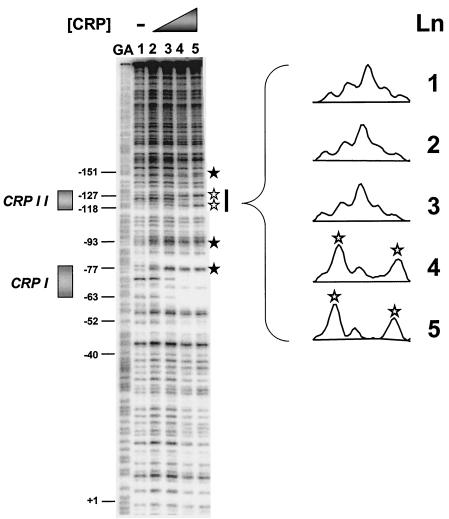
Previously, we used electrophoretic mobility shift analysis to demonstrate the existence of one CRP binding site in the acs regulatory region (13). To pinpoint this binding site, we performed DNase I footprint analysis. We incubated the acs promoter fragment pacs444, an AatII-HindIII fragment that carries acs sequences from position −379 to position +65 (Table 1), with increasing concentrations of purified CRP in the presence of cAMP (Fig. 3). Surprisingly, we observed two regions of protection, each with associated hypersensitive sites. At the lowest CRP concentration (Fig. 3, compare lane 1 with lane 2), two hypersensitive sites appeared (−77 and −93). With increasing CRP concentration (Fig. 3, lanes 3 and 4), protection appeared between positions −60 and −83. These results support the existence of a site (CRP I) centered at −69.5, as predicted by sequence analysis (Fig. 1). At higher CRP concentrations (Fig. 3, lanes 3 to 5), hypersensitive sites appeared at positions −120, −130, and −151, followed by a second region of protection in the region between −118 and −127 (inset). These results suggest the existence of a second, lower-affinity upstream DNA site for CRP (CRP II). Sequence inspection revealed a site centered at position −122.5 that resembled the consensus CRP sequence (Fig. 1).
FIG. 3.
CRP binds specifically to two sites within the acs regulatory region. (A) End-labeled pacs444 AatII-HindIII fragment (carrying acs promoter sequences from positions −379 to +65) was incubated with increasing concentrations of CRP protein and subjected to DNase I footprint analysis. The concentration of CRP in each reaction was as follows: lane 1, no protein; lane 2, 30 nM; lane 3, 125 nM; lane 4, 250 nM; lane 5, 500 nM. The gel was calibrated with Maxam-Gilbert G+A sequencing reactions of the labeled fragment (GA). The locations of CRP I and CRP II are shown by boxes, and hypersensitive sites are indicated by stars. The region encompassing the CRP II signature is designated by the thick bar to the right of the figure. (B) A graphic representation of the CRP II region as quantified with ImageQuant (version 5.2) software (Molecular Dynamics). The hypersensitive sites are indicated by stars.
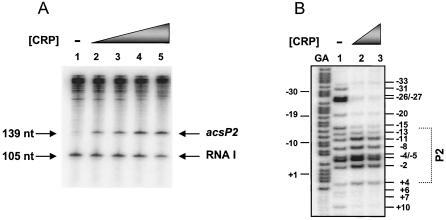
Using an in vitro transcription assay, we determined that CRP activates transcription of acs. Alone, RNAP did not transcribe acs efficiently (Fig. 4A, lane 1), while producing a robust 105-nucleotide band that corresponds to the RNA 1 control. In the presence of CRP, however, RNAP transcribed from P2 quite efficiently (Fig. 4A, lanes 2 to 5), producing a 139-nucleotide band with no change in the intensity of the RNA 1 control.
FIG. 4.
CRP activates transcription in vitro by focusing RNAP to P2. (A) CRP is required for transcription of P2 in vitro. In vitro multiround transcription assays of the P2 promoter carried by plasmid pSRacs444. Reactions contained purified RNA polymerase and increasing concentrations of purified CRP. The concentration of CRP was as follows: lane 1, no protein; lane 2, 25 nM; lane 3, 50 nM; lane 4, 100 nM; lane 5, 300 nM. Transcription initiates from P2 and terminates at a strong transcriptional terminator within the vector backbone (pSR), producing a single 139-nucleotide (nt) transcript. The 105-nucleotide RNA I transcript, encoded by pSR, served as an internal transcriptional control. Arrows indicate the P2 and RNA I transcripts. (B) Potassium permanganate footprint analysis of complexes formed at the acs promoter region. The figure shows the cleavage products produced when the end-labeled pacs444 AatII-HindIII fragment was incubated with purified RNA polymerase and purified CRP and then subjected to potassium permanganate footprint analysis. All reactions contained 50 nM RNAP polymerase and the following CRP concentration: lane 2, no protein; lane 3, 25 nM; lane 4, 100 nM. The gel was calibrated with Maxam-Gilbert G+A sequencing reactions of the labeled fragment (GA). The location of the P2 promoter is shown.
Although RNAP alone cannot transcribe from P2, it can bind and melt DNA in the region that includes P2. Note that permanganate footprint analysis revealed a long stretch of modified and cleaved residues from −33 to +10 (Fig. 4B, lane 1). In contrast, we observed no cleaved residues in the absence of RNAP (data not shown). Permanganate footprint analysis modifies and cleaves T's within single-stranded DNA, such as those present in a transcription open complex. Open complexes tend to be 10 to 14 bp in length; thus, these results suggest the potential for multiple open complexes. Permanganate footprint analysis also showed that CRP favored open-complex formation between nucleotides +4 and −15 (Fig. 4B, lanes 2 and 3), a region that includes the P2 −10 element (TATTAT, Fig. 1). Taken together, these results suggest that CRP facilitates transcription by “focusing” RNAP to P2.
Both CRP I and CRP II contribute to acs transcription in vivo.
To assess the regulatory role of each DNA site for CRP (CRP I and CRP II), we generated mutations predicted, on the basis of analogous mutations in the site for CRP within the lac promoter that reduce affinity to as much as 1/70th that of the wild-type site (6), to reduce the ability of CRP to bind. We incorporated each mutation into an acs::lacZ fusion (λCB12, Table 1) that encompasses the entire acs regulatory region, as described above (Fig. 1B). We then constructed a set of lysogens in a strain wild type for crp (AJW678, Table 1), with each lysogen carrying a single copy of either λCB12 (AJW1941) or one of the mutant derivatives (Table 1).
For the proximal, higher-affinity site CRP I, we generated substitutions in the upstream half-site (G75C, T76A, and G77C) or inserted 1 bp into the spacer (+1A) (Fig. 5A). Electrophoretic mobility shift assays showed that each mutation significantly diminished the ability of CRP to bind (data not shown). Compared to the wild-type fusion (λCB12), all four mutant fusions yielded significantly lower activity (Fig. 5B). Substitution at the critical G75 position or insertion into the spacer yielded ≈6-fold-lower activity, only slightly higher than the activity exhibited by a λCB12 lysogen of a strain deleted for crp (13). This result suggests the existence of low-level basal CRP-independent activity and clearly supports the hypothesis that optimal CRP-dependent activation requires CRP I.
FIG. 5.
acs transcription requires CRP I, while optimal transcription also involves CRP II. (A) Schematic of the acs promoter region showing the sequences of CRP I and CRP II. The italic letters and numbers above the sequences indicate site-directed mutations. (B) β-Galactosidase activity of acs::lacZ transcriptional fusions, either wild type or mutant for CRP I (G75C, T76A, G77C, +1A) or CRP II (G126C). Wild-type strain AJW678 was lysogenized with a hybrid λ that carried either the wild-type acs::lacZ fusion (λCB12) or one of its mutant derivatives (λCB15, λCB20, λCB21, λCB21a, or λCB36). The resultant strains were grown in TB, samples were harvested during the transition from exponential growth to stationary phase, the β-galactosidase activity was determined, and the activity was expressed as a percentage of the wild-type value. Each value represents the mean ± SEM of at least three independent measurements. Fold changes in relation to the wild type are noted below the histogram. (C) β-Galactosidase activity of acs::lacZ transcriptional fusions, either wild type (λCB12), mutant (G75C) for CRP I (λCB20), or mutant (G126C) for CRP II (λCB26) carried by cells with crp deleted (strains AJU2163, AJW2181, or AJW2182, respectively) and transformed. The resultant transformants were grown in TB, samples were harvested during the transition from exponential growth to stationary phase, the β-galactosidase activity was determined, and the activity was expressed as a percentage of the wild-type value. Each value represents the mean ± SEM of at least three independent measurements.
For the upstream, lower-affinity site CRP II, we generated the substitution G126C in the upstream half-site (Fig. 5A). Electrophoretic mobility shift assays did not detect CRP binding to CRP II (data not shown). However, this substitution is analogous to G75C, the most severe mutation generated in CRP I. Therefore, we predict that it severely impacted the ability of CRP to bind. Compared to the wild-type fusion (λCB12), this mutant fusion yielded 1.7-fold lower activity (Fig. 5B). This moderate decrease in transcription, together with the fact that −10 element and CRP I substitutions resulted in significant decreases in transcription, suggests that CRP bound to CRP II functions as a coactivator of the CRP bound at CRP I.
To confirm that the G126C mutation in CRP II does not exert a CRP-independent effect upon transcription, we generated Δcrp cells carrying fusions to the wild-type, G75C mutant, and G126C mutant promoters. In the absence of CRP, the G126C mutant promoter yielded activity almost identical to that of its wild-type parent and the G75C mutant (Fig. 5C). This lack of a difference in promoter activities supports the notion that the loss of activity due to G126C is CRP dependent. Complementation with wild-type CRP yielded activities similar to that observed in the wild-type background. Strains carrying the wild-type fusion regained wild-type levels of transcription, those carrying the G75C mutant fusion gained no additional activity, and the G126C mutant fusion regained 50 to 60% of wild-type activity. These results further support the importance of CRP binding to CRP I and the contribution of CRP binding to CRP II.
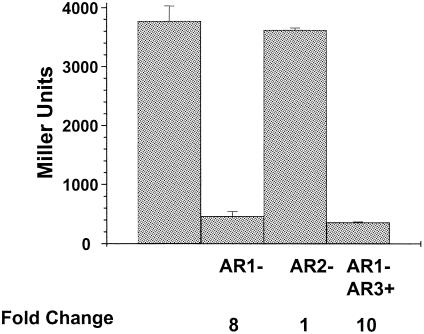
CRP-dependent activation requires activating region I.
Given the location of CRP I (−69.5) and CRP II (−122.5), we predicted that CRP activates acs transcription by class I interactions. To test this prediction, we constructed a strain deleted for crp and lysogenized with λCB12 (AJW2163). We transformed this strain with plasmids that express wild-type CRP or its mutant variants. The variant CRP H159L possesses a defective AR1 and therefore activates class I and class II promoters poorly. The variant CRP K101E possesses a defective AR2 and thus activates class II promoters poorly. The double mutant CRP H159L/K52N is defective in AR1 and possesses AR3, a nonnative activation region that compensates for the lack of AR1 in class II interactions but not in class I interactions (reviewed in reference 5). Figure 6 shows that the AR1 substitution (regardless of AR3 status) resulted in an 8- to 10-fold reduction in β-galactosidase activity, while the AR2 substitution exerted no significant effect. These results support the hypothesis that CRP activates acs transcription by means of class I interactions and not via class II interactions.
FIG. 6.
CRP-dependent activation requires activating region I. β-Galactosidase activity of a λCB12 lysogen deleted for crp (strain AJW2163) and transformed with plasmids expressing either wild-type CRP, CRP H159L, CRP K101E, or CRP H159L/K52N was determined. The resultant transformants were grown in TB, samples were harvested at the transition from exponential to stationary phase, and the β-galactosidase activity was determined and expressed in Miller units. Each value represents the mean ± SEM of at least three independent measurements. Changes in relation to the wild type are noted below the histogram.
Multiple α-CTD residues influence transcription.
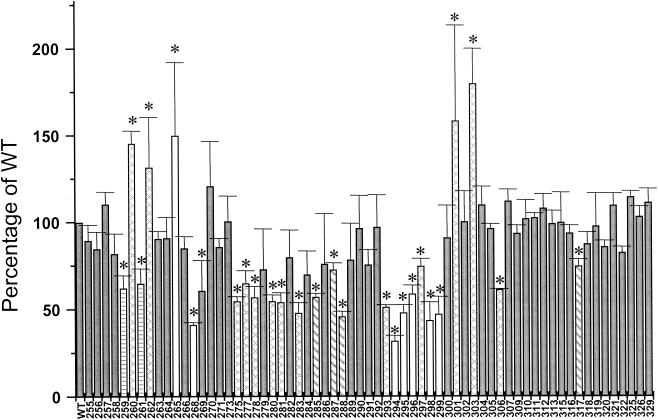
Class I interactions also require specific, surface-exposed α-CTD determinants (reviewed in reference 5). To identify α-CTD residues involved in acs transcription, we used a collection of α-subunit mutants containing single alanine substitutions at each nonalanine position in the α-CTD (residues 255 to 329). With this collection of mutants, we transformed the λCB12 lysogen AJW1941. We expressed the β-galactosidase activity of each transformant as a percentage of wild-type activity and considered those altered by at least 25% significant. By these criteria, we identified 22 residues whose substitution resulted in decreased activity (D259, E261, N268, C269, I275, Y277, I278, D280, L281, Q283, T285, V287, E288, P293, N294, L295, G296, K297, K298, S299, V306, and R317) and five residues whose substitution resulted in increased activity (L260, L262, R265, T301, and I303) (Fig. 7). These results suggest that multiple, distinct regions of the α-CTD contribute to acs transcription (Table 2).
FIG. 7.
Effect of single alanine substitutions within α-CTD. The relative β-galactosidase activity of a λCB12 lysogen wild type for crp (AJW1941) and transformed with a set of plasmids expressing a library of alanine-substituted variants of the α-CTD of RNAP (residues 255 to 329) was determined. Residues 267, 272, 274, 308, 324, and 327 are naturally alanines. The resultant transformants were grown in TB, samples were harvested during the transition from exponential growth to stationary phase, and the β-galactosidase activity was determined. Activities are expressed as a percentage of that of the transformant containing a plasmid expressing wild-type α. Each value represents the mean ± SEM of at least two independent measurements. All alanine substitutions that significantly altered expression of the acs::lacZ fusion are marked with an asterisk above the bar. Horizontally striped bars, the 261 determinant; open bars, the 265 determinant; diagonally striped bars, the 287 determinant; cross-hatched bars, residues outside a known determinant.
TABLE 2.
α-CTD substitutions that affect acs transcription
| α-CTD determinant | Substitutions that decreased acs transcription | Substitutions that increased acs transcription | Substitutions adjacent to known determinants |
|---|---|---|---|
| 287 | T285A, V287A, E288A | ||
| 261 | D259A, E261A | L260A, L262A (both increased transcription) | |
| 265 | N268A, N294A, G296A, K298A, S299A | R265A | P293A, K297A (decreased transcription); T301 (increased transcription) |
| Other residues | C269A, I275A, Y277A, I278A, D280A, L281A, Q283A, V306A | I303A |
DISCUSSION
Previously, we demonstrated that CRP plays a critical role in the activation of acs transcription (13). In this study, we determined that P2 can act as the major acs promoter and examined the mechanism by which CRP controls its transcription. In vitro, RNAP alone melted an extensive region in the vicinity of P2 yet failed to transcribe. In contrast, CRP “focused” RNAP to form a single, productive, open complex at P2. In vitro, we defined two DNA sites for CRP, the higher-affinity CRP I site (centered at −69.5) and the lower-affinity CRP II site (centered at −122.5). In vivo, we showed that acs activation absolutely requires a functional CRP I, while optimal transcription also requires CRP II. Gaston et al. (9) demonstrated that a DNA site for CRP located at −69.5 resulted in weak activation compared to the preferred −61.5 site. The addition of a UP element, however, renders the −69.5 position optimal for CRP activation (17). P2 lacks a good consensus UP element proximal to the CRP I site (7). CRP bound to the lower-affinity upstream CRP II site, however, may substitute for the UP element to enhance activation of P2 from the suboptimal −69.5 site.
CRP I and CRP II both reside in classic class I positions (5). Therefore, we hypothesized that class III CRP-dependent activation of acs might consist of simple class I interactions between RNAP and each of two CRP dimers (11, 16, 23), and the evidence supported this conclusion. In vivo, acs transcription required AR1, a surface patch of CRP residues that contacts its α-CTD counterpart, the 287 determinant. Notably, activation did not involve AR2 or AR3, surfaces important for activation of class II promoters (5). While our experiments clearly demonstrated a requirement for AR1, they did not show unequivocally that both dimers require a functional activating region. Using an alanine scanning library of the α-CTD, we showed that acs transcription involves residues within the 287 and 265 determinants. Importantly, activation also involved residues within or adjacent to the 261 determinant, a surface patch known to be important for activation of class I but not class II promoters (Table 2) (20).
Four of the alanine substitutions that significantly increased acs promoter activity (L260A, L262A, R265A, and T301A) reside in or around previously defined determinants (Table 2). R265, the surface-exposed eponymous residue of the 265 determinant, interacts directly with UP elements (1). T301, a surface-exposed residue, resides immediately adjacent to this determinant. In contrast, residues L260 and L262, located adjacent to the 261 determinant, are generally buried, suggesting that they alter the structure of the α-CTD.
Relative to the extensively characterized CRP-dependent class I promoters lacP2 and CC(61.5), we observed some differences in the identity and involvement of residues within determinants. For example, the R265A substitution diminishes transcription from lacP2 and CC(61.5) but enhanced transcription from acs (20). This and other differences probably result from architectural differences between promoters. At acs, however, they may also reflect the involvement of proteins other than CRP and RNAP.
Intriguingly, several substitutions that either significantly decreased (C269A, I275A, Y277A, I278A, D280A, L281A, Q283A, and V306A) or significantly increased (I303A) acs promoter activity did not correspond to α-CTD surfaces known to be involved in class I activation (Table 2). This observation suggests that the α-CTD plays a more complex role at acs than it does at simple class I promoters, possibly making additional interactions with proteins other than CRP. Candidate proteins include the nucleoid proteins FIS (factor for inversion stimulation) and IHF (integration host factor), both of which participate directly in the regulation of acs transcription (D. F. Browning, C. M. Beatty, S. J. W. Busby, and A. J. Wolfe, unpublished data).
Acknowledgments
We thank Rick Gourse for providing the α-CTD alanine scan and David Keating and Karen Visick for critical reading of the manuscript.
This work was supported by grants MCB-9982762 from the National Science Foundation and LU#11200 from the Loyola University Chicago Potts Foundation (A.J.W.) and by WT066685 from the Wellcome Trust (S.J.W.B.).
REFERENCES
- 1.Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297:1562-1566. [DOI] [PubMed] [Google Scholar]
- 2.Berg, P. 1956. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 222:991-1013. [PubMed] [Google Scholar]
- 3.Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 4.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regualtory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 6.Ebright, R. H., P. Cossart, B. Gicquel-Sanzey, and J. Beckwith. 1984. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein in E. coli. Nature 311:232-235. [DOI] [PubMed] [Google Scholar]
- 7.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 95:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 9.Gaston, K., A. Bell, A. Kolb, H. Buc, and S. Busby. 1990. Stringent spacing requirements for transcription activation by CRP. Cell 62:733-743. [DOI] [PubMed] [Google Scholar]
- 10.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 11.Joung, J. K., L. U. Le, and A. Hochschild. 1993. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc. Natl. Acad. Sci. USA 90:3083-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari, S., C. M. Beatty, Browning, D. F., Busby, S. J. W., E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl-coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari, S., E. Simel, and A. J. Wolfe. 2000. Sigma 70 is the principal sigma factor responsible for the transcription of acs, which encodes acetyl-coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:551-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari, S., R. Tishel, M. Eisenbach, and A. J. Wolfe. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl-coenzyme A synthetase in Escherichia coli. J. Bacteriol. 177:2878-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langdon, R. C., and A. Hochschild. 1999. A genetic method for dissecting the mechanism of transcriptional activator synergy by identical activators. Proc. Natl. Acad. Sci. USA 96:12673-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law, E. C., N. J. Savery, and S. J. W. Busby. 1999. Interactions between the Escherichia coli cAMP receptor protein and the C-terminal domain of the alpha subunit of RNA polymerase at class I promoters. Biochem. J. 337:415-423. [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a molecular laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Savery, N., V. Rhodius, and S. Busby. 1996. Protein-protein interactions during transcription activation: the case of the Escherichia coli cyclic AMP receptor protein. Phil. Trans. R. Soc. London B 351:543-550. [DOI] [PubMed] [Google Scholar]
- 20.Savery, N. J., G. S. Lloyd, S. J. W. Busby, M. S. Thomas, R. H. Ebright, and R. L. Gourse. 2002. Determinants of the C-Terminal domain of the Escherichia coli RNA polymerase α subunit important for transcription at class I cyclic AMP Receptor protein-dependent promoters. J. Bacteriol. 184:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 22.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 23.Tebbutt, J., V. A. Rodius, C. L. Webster, and S. J. Busby. 2002. Architectural requirements for optimal activation by tandem CRP molecules at a class I CRP-dependent promoter. FEMS Microbiol. Lett. 210:55-60. [DOI] [PubMed] [Google Scholar]
- 24.West, D., R. Williams, V. Rhodius, A. Bell, N. Sharma, C. Zou, N. Fujita, A. Ishihama, and S. Busby. 1993. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol. Microbiol. 10:789-797. [DOI] [PubMed] [Google Scholar]