Abstract
Frequent unintended secondary mutations occurred in extraintestinal pathogenic Escherichia coli strains CP9, CFT073, and RS218 during suicide plasmid-mediated, putatively specific deletions of hlyA, papG allele III, and iha. Pulsed-field gel electrophoresis and PCR analyses demonstrated genomic alterations and/or unintended loss of defined virulence genes (papG, the F7-2 papA allele, iutA, sat, hlyD, and cnf). Caution is warranted when attributing the observed phenotypic changes to the intended mutation.
Background.
Bacteria differing only for a particular gene are often used to assess the phenotypic significance of specific genes (5-7, 10, 16, 21, 22). One technique for producing isogenic Escherichia coli mutants involves introducing a replication-defective suicide plasmid containing a modified version of the target gene into a wild-type strain. Derivatives with a mutant allele exchanged for a wild-type allele are isolated through serial selection and screening steps (8, 23). Mutant validation is usually limited to confirming the intended allelic exchange. Here we describe inapparent secondary mutations in allelic exchange mutants created in different laboratories and wild-type strains by contemporary suicide plasmid methods.
Strains.
CP9 (O4:K10/54/96:H5), CFT073 (O6:K2:H1), and RS218 (O18:K1:H7), wild-type pathogenic E. coli strains (1, 11, 13, 14, 23, 26-28), contain established or putative virulence genes, such as pap (P fimbriae), hly (hemolysin), cnf1 (cytotoxic necrotizing factor 1), iha (iron-regulated gene homologue adhesin), sat (secreted autotransporter toxin), iutA (aerobactin system), and malX (pathogenicity-associated island [PAI] marker) (20). UPEC76 is a double pap mutant of CFT073 (23).
Mutagenesis.
The two CP9 hlyA genes, encoded on separate PAIs (18), were targeted for mutation (in the laboratory of H.A.L., Ohio State University, Columbus, Ohio) by using pWAM1746, a derivative of the sucrose-counterselectable suicide vector pCVD442 (24). The hlyA::Kanr mutation from pWAM1746 was introduced into one copy of hlyA in CP9. In a second round of allelic exchange targeting the second CP9 hlyA gene, selection for mutants with two copies of hlyA::Kanr yielded derivatives such as WEX139 (Table 1). Additionally, the Kanr gene in pWAM1746 was replaced with a chloramphenicol resistance (Chlr) gene; this construct was used to generate hlyA::Kanr hlyA:Chlr derivatives (e.g., WEX404 in Table 1). Putative double hlyA mutants were screened for antibiotic resistance, hemolysin, and CNF1 production (25) and were examined in Southern blots (30) for hlyA, cnf1, and pCVD442 sequences (8).
TABLE 1.
Characteristics of selected E. coli parent strains and their putatively isogenic derivatives
| Vector-strain combination | Parent (laboratory) | Strain(s)a | Immediate parent | Intended knockout | Secondary mutation(s)b | PFGE profilec
|
|
|---|---|---|---|---|---|---|---|
| XbaI | AvrII | ||||||
| NAd | CP9 (H.A.L.) | CP9 | NA | NA | NA | 1 | ND |
| 1 | WEX135 | CP9 | cnfl | None | 1 | ND | |
| 2e | WEX139 | CP9 | hlyA (2nd copy) | cnf1 | 1 | ND | |
| 3e | WEX404 | CP9 | hlyA (2nd copy) | cnf1, papG allele III, PFGE shift | 2 | ND | |
| NA | CP9 (J.R.J.) | CP9Nalr | NA | NA | NA | 1 | ND |
| 4 | JJ969-JJ971 | CP9Nalr | papG allele III | None | 1 | ND | |
| 5 | JJ972-JJ975 | CP9Nalr | papG allele I | None | 1 | ND | |
| 6 | JJ985-JJ987 | JJ972 | papG alleles I + III | None | 1 | ND | |
| 6 | JJ988, JJ989 | JJ972 | papG alleles I + III | PFGE shift | 2 | ND | |
| NA | CFT073 (P.I.T.) | CP9Nalr | NA | NA | NA | 1 | 1 |
| 7 | JJ1079, JJ1091, JJ1094 | CP9Nalr | iha | None | 1 | 1 | |
| 7 | JJ1074-5, JJ1078, JJ1080 | CP9Nalr | iha | F7-2 papA allele, iha flanking regions, sat, iutA, PFGE shift | 3 | 3 | |
| 7 | JJ1073, JJ1076-7 | CP9Nalr | iha | hlyD, F7-2 papA allele, iha flanking regions, sat, iutA, PFGE shift | 2 | 2 | |
Mutants analogous to JJ988 and JJ989 were obtained with attempted deletion of papG allele III in RS218Nal (vector -strain combination 8), and mutants analogous to JJ1074-5, JJ1078, and JJ1080 were obtained with attempted deletion of iha in UPEC76 (vector-strain combination 9).
As detected by PCR in extended virulence marker screening (with or without Southern blot confirmation) and/or PFGE.
PFGE profile designations are specific to the particular parent strain (always designated as pattern 1) and its derivatives from a particular laboratory and do not apply across parent strains or laboratories. ND, not done.
NA, not applicable.
Different hlyA suicide plasmid constructs were used to generate WEX139 and WEX404.
The cnf1 gene of CP9 was targeted for allelic exchange (by H.A.L.) by using a pSC101-based suicide plasmid and was extensively confirmed (25).
To delete papG alleles I and III in strain CP9, in-frame deleted versions of these genes were ligated separately into suicide vector pCVD442 and the resulting plasmids were introduced into a nalidixic acid-resistant CP9 derivative (CP9Nalr) in the laboratory of J.R.J. (University of Minnesota, Minneapolis). Dual antibiotic selection yielded the desired integrants. After nonselective growth, derivatives having resolved the plasmid were selected on 5% sucrose agar without sodium chloride. PCR using flanking and internal papG primers confirmed allele exchange. A double papG mutant was created in a papG allele I single mutant by using the papG allele III suicide plasmid, described above.
papG allele III of a Nalr RS218 derivative (RS218Nalr) was deleted (by J.R.J) as described for CP9Nalr.
iha was deleted (in the laboratory of P.I.T., University of Washington, Seattle) from CFT073Nalr, a Nalr CFT073 derivative. An in-frame-deleted version of iha was ligated into pCVD442, yielding the construct used to derive allelic exchange iha mutants of CFT074Nalr, as described above for papG in CP9Nalr. Exchange was confirmed by PCR with internal and flanking iha primers. iha was similarly deleted from UPEC76.
Extended virulence genotypes.
Isolates were tested in duplicate for 35 virulence-associated markers by multiplex PCR (19, 20).
PFGE.
XbaI- with or without AvrII-generated macrorestriction profiles were examined by pulsed-field gel electrophoresis (PFGE) (performed according to the 1998 Centers for Disease Control and Prevention training course “Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis”). Altered profiles were confirmed in duplicate.
Findings.
In six of the nine unique (host strain-target gene-suicide vector) combinations, unintended nonparental genotype alterations were observed (Table 1 and Fig. 1 to 3).
FIG. 1.
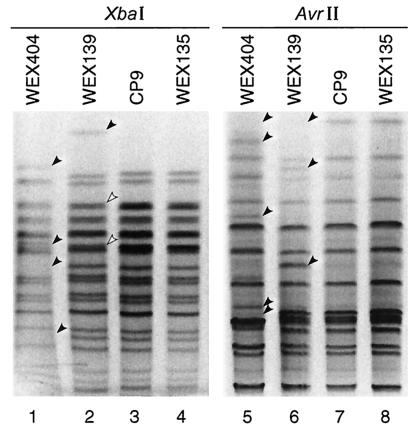
PFGE profiles of strain CP9 and selected derivatives. Lanes 1 to 4 contain XbaI digests, and lanes 5 to 8 contain AvrII digests. Lanes: 1 and 5, WEX404 (hlyA cnf1 papG allele III); 2 and 6, WEX139 (hlyA cnf1); 3 and 7, CP9 (parent); and 4 and 8, WEX135. Arrowheads indicate positions of profile differences between parent and derivatives. Solid arrowheads indicate complete loss or gain of band, and open arrowheads indicate diminished intensity of band.
FIG. 3.
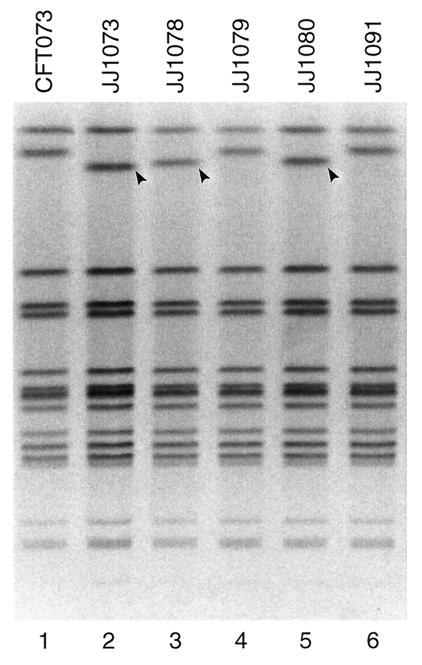
AvrII PFGE profiles of strains CFT073Nalr and selected derivatives. Lanes: 1, CFT073Nalr; 2, JJ1073 (iha iutA hlyD mutant of CFT073Nalr); 3 and 5, JJ1078 and JJ1080, respectively (both, iha iutA mutants of CFT073Nalr); 4 and 6, JJ1079 and JJ1091, respectively (both, iha mutants of CFT073Nalr). Arrowheads indicate positions of profile differences between parent and derivatives.
cnf1 was deleted from strain CP9 without detectable secondary mutations (WEX135 in Table 1 and Fig. 1), but WEX139 (intentionally deleted of hlyA) neither produced CNF1 nor contained cnf1 by Southern hybridization (data not shown) or PCR analysis (Table 1). Also, its PFGE profiles were nonparental (Fig. 1).
Forty-six (6%) of 800 second-round derivatives (e.g., WEX404), created with a Chlr derivative of pWAM1746, were phenotypically Kanr Chlr Amps Hly− as expected. However, like WEX139, each was CNF1− and contained no cnf1 by Southern hybridization. These isolates also lost papG allele III (Table 1) and exhibited altered PFGE profiles (Fig. 1).
papG alleles I and III were deleted individually from CP9Nalr without detectable secondary mutations (Table 1). However, deletion of papG allele III from an allele I single mutant (JJ972), although producing several apparently isogenic double papG mutants (e.g., JJ985 to -987), also yielded derivatives (e.g., JJ988 to -989) with altered XbaI PFGE profiles (Fig. 2) despite retaining their virulence genotype (Table 1). PFGE alterations with retained virulence genotyping also occurred with attempted deletion of papG allele III from RS218Nalr (data not shown).
FIG. 2.
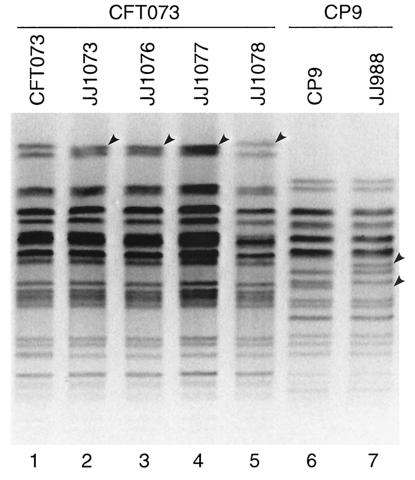
XbaI PFGE profiles of strains CFT073Nalr, CP9Nalr, and selected derivatives. Lanes: 1, CFT073Nalr; 2 to 4, JJ1073, JJ1076, and JJ1077, respectively (all iha iutA hlyD mutants of CFT073Nalr); 5, JJ1078 (iha iutA mutant of CFT073Nalr); 6, CP9Nalr; 7, JJ988 (double papG mutant of CP9Nalr). Arrowheads indicate positions of profile differences between parents and respective derivatives.
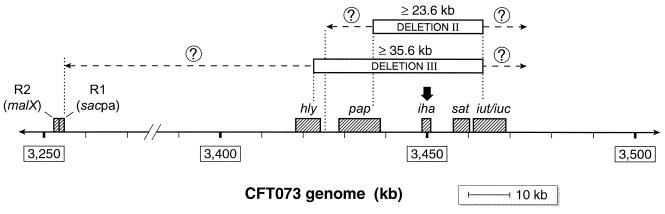
Attempted deletion of iha in strain CFT073Nalr yielded iha mutants with the parental PFGE profile (e.g., JJ079, etc.); these accounted for approximately 25% of colonies from plates containing no salt or sucrose. Except for iha, such derivatives exhibited preserved virulence profiles, including iha flanking sequences. However, an approximately equal number of derivatives (e.g., JJ1074, etc.) also lost iha flanking sequences, iutA, sat, and the F7-2 papA (structural subunit) allele and exhibited altered PFGE profiles (Table 1 and Fig. 2 and 3). Additional isolates (e.g., JJ1073) were nonhemolytic, having lost hlyD in addition to the inadvertent deletions described above, and exhibited yet different PFGE profiles (Table 1 and Fig. 2 and 3). Downward shifts in the second-highest band in the AvrII profiles were more pronounced for the latter than for the former group of mutants, consistent with larger deletions (Fig. 3). The orientation of the corresponding genes within the pheV PAI of CFT073 suggested possible large deletions flanking iha, with minimum estimated losses of 26.6 and 35.6 kb, respectively, for deletions II and III (Fig. 4).
FIG. 4.
Map of the regions of the CFT073 genome adjacent to iha. Positions of relevant genes (hatched boxes) are shown according to their coordinates in the CFT073 genome. Open boxes represent putative deletions involving iha (downward arrow) and flanking regions. Deletion I iha mutants (not shown) exhibit only an internal deletion within iha. Deletion II iha mutants also have lost the F7-2 papA allele, iha flanking sequences, sat, and iutA. Deletion III mutants have lost all these markers plus hlyD. All mutants retain R2 (malX) and R1 (sacpa). Dashed arrows indicate possible extension of deletions, and dotted vertical lines indicate known minimal or maximal boundaries of deletions.
Attempted excision of iha from UPEC76 also yielded iutA iha double mutants (not shown).
Implications of findings.
The high frequency of unanticipated, inadvertent, secondary mutations involving shifts in PFGE patterns and loss of other known virulence loci when often-employed suicide vectors are used is of concern, because the central tenet of allelic exchange is that the resulting phenotypic differences are specifically attributable to the target mutation. However, if additional mutations result, they, and not the intended mutation, may cause the differences. This is particularly problematic when secondary mutations involve known or putative virulence genes and the phenotype of interest is virulence (10).
The high frequency of secondary mutations using unrelated strains and a diversity of target alleles suggests that inadvertent mutations may be common with pCVD442 and, perhaps, other allelic exchange systems. It should be noted that we targeted genes within PAIs, which are structures with a propensity for spontaneous deletions and rearrangements (3, 4, 12, 29, 32), and the unintentionally deleted genes occurred within the same PAIs. We speculate that aberrant integration and/or excision of plasmids caused changes in neighboring DNA, analogous to perturbations in genomic architecture in E. coli K-12 when lysogenized by a Shiga toxin 2-encoding bacteriophage (17). The patterns of virulence gene loss support this hypothesis with CFT073, for which the structure of the relevant PAI is defined (Fig. 4).
Exclusion of inadvertent secondary mutations, short of whole-genome sequencing (2, 32) or other methods that detect small interchromosomal differences (31), poses challenges. That PFGE detected all secondary mutants recommends this modality. Alternatively, multiple independent deletants for the same target gene could be prepared, possibly even in different wild-type backgrounds (25), subjected to confirmation of retention of a panel of non-target loci, and then tested in various systems to confirm uniformity of effect.
Complementation of presumed isogenic in-frame mutants would also help corroborate specific attribution of effect (10), but this approach too is imperfect, because gene regulation and copy number may significantly influence phenotype (9, 15, 25), although low-copy-number vectors may help (15). Alternatively, the wild-type allele could be restored to the genome at the native locus by reverse allelic exchange. These considerations illustrate the challenges inherent in attempting to define causal relationships in complex biological systems, the use of contemporary molecular methods notwithstanding.
Acknowledgments
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.), National Institutes of Health grants DK-47504 (J.R.J.) and AI47499 (P.I.T.), National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.), and the National Kidney Foundation (H.A.L.).
Thomas A. Russo provided CP9, Harry L. T. Mobley provided CFT073 and UPEC76, and Kwang Sik Kim provided RS218. Adam L. Stell and Sheldon N. Finver provided expert technical assistance. Dave Prentiss (VA Medical Center) helped prepare the figures.
REFERENCES
- 1.Badger, J. L., C. A. Wass, S. J. Weissman, and K. S. Kim. 2000. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect. Immun. 68:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, C. A., and C. K. Rode. 1996. Pathogenicity island evaluation in Escherichia coli K1 by crossing with laboratory strain K-12. Infect. Immun. 64:3218-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, M. P. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar− agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dattelbaum, J. D., C. V. Lockatell, D. E. Johnson, and H. L. T. Mobley. 2003. UreR, the transcriptional activator of the Proteus mirabilis urease gene cluster, is required for urease activity and virulence in experimental urinary tract infections. Infect. Immun. 71:1026-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dozois, C. M., F. Daigle, and R. R. Curtiss. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkow, S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10:S274-S276. [DOI] [PubMed] [Google Scholar]
- 11.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. T. Mobley. 2000. Identification of Sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-56. [DOI] [PubMed] [Google Scholar]
- 12.Hacker, J., S. Knapp, and W. Goebel. 1984. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinants of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman, J. A., C. Wass, M. F. Stins, and K. S. Kim. 1999. The capsule supports survival but not traversal of Escherichia coli K1 across the blood-brain barrier. Infect. Immun. 67:3566-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, S.-H., Z.-S. Wan, Y.-H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071-1078. [DOI] [PubMed] [Google Scholar]
- 15.Hull, R. A., B. Nowicki, A. Kaul, R. Runyan, C. Svanborg, and S. I. Hull. 1994. Effect of pap copy number and receptor specificity on virulence of fimbriated Escherichia coli in a murine urinary tract colonization model. Microb. Pathog. 17:79-86. [DOI] [PubMed] [Google Scholar]
- 16.Hytönen, J., S. Haataja, and J. Finne. 2002. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 71:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iguchi, A., R. Osawa, J. Kawano, A. Shimizu, J. Terajima, and H. Watanabe. 2003. Effects of lysogeny of Shiga toxin 2-encoding bacteriophages on pulsed-field gel electrophoresis fragment pattern of Escherichia coli K-12. Curr. Microbiol. 46:224-227. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R., T. A. Russo, F. Scheutz, J. J. Brown, L. Zhang, K. Palin, C. Rode, C. Bloch, C. F. Marrs, and B. Foxman. 1997. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“Class I”) and PrsGJ96 (“Class III”) Gal(α1-4)Gal-binding adhesins. J. Infect. Dis. 175:983-988. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. R., C. Van der Schee, M. A. Kuskowski, W. Goessens, and A. Van Belkum. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from The Netherlands. J. Infect. Dis. 186:1852-1856. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, S., P. B. Killoran, F. C. Fang, and L. W. Riley. 2002. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect. Immun. 70:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mobley, H. L. T., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of αGal(1-4)-βGal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 24.Moxley, R. A., E. M. Berberov, D. H. Francis, J. Xing, M. Moayeri, R. A. Welch, D. R. Baker, and R. G. Barletta. 1998. Pathogenicity of an enterotoxigenic Escherichia coli hemolysin (hlyA) mutant in gnotobiotic piglets. Infect. Immun. 66:5031-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rippere-Lampe, K. E., A. D. O'Brien, R. Conran, and H. A. Lockman. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69:3954-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo, T. A., J. J. Brown, S. T. Jodush, and J. R. Johnson. 1996. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence in an extraintestinal isolate of Escherichia coli. Infect. Immun. 64:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo, T. A., S. T. Jodush, J. J. Brown, and J. R. Johnson. 1996. Identification of two previously unrecognized genes (guaA, argC) important for uropathogenesis. Mol. Microbiol. 22:217-229. [DOI] [PubMed] [Google Scholar]
- 29.Russo, T. A., S. Wenderoth, U. B. Carlino, J. M. Merrick, and A. J. Lesse. 1998. Identification, genomic organization, and analysis of the group III capsular polysaccharide genes kpsD, kpsM, kpsT, and kpsE from an extraintestinal isolate of Escherichia coli (CP9, O4/K54/H5). J. Bacteriol. 180:338-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sokurenko, E. V., V. Tchesnokova, A. T. Yeung, C. A. Oleykowski, E. Trintchina, K. T. Hughes, R. A. Rashid, J. M. Brint, S. L. Moseley, and S. Lory. 2001. Detection of simple mutations and polymorphisms in large genomic regions. Nucleic Acids Res. 29:E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch, R., V. Burland, G. R. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]