Abstract
Salmonella enterica serovar Typhimurium encounters numerous host environments and defense mechanisms during the infection process. The bacterium responds by tightly regulating the expression of virulence genes. We identified two regulatory proteins, termed RtsA and RtsB, which are encoded in an operon located on an island integrated at tRNAPheU in S. enterica serovar Typhimurium. RtsA belongs to the AraC/XylS family of regulators, and RtsB is a helix-turn-helix DNA binding protein. In a random screen, we identified five RtsA-regulated fusions, all belonging to the Salmonella pathogenicity island 1 (SPI1) regulon, which encodes a type III secretion system (TTSS) required for invasion of epithelial cells. We show that RtsA increases expression of the invasion genes by inducing hilA expression. RtsA also induces expression of hilD, hilC, and the invF operon. However, induction of hilA is independent of HilC and HilD and is mediated by direct binding of RtsA to the hilA promoter. The phenotype of an rtsA null mutation is similar to the phenotype of a hilC mutation, both of which decrease expression of SPI1 genes approximately twofold. We also show that RtsA can induce expression of a SPI1 TTSS effector, slrP, independent of any SPI1 regulatory protein. RtsB represses expression of the flagellar genes by binding to the flhDC promoter region. Repression of the positive activators flhDC decreases expression of the entire flagellar regulon. We propose that RtsA and RtsB coordinate induction of invasion and repression of motility in the small intestine.
Salmonella serovars cause a range of human diseases from self-limiting gastroenteritis to life-threatening systemic infections (62). The disease process is initiated by bacterial interaction with and invasion of the intestinal epithelium. Analyses of the early steps in colonization and invasion by Salmonella enterica serovar Typhimurium with the mouse model of infection and in vitro tissue culture models have significantly increased our understanding of these events (20). Serovar Typhimurium invades intestinal epithelial cells by using a type III secretion system (TTSS) encoded by Salmonella pathogenicity island 1 (SPI1). The SPI1 TTSS forms a needle-like structure that injects effector proteins directly into the cytosol of host cells (45, 46, 79). The various effector proteins are implicated in a number of physiological responses, including actin rearrangement that promotes invasion (89), fluid accumulation and transepithelial migration of polymorphonuclear leukocytes (83), and necrosis of Peyer's patch macrophages (12, 63).
The expression of the SPI1 TTSS is controlled in response to a specific combination of environmental signals that presumably act as a cue that the bacteria are in the appropriate anatomic location (7, 71, 75). In the laboratory, the system is active when cells are grown under SPI1-inducing conditions, i.e., high osmolarity and low oxygen. Regulation is mediated primarily via control of the level of HilA, a member of the OmpR/ToxR family of transcriptional regulators encoded on SPI1 (6, 49, 54). HilA directly activates expression of the prg/org and inv/spa operons in SPI1 by binding just upstream of the −35 sequences of PprgH and PinvF (54). Activation of PinvF increases production of InvF, a member of the AraC family of transcriptional regulators (44). InvF then induces expression of effector proteins encoded both within and outside SPI1, including the products of sicA (SPI1), sopE (SopEφ in strain SL1344), and sopB (sigD) (SPI5) (54). Activation of these promoters by InvF requires SicA, a TTSS chaperone (19, 21, 24), which has been suggested to stabilize a complex between InvF, RNA polymerase, and DNA (22).
Two SPI1-encoded proteins, HilC (SirC or SprA) and HilD, both members of the AraC/XylS family of transcriptional regulators, induce expression of hilA (26, 67, 71). Loss of HilD decreases expression of hilA ∼10-fold under SPI1-inducing conditions, whereas a mutation of hilC reduces expression of hilA ∼2-fold (26, 67, 71). HilC and HilD bind to the hilA promoter region, and it is believed that this binding induces the expression of hilA (64, 71, 72). Recent data suggest that HilD acts as a direct activator of hilA (11) as opposed to a derepressor as previously inferred (71, 72). HilC and HilD also induce expression of the inv/spa operon independent of HilA (2, 26, 67). This induction is due to activation of a second promoter located 5′ to the known PinvF (2).
Genetic studies have identified a number of regulatory proteins encoded outside SPI1 that control expression of hilA. These include the two-component regulatory systems PhoPQ (9, 66), PhoBR (56), OmpR/EnvZ (55), and SirA/BarA (1, 43, 67). With the exception of OmpR/EnvZ, which alters hilA expression via hilC (55), it is not known whether control of hilA is direct or indirect. Other proteins reported to affect hilA expression include Hha (29), Lon (81), Fis (85), integration host factor (IHF) (28), FadD (56), FliZ (25, 42, 56), and HilE (28). With the exceptions of Hha, which binds to the hilA promoter region (29), and HilE, which interacts with HilD (8), it remains mechanistically unclear how these proteins control expression of hilA. The csrAB genes encode a protein-RNA pair that act as both positive and negative regulators of hilA expression (3, 4, 69). A csrA null mutation decreased the steady-state levels of hilC and hilD mRNAs (3), suggesting that the control of hilA is via these two regulators.
Serovar Typhimurium produces peritrichous flagella required for motility. There are more than 40 genes involved in flagellar biosynthesis. These genes are controlled by a regulatory cascade, which is initiated by the production of FlhDC. These regulatory proteins induce expression of the class 2 flagellar genes, including fliA, which encodes an alternative sigma factor required for transcription of the class 3 flagellar genes. This cascade serves to control the timing of gene expression to coincide with assembly of the flagellar apparatus (16, 57). Serovar Typhimurium can produce two immunologically distinct flagellin proteins, FliC or FljB. The production of these proteins is phase variable and mediated by DNA inversion (57).
A number of regulators affect expression of the flagellar regulon. The two-component regulators SirA/BarA act to repress expression of most flagellar operons, including flhDC (35). Interestingly, this repression was observed only during growth in motility agar (35). It remains unclear whether the repressing effects of SirA are via direct interaction with the promoter region of flhDC or whether this repression is mediated through some other regulator. It has been shown in Escherichia coli that CsrA binds the flhDC transcript, protecting it from degradation (84). Other regulators implicated in control of flhDC include catabolite gene activator protein (48, 74), LrhA (51) and HNS, via HdfR (47).
Here we describe two regulatory proteins, one of which induces expression of the SPI1 TTSS (RtsA) whereas the other represses expression of flagellar genes (RtsB). We show that RtsA is capable of directly binding to the promoter region of hilA, suggesting that RtsA can directly induce expression of hilA. We also show that RtsB can directly bind just downstream of the flhDC promoter, suggesting that RtsB directly represses expression of flhDC and thus of the entire flagellar regulon.
MATERIALS AND METHODS
Media, reagents, and enzymatic assays.
Luria-Bertani medium (LB) was used in all experiments for growth of bacteria, and SOC was used for the recovery of transformants (58). MacConkey medium was prepared according to the manufacturer's directions and supplemented with 1% lactose and the appropriate antibiotics (Difco). Motility agar contained 0.3% Bacto agar, 1% tryptone, and 0.5% NaCl supplemented with either 0.2% glucose or 0.2% l-arabinose. Bacterial strains were routinely grown at 37°C, except for strains containing the temperature-sensitive plasmid pCP20 or pKD46, which were grown at 30°C. Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 25 μg/ml. The β-galactosidase chromogenic indicator 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at a concentration of 80 μg/ml. Enzymes were purchased from Invitrogen or New England Biolabs and were used according to the manufacturer's recommendations. Primers were purchased from IDT Inc. β-Galactosidase assays were performed with a microtiter plate assay as previously described (76) on strains grown under the indicated conditions. β-Galactosidase activity units are defined as (micromoles of o-nitrophenol [ONP] formed minute−1) × 103/(optical density at 600 nm [OD600] × milliliters of cell suspension) and are reported as means ± standard deviations, where n is ≥2. Cultures requiring SPI1-inducing conditions were grown statically in LB with 1% NaCl. Cultures requiring SPI1-repressing conditions were grown in LB without NaCl and were highly aerated.
Strain and plasmid construction.
Bacterial strains and plasmids are described in Table 1. All S. enterica serovar Typhimurium strains created for this study are isogenic derivatives of strain 14028 (American Type Culture Collection) and were constructed by using P22 HT105/1 int-201 (P22)-mediated transduction (58). Random MudJ transcriptional fusions to the lac operon were created by using strain TT10286 and transitory cis complementation as previously described (40). Sequencing of the RtsAB-regulated MudJ fusions was performed by utilizing the semi-random-primed sequencing method previously described (17, 39). The Pi-dependent plasmids used in this study were maintained in DH5αλpir. All plasmids were passaged through a restriction-negative, modification-positive Pi+ serovar Typhimurium strain (JS198) prior to transformation into derivatives of strain 14028.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotypea or relevant characteristics | Deletion or cloned end pointsb | Source or referencec |
|---|---|---|---|
| Strains | |||
| 14028 | Wild-type serovar Typhimurium | ATCCd | |
| JS135 | zii-8104::Tn10dTc | 78 | |
| JS247 | Φ(tetA+-rtsA+B+)3 | 4561769-4561768 | |
| JS248 | ΔrtsA5 | 4561755-4560884 | |
| JS249 | ΔrtsB6 | 4560868-4560602 | |
| JS250 | ΔrtsAB7 | 4561769-4560602 | |
| JS251 | ΔhilA112::Cm | 3019885-3021480 | |
| JS252 | ΔhilC113::Cm | 3012135-3012976 | |
| JS253 | ΔhilD114::Cm | 3017865-3018730 | |
| JS254 | ΔinvF100::Cm | 3043931-3043290 | |
| JS255 | ΔhilC-A2914::Cm | 3012135-3021480 | |
| JS256 | ΔhilC-D2915::Cm | 3012135-3018730 | |
| JS257 | ΔslrP100::Cm | 866973-869216 | |
| JS258 | Φ(tetRA-rtsA+B+)3 PhilD111::MudJ | ||
| JS259 | Φ(tetRA-rtsA+B+)3 invC101::MudJ | ||
| JS260 | Φ(tetRA-rtsA+B+)3 sopA100::MudJ | ||
| JS261 | Φ(tetRA-rtsA+B+)3 sopB101::MudJ | ||
| JS262 | Φ(tetRA-rtsA+B+)3 icgA1::MudJ | ||
| JS263 | Φ(tetRA-rtsA+B+)3 yhgF:231:MudJ | ||
| JS264 | zii-8104::Tn10dTc PhilD111::MudJ | ||
| JS265 | zii-8104::Tn10dTc invC101::MudJ | ||
| JS266 | zii-8104::Tn10dTc sopA100::MudJ | ||
| JS267 | zii-8104::Tn10dTc sopB101::MudJ | ||
| JS268 | zii-8104::Tn10dTc icgA1::MudJ | ||
| JS269 | zii-8104::Tn10dTc yhgF231::MudJ | ||
| JS270 | ΔrtsAB7 ΔhilA112::Cm | ||
| JS271 | ΔrtsAB7 invC101::MudJ | ||
| JS272 | ΔrtsAB7 ΔhiD114::Cm | ||
| JS273 | ΔrtsAB7 Φ(hilD-lac+)114 | ||
| JS274 | ΔrtsAB7 Φ(hilC-lac+)113 | ||
| JS275 | ΔrtsAB7 Φ(hilA-lac+)112 | ||
| JS276 | ΔrtsAB7 Φ(invF-lac+)100 | ||
| JS277 | ΔrtsAB7 flhC5456::MudJ | ||
| JS278 | ΔrtsAB7 fliC5050::MudJ | ||
| JS279 | Φ(hilA-lac+)112 | ||
| JS280 | ΔrtsA5 Φ(hilA-lac+)112 | ||
| JS281 | ΔrtsB6 Φ(hilA-lac+)112 | ||
| JS282 | Φ(invF-lac+)100 | ||
| JS283 | ΔrtsA5 Φ(invF-lac+)100 | ||
| JS284 | ΔrtsB6 Φ(invF-lac+)100 | ||
| JS285 | sopA100::MudJ | ||
| JS286 | ΔrtsA5 sopA100::MudJ | ||
| JS287 | ΔrtsB6 sopA100::MudJ | ||
| JS288 | ΔrtsAB7 sopA100::MudJ | ||
| JS289 | sopB101::MudJ | ||
| JS290 | ΔrtsA5 sopB101::MudJ | ||
| JS291 | ΔrtsB6 sopB101::MudJ | ||
| JS292 | ΔrtsAB7 sopB101::MudJ | ||
| JS293 | invC101::MudJ | ||
| JS294 | ΔrtsA5 invC101::MudJ | ||
| JS295 | ΔrtsB6 invC101::MudJ | ||
| JS296 | icgA1::MudJ | ||
| JS297 | ΔrtsA5 icgA1::MudJ | ||
| JS298 | ΔrtsB6 icgA1::MudJ | ||
| JS299 | ΔrtsAB7 icgA1::MudJ | ||
| JS300 | ΔrtsAB7 ΔhilC113::Cm Φ(hilA-lac+)112 | ||
| JS301 | ΔrtsAB7 ΔhiD114::Cm Φ(hilA-lac+)112 | ||
| JS302 | ΔrtsAB7 ΔhilC-D2915::Cm Φ(hilA-lac+)112 | ||
| JS303 | ΔrtsAB7 ΔhilA112::Cm Φ(invF-lac+)100 | ||
| JS304 | ΔrtsAB7 ΔhilC-D2915::Cm Φ(invF-lac+)100 | ||
| JS305 | ΔrtsAB7 ΔhilC-A2914::Cm Φ(invF-lac+)100 | ||
| JS306 | ΔrtsAB7 sopA100::MudJ | ||
| JS307 | ΔrtsAB7 ΔhilA112::Cm sopA100::MudJ | ||
| JS308 | ΔrtsAB7 ΔinvF100::Cm sopA100::MudJ | ||
| JS309 | ΔrtsAB7 sopB101::MudJ | ||
| JS310 | ΔrtsAB7 ΔhilA112::Cm sopB101::MudJ | ||
| JS311 | ΔrtsAB7 ΔinvF100::Cm sopB101::MudJ | /PICK> | |
| JS312 | ΔrtsAB7 invC101::MudJ | ||
| JS313 | ΔrtsAB7 ΔhilA112::Cm invC101::MudJ | ||
| JS314 | ΔrtsAB7 ΔinvF100::Cm invC101::MudJ | ||
| JS315 | ΔrtsAB7 icgA1::MudJ | ||
| JS316 | ΔrtsAB7 ΔhilA112::Cm icgA1::MudJ | ||
| JS317 | ΔrtsAB7 ΔinvF100::Cm icgA1::MudJ | ||
| JS318 | ΔrtsAB7 Φ(slrP-lac+)100 | ||
| JS319 | ΔrtsAB7 ΔhilC-D2915::Cm Φ(slrP-lac+)100 | ||
| JS320 | ΔrtsAB7 ΔinvF100::Cm Φ(slrP-lac+)100 | ||
| JS321 | ΔrtsAB7 ΔhilA112::Cm Φ(slrP-lac+)100 | ||
| JS322 | ΔrtsA5 ΔhilC-D2915::Cm Φ(hilA-lac+)112 | ||
| JS323 | ΔrtsA5 ΔhilC-D2915::Cm Φ(slrP-lac+)100 | ||
| JS324 | Φ(rtsA-lac+)5 | ||
| JS325 | Φ(rtsB-lac+)6 | ||
| TT10286 | LT2 hisD9953::MudJ his-9944::MudI | 40 | |
| TH1077 | LT2 fliC5050::MudJ | 34 | |
| TH4054 | LT2 flhC5456::MudJ | 18 | |
| DH5αλpir | E. coli K-12 endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lac-argF)U169 deoR φ80 Δ(lac)M15 λpir+ | Lab stock | |
| MG1655 | E. coli K-12 F− λ−ilvG rfb-50 rph-1 | Lab stock | |
| Plasmids | |||
| pKD46 | bla PBADgam bet exo pSC101 oriTS | 23 | |
| pCP20 | bla cat cI857 λPRflp pSC101 oriTS | 15 | |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 23 | |
| pKD13 | bla FRT ahp FRT PS1 PS4 oriR6K (for creating in-frame deletions) | 23 | |
| pCE36 | ahp FRT lacZY+ this oriR6K | 27 | |
| pCE37 | ahp FRT lacZY+ this oriR6K | 27 | |
| pBAD30 | bla araC PBAD pACYC184 ori | 36 | |
| pCE46 | bla araC λAttL1 ccdB+cat λAttL2 pACYC184 ori | ||
| pRtsA | bla PBAD λAttB1 rtsA+ λAttB2 pACYC184 ori | 4561766-4560885 | |
| pRtsB | bla PBAD λAttB1 rtsB+ λAttB2 pACYC184 ori | 4560890-4560595 | |
| pRtsAB | bla PBAD λAttB1 rtsA+B+ λAttB2 pACYC184 ori | 4561766-4560595 | |
| pCE81 | bla PBAD λAttB1 myc-rtsA λAttB2 pACYC184 ori | 4561737-4560885 | |
| pCE82 | bla PBAD λAttB1 myc-rtsB λAttB2 pACYC184 ori | 4560850-4560595 | |
| pLS118 | bla PBADhilD-myc-His pACYC184 ori | 72 | |
| pLS119 | bla PBADhilC-myc-His pACYC184 ori | 72 |
Unless otherwise noted, all strains are isogenic derivatives of 14028.
Numbers indicate the base pairs (inclusive) that are deleted (for strains) or cloned (for plasmids), as defined in the S. enterica serovar Typhimurium LT2 genome sequence in the National Center for Biotechnology Information database.
This study, unless otherwise noted.
ATCC, American Type Culture Collection.
Standard recombinant DNA methods were used for the construction of plasmids (70). The rtsA, rtsB, and rtsAB genes were cloned by using the Gateway PCR method (Invitrogen) (38). Primers flanked with the λattB sites were used to PCR amplify rtsA (RtsA1, GGGGACAAGTTTGTACAAAAAAGCAGGCTTACACGCACATTTAATAAAAGG; RtsA2, GGGGACCACTTTGTACAAGAAAGCTGGGTCAGTATTAACATATTGATACG), rtsB (RtsB1, GGGGACAAGTTTGTACAAAAAAGCAGGCTTATTCCTCTCGTCATCAATATG; RtsB2, GGGGACCACTTTGTACAAGAAAGCTGGGTCAAATTACGTAATATCGACTG), and rtsAB (RtsA1and RtsB2). The PCR products were gel purified and cloned into plasmid pDONR201 (38) with a mixture of λInt and IHF, which catalyzes a site-specific recombination between the λattB-flanked PCR products and the λattP sites located on the plasmid. This resulted in plasmids that contained the gene of interest flanked by λattL sites (38). The inserts were then sequenced to ensure that they did not contain mutations. To create plasmids in which expression of these genes was l-arabinose inducible, we cloned a λattR-flanked ccdB+ cat cassette into a blunt-ended EcoRI site of pBAD30, creating pCE46 (36, 38). λInt, λXis, and IHF were used to recombine the λattL-flanked rtsA, rtsB, and rtsAB fragments into pCE46 to create the plasmids pRtsA, pRtsB, and pRtsAB (38). To create c-Myc epitope-tagged versions of RtsA and RtsB, we utilized a two-step overlapping PCR method in conjunction with Gateway cloning. The Myc tag was located in the 5′ primer. The primers were used to PCR amplify rtsA (myc-RtsA, GATAGAACCATGGAACAAAAATTAATTTCTGAAGAAGATTTACTAAAAGTATTTAATCCC; RtsA2) or rtsB (myc-RtsB, GATAGAACCATGGAACAAAAATTAATTTCTGAAGAAGATTTACAGTATAAGAACAAAGCA; RtsB2). A primer with homology to the Myc tag (AttBmyc, GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAGAAGATAGAACCATGGAAC) was then used to add the AttB1 sequences to the 5′ end of rtsA or rtsB in separate PCR amplifications. The resulting PCR products were cloned by using a single-step reaction into pCE46 according to the directions of the manufacturer (Invitrogen).
Construction of chromosomal deletion-insertions and lac fusions.
Deletions of the hilA, hilC, hilD, invF, hilC-D, hilC-A, and slrP genes and concomitant insertion of a chloramphenicol resistance cassette were carried out by lambda Red-mediated recombination (23, 88) as described previously (27). PCR products containing the antibiotic resistance cassette flanked by 30 bp of homology to the gene of interest in the 5′ and 3′ ends were transformed into a serovar Typhimurium strain containing pKD46, which is temperature sensitive and carries the λ red, gam, and bet genes under l-arabinose control (23). The endpoints of each deletion are indicated in Table 1. The tetRA genes from Tn10 were inserted upstream of rtsAB by using this system. The tetRA cassette was amplified by using rtsA-tet1 and rtsA-tet2, which have 15 nucleotides of homology to tetRA on the 3′ end and 30 nucleotides of homology to the DNA 10 bp upstream of the rtsA open reading frame (ORF). In all cases, the appropriate insertion of the antibiotic resistance marker was checked by P22 linkage to known markers and/or by PCR analysis. In each case, the constructs resulting from this procedure were moved into a clean wild-type background (14028) by P22 transduction. In-frame deletions were created or antibiotic resistance cassettes were removed by using the temperature-sensitive plasmid pCP20 carrying the FLP recombinase (15). Mutations constructed with the pKD3 template plasmid (23) were converted to transcriptional lac+ fusions by using an FLP/FRT-mediated site-specific recombination method as previously described (27).
Analysis of Salmonella secreted proteins.
Overnight cultures of the indicated strains were diluted 1/20 into 10 ml of LB-ampicillin-0.2% l-arabinose and grown with shaking at 225 rpm on a platform shaker for 4 h at 37°C. Ten milliliters of culture supernatant was centrifuged two times at 5,000 × g. The culture supernatant was then filter sterilized by using a 0.2-μm-diameter syringe filter. Proteins were precipitated with ice-cold trichloroacetic acid at a final concentration of 10% after incubation on ice for 30 min. The proteins were pelleted by centrifugation at 15,000 × g for 30 min at 4°C. The supernatant was then removed, and the pellets were washed with 5 ml of cold acetone. The samples were then centrifuged for another 20 min at 15,000 × g. The supernatant was removed, and the pellets were allowed to air dry. The pellets were then resuspended in 50 μl of 50 mM Tris (pH 8), and 20 μl of 4× sodium dodecyl sulfate (SDS) loading buffer (5) was added. The samples (15 μl total) were then separated by SDS-12.5% polyacrylamide gel electrophoresis (PAGE) (5). The proteins were stained with GelCode Blue according to the directions of the manufacturer (Pierce).
Gel shift assays.
Whole-cell extracts for gel shift assays were prepared by subculturing overnight cultures 1/100 in LB and growing them to an OD600 of 0.5, at which time 0.2% l-arabinose was added and cultures were grown for an additional 3 h at 37°C. Cultures were then centrifuged, and the pellets were resuspended in 10 mM Tris-Cl (pH 8)-50 mM KCl-10% glycerol-1 mM dithiothreitol-0.5 mM EDTA. Samples were lysed by passage through a French press. Lysates were clarified by centrifugation at 16,000 × g for 30 min at 4°C. The protein concentration in each sample was determined by using a bicinchoninic acid assay (Pierce). Binding reaction mixtures contained approximately 0.1 ng of 32P-labeled DNA, 50 μg of herring sperm DNA per ml, 10 mM Tris-Cl (pH 8), 50 mM KCl, 100 μg of bovine serum albumin per ml, 10% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA, and the appropriate concentration of whole-cell extract in a final volume of 20 μl. To determine if RtsA or RtsB directly bound to the DNA, 100 ng of anti-Myc antibody (Invitrogen) were added to the appropriate reaction mixtures. Binding reaction mixtures were incubated for 20 min at room temperature and then subjected to electrophoresis on a 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA at room temperature (5). Gels were dried on filter paper in a vacuum drier and visualized with a Fuji phosphorimager (FLA-3000) and Image Reader software.
RESULTS
Identification of rtsAB.
The completed S. enterica serovar Typhimurium LT2 genome reveals a large number of putative genes with no known function. We identified two regulatory genes that are present in most Salmonella serovars but absent from E. coli K-12 (10, 59, 65). In serovar Typhimurium, these regulators are located on a recently described 15-kb island inserted near the tRNAPheU gene (37). The tRNAPheU island in serovar Typhimurium contains one previously identified gene, phoN, which encodes a nonspecific acid phosphatase. It also contains several unknown ORFs, including an operon that encodes a homolog of dimethyl sulfoxide reductase, an operon encoding a putative acetyltransferase and a transcriptional regulator belonging to the CopG family, an IS200 element, and a regulatory operon encoding two putative transcriptional regulators that we have named rtsAB (for regulator of TSS; STM4315 and STM4314, respectively). Sequence analysis of the regulatory operon revealed that RtsA belongs to the AraC/XylS family of transcriptional regulators and is homologous to both HilC (37% identical and 55% similar overall) and HilD (34% identical and 55% similar overall). RtsB is a small protein that contains a putative helix-turn-helix DNA binding motif.
Identification of RtsAB-regulated genes.
To identify RtsAB-regulated genes, we created a strain in which expression of rtsAB was conditional. The strain JS247 contains the tetRA genes from Tn10 integrated 10 bp upstream of the rtsA ORF by the λ Red recombinase method (23, 88). This construct uncoupled expression of rtsAB from the putative rtsA promoter and placed expression of rtsAB under the control of the tetA promoter (68). In the absence of tetracycline, TetR represses expression of tetA and thus rtsAB. In the presence of tetracycline, expression of both tetA and rtsAB is induced 5.5-fold (data not shown).
We generated 50,000 independent MudJ transcriptional lac fusions in the Φ(tetA+-rtsA+B+)3 strain. The resulting colonies were replica plated onto LB-X-Gal and LB-X-Gal-tetracycline or on MacConkey-lactose and MacConkey-lactose-tetracycline. Using this method, we identified 39 lac fusions with altered levels of expression in the presence of tetracycline. Two classes of false positives were anticipated in this screen. First, the lac+ fusion could be controlled directly by the tetRA promoters. These fusions would be linked to the Φ(tetA+-rtsA+B+)2 construct by P22 transduction. None of the MudJ fusions fell into this class. Second, the expression of the fusion could be altered by the presence of tetracycline independent of the effect of tetracycline on the expression of rtsAB. To eliminate these fusions, we transduced each MudJ into a strain containing zii-8104::Tn10dTc, which does not affect the expression of rtsAB (78), and screened for those fusions in which expression was not altered by tetracycline. Six fusions had increased expression in the Φ(tetA+-rtsA+B+)3 background but were unaffected by tetracycline in the zii-8104::Tn10dTc strain. These MudJ fusions were considered candidates for RtsA-or RtsB-regulated genes and were studied further.
Sequence analysis of potential RtsAB-regulated genes.
To identify the genes regulated by either RtsA or RtsB, the insertion site of each MudJ was sequenced. Sequence analysis revealed that two of the fusions were to genes located on SPI1. One fusion appeared to be to the hilD promoter, although the fusion joint was located 39 bp upstream of the hilD translational start site. The other fusion was to invC, which is directly induced by binding of HilA to the invF promoter. We also isolated fusions to sopA and sopB (sigD), which encode TTS effectors not encoded on SPI1. It is known that InvF and SicA act in concert to activate expression of sopB (21). The regulation of sopA remains unclear. The fifth gene identified was STM4261, a large gene located on SPI4, which we have termed icgA (for invasion-coregulated gene A) because it was previously shown to be regulated by SirA in a HilA-dependent manner (1). Based on sequence analysis, icgA appears to lie in an operon with genes that contain homology to an ABC-like transport apparatus. IcgA itself shows homology to both putative RTX pore-forming toxins and putative adhesins. This suggests that IcgA may be a type 1 secreted toxin or adhesin. We also isolated a fusion to yhgF, a gene with no known function. Sequence analysis of YhgF showed that it contains a putative S1 RNA binding domain and similarity to RNase R (13, 14). YhgF also contains similarity to the Tex protein of Bordetella pertussis, which has been implicated in control of toxin expression (30).
Requirements for induction of the RtsAB-regulated genes.
Our screen identified six genes that were induced by increased production of RtsAB. We wanted to determine if the RtsA and RtsB regulators could act independently of one another and, if so, which of the regulators was required for induction. Therefore, we constructed strains that had the chromosomal rtsAB genes deleted and contained plasmids in which the expression of rtsA, rtsB, or both was arabinose inducible (36). We transduced each of the fusions into the four plasmid-bearing strains (carrying pBAD30, pRtsA, pRtsB, and pRtsAB) and measured the β-galactosidase activity of each strain after growth for 3 h in l-arabinose. RtsA alone was required for increased expression of hilD, sopA, sopB, invC, and icgA. Indeed, the sopA, sopB, and invC fusions were induced 50- to 80-fold by RtsA, while expression of hilD was induced ∼5- to 7-fold. Induction of these genes was independent of RtsB (data not shown). RtsB induced expression of yhgF approximately sevenfold, while RtsA had no effect (data not shown). Simultaneous production of RtsA and RtsB induced expression of all genes to the same levels observed with the single regulator (data not shown). Thus, RtsA and RtsB act independently to control expression of these genes.
RtsA induces secretion of SPI1 TTS effectors in a HilA-dependent manner.
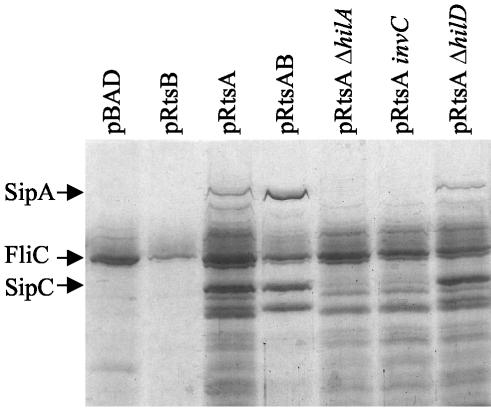
Our screen for RtsA-regulated genes identified five genes known to be part of the SPI1 regulon. To determine if RtsA increased expression of the entire SPI1 TTSS, we assayed the secretion of SPI1 TTS effectors into culture supernatants. Plasmid-bearing strains were grown under SPI1-repressing conditions with 0.2% l-arabinose. The resulting cell-free supernatant was trichloroacetic acid precipitated, and the proteins were separated by SDS-PAGE. As shown in Fig. 1, the strain containing the pBAD vector alone secreted few proteins into the culture supernatant, with the flagellin filament FliC being the predominant protein. The pRtsB strain resembled the vector control except that the band corresponding to the flagellin subunit FliC was decreased in this background. This phenotype was repeatable and is addressed below. In contrast, a strain expressing RtsA had increased amounts of SPI1 TTS effector proteins in the culture supernatant. The strain containing pRtsAB had both increased SPI1 effector proteins and decreased flagellin protein FliC in the culture supernatant. In order to ensure that the proteins found in the culture supernatants were dependent upon the presence of the SPI1 TTS apparatus, we assayed protein secretion in strains containing pRtsA and hilA, hilD, or invC mutations. Mutation of either hilA or invC blocked secretion of the SPI1 TTS proteins. In contrast, a hilD mutation did not appear to decrease RtsA-dependent induction of the SPI1 TTSS (Fig. 1). Thus, production of RtsA induces expression of the entire SPI1 TTSS. This induction is dependent upon the master regulator HilA but is independent of HilD.
FIG. 1.
Effect of RtsA and RtsB on secretion of proteins into culture supernatants. The strains are ΔrtsAB7 and contain the plasmid and mutations specified. Overnight cultures were subcultured into LB (without salt)-ampicillin-0.2% l-arabinose and grown with aeration to an OD600 of ∼0.8. Culture supernatants were prepared as described in Materials and Methods. An equivalent amount of sample from the supernatant of each strain was separated by SDS-12.5% PAGE. The gel was stained for total protein with GelCode Blue. Proteins of greater than ∼25 kDa are shown. The strains used were plasmid-containing derivatives of JS250, JS270, JS271, and JS272.
RtsA induces expression of hilA, invF, hilC, and hilD.
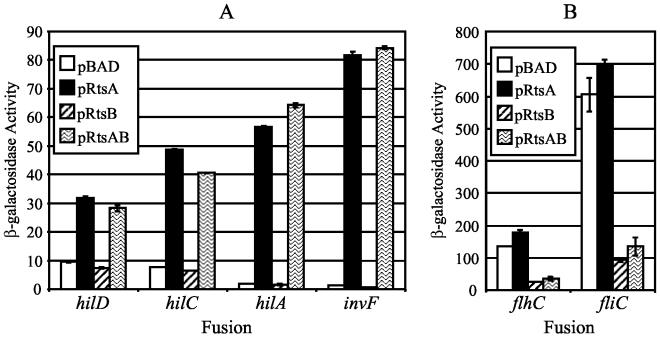
To understand the mechanism of RtsA induction of the invasion genes, we constructed lac fusions to the major SPI1 regulatory genes by using an FLP-mediated method that we recently described (27). These included fusions to hilA, hilC, and invF and a new fusion to hilD, in which the fusion joint is downstream of the translational start site. These fusions were introduced into strains containing ΔrtsAB and either pBAD, pRtsA, pRtsB, or pRtsAB. As shown in Fig. 2A, the expression of hilD and hilC was induced approximately five- to sevenfold by production of RtsA but not RtsB. Thus, RtsA can induce expression of hilC and hilD, both of which encode positive regulators of hilA. Figure 2A also shows that the hilA and invF fusions are induced approximately 40- to 60-fold by RtsA but not RtsB. Thus, RtsA can induce expression of both hilA and invF, and this induction is approximately 10-fold higher than RtsA induction of hilC and hilD.
FIG. 2.
Effect of RtsA, RtsB, or RtsAB on the expression of the SPI1 regulatory genes (A) and the flagellar genes (B). The strains are ΔrtsAB7 and contain pBAD30, pRtsA, pRtsB, or pRtsAB and a lacZ transcriptional fusion to the specified gene. Overnight cultures were subcultured into LB (without salt)-ampicillin-0.2% l-arabinose and grown to an OD600 of ∼0.6. β-Galactosidase activity units are defined as (micromoles of ONP formed minute−1) × 103/(OD600 × milliliters of cell suspension) and are reported as means ± standard deviations, where n = 4. The strains used were plasmid-containing derivatives of JS273 through JS278.
A functional chromosomal copy of rtsA is required for maximal expression of the invasion genes.
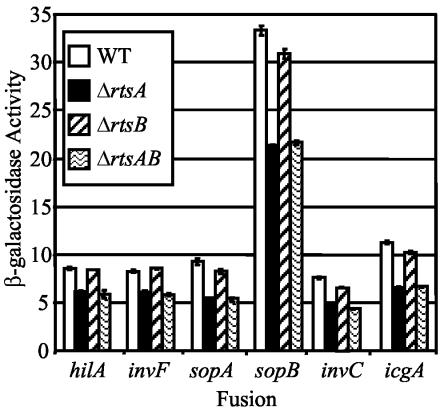
Production of RtsA under normally SPI1-repressing conditions results in the induction of several genes, which belong to the SPI1 TTSS regulon. In order to determine if RtsA is required for expression of the SPI1 genes under SPI1-inducing conditions, we constructed strains with in-frame deletions in rtsA, rtsB, or both. We then introduced hilA-lac, invF-lac, sopA100::MudJ, sopB101::MudJ, invC101::MudJ, and icgA1::MudJ fusions into wild-type and mutant strains and assayed the β-galactosidase activities of the resulting strains. Figure 3 shows that deletion of rtsA or rtsAB, but not rtsB alone, decreased expression of each of the fusions approximately 1.5- to 2-fold. This is similar to the effect seen in a hilC mutant (reference 55 and data not shown). This suggests that RtsA is normally required for maximal expression of the SPI1 TTSS. Under these laboratory conditions, deletion of rtsA confers a modest decrease in expression of the SPI1 regulon. Therefore, we chose to perform subsequent genetic analysis by ectopic production of RtsA and/or RtsB.
FIG. 3.
Effect of ΔrtsA5, ΔrtsB6, or ΔrtsAB7 mutations on the expression of RtsA-regulated genes. Stationary-phase cultures were subcultured 1/100 into LB-1% NaCl and grown statically overnight at 37°C, at which point β-galactosidase activities were determined. β-Galactosidase activity units are defined as (micromoles of ONP formed minute−1) × 103/(OD600 × milliliters of cell suspension) and are reported as means ± standard deviations, where n = 4. The strains used were plasmid-containing derivatives of JS271, JS275, JS276, and JS279 through JS299. WT, wild type.
RtsA induction of hilA does not require HilC or HilD.
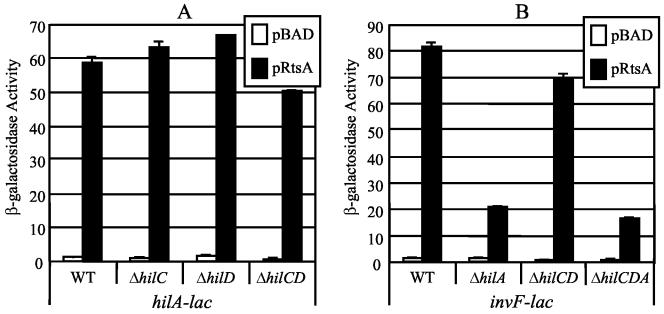
It is possible that RtsA increases expression of hilC and hilD, whose products then act to induce expression of hilA (26, 67, 71). A second possibility is that RtsA induces expression of hilA independent of HilC and HilD. To distinguish between these models, we constructed strains that had hilC, hilD, or both genes deleted. As shown in Fig. 4A, production of RtsA results in increased expression of hilA in the absence of either hilC or hilD. Deletion of both regulators decreased the absolute level of expression from the hilA-lac fusion but did not alter the fold induction of hilA by RtsA, which remained approximately 50- to 60-fold. This suggests that increased expression of hilA by RtsA is independent of the increase in hilC and hilD expression. This is consistent with the observation that RtsA-dependent secretion of TSS effector proteins was independent of HilD (Fig. 1).
FIG. 4.
Effect of mutations in SPI1 regulatory genes on RtsA-induced expression of hilA-lac+ (A) and invF-lac+ (B) fusions. The hilA-lac+ or invF-lac+ transcriptional fusion strains are ΔrtsAB; contain pBAD30, pRtsA, pRtsB, or pRtsAB; and have the indicated regulatory gene(s) deleted. The hilC, hilD, and hilA mutations are simple deletion-insertions of a chloramphenicol cassette. The ΔhilC-D mutation deletes hilC to hilD, including the prgHIJK operon, and the ΔhilC-A deletion removes hilC, prgHIJK, hilD, and hilA. Overnight cultures were subcultured into LB (without salt)-ampicillin-0.2% l-arabinose and grown to an OD600 of ∼0.6 before assay of β-galactosidase. β-Galactosidase activity units are defined as (micromoles of ONP formed minute−1) × 103/(OD600 × milliliters of cell suspension) and are reported as means ± standard deviations, where n = 2. The strains used were plasmid-containing derivatives of JS275, JS276, and JS300 through JS305. WT, wild type.
RtsA directly binds the hilA promoter region.
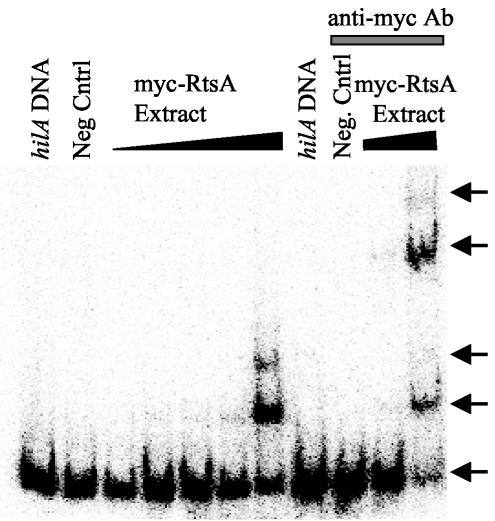
Does RtsA increase expression of hilA by directly interacting with the hilA promoter region or by altering the expression of another regulator of hilA? RtsA belongs to the AraC/XylS family of transcriptional regulators and contains similarity to HilC and HilD. To determine if RtsA binds to the hilA promoter region, we first constructed an antibody epitope-tagged version of RtsA, Myc-RtsA. This fusion protein is functional as evidenced by its ability to activate expression of a hilA-lac fusion (data not shown). We performed gel shift assays with the minimal DNA required for HilC and HilD activation of the hilA promoter (64, 71, 72). This region of DNA contains both HilC and HilD binding sites (64, 72). As shown in Fig. 5, increasing protein concentrations of whole-cell extracts from an E. coli strain (which lacks hilC, hilD, and rtsA) producing a Myc-RtsA fusion protein resulted in the altered migration of hilA DNA through the gel matrix. There was also a supershift, suggesting the presence of multiple RtsA binding sites similar to those seen with HilC binding to the hilA promoter region (64, 72). The addition of the anti-Myc antibody to the binding reactions containing Myc-RtsA causes a further retardation of the hilA DNA, showing that the shift is mediated directly by RtsA. Production of wild-type RtsA also shifts the hilA promoter region (data not shown). Thus, RtsA binds to the hilA promoter, suggesting that it directly activates expression.
FIG. 5.
Gel shift analysis of RtsA binding to the hilA promoter region. Binding reaction mixtures contained ∼0.1 ng of 32P-labeled hilA promoter region DNA corresponding to −189 to +19 bp from the start site of transcription. Increasing amounts of the Myc-RtsA whole-cell extract from 2 to 1,000 ng were included in the indicated reaction mixtures. The hilA DNA lane has no extract. The negative control (Neg Cntrl) lane contains 1,000 ng of whole-cell extract of the vector control strain. One hundred nanograms of anti-Myc antibody (Ab) was added to the indicated reaction mixtures. The strain used was MG1655 with either pBAD or pCE81.
RtsA can induce expression of invF independently of HilA, HilC, and HilD.
RtsA induces expression of the invF-invC operon. Transcription of invF is known to be regulated by HilA, HilC, and HilD. To determine if RtsA-mediated induction was dependent on these regulators, we constructed invF-lac fusion strains with pBAD or pRtsA and either hilA, hilCD, or hilCDA deletions. Figure 4B shows that loss of HilC and HilD caused only a slight decrease in invF transcription. A deletion of hilA or hilCDA decreased but did not abolish RtsA induction of the invF-lac+ fusion. This suggests that RtsA can induce expression of invF independent of HilA, similar to what has been observed for both HilC and HilD (2, 26, 67). Thus, RtsA-dependent induction of invF is apparently a combination of direct and indirect effects.
HilA and/or InvF is required for maximal RtsA induction of the MudJ fusions.
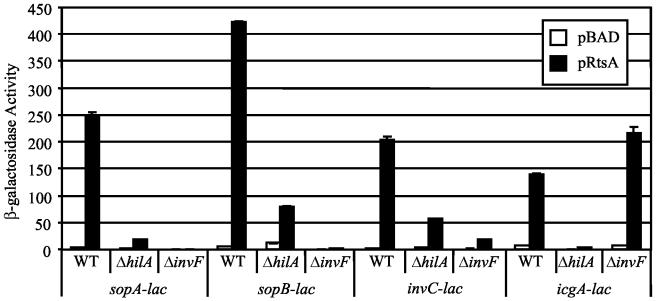
RtsA induces expression of hilA and invF, whose products can directly or indirectly regulate expression of several genes encoded both within and outside SPI1. This information prompted us to test whether HilA and InvF were required for RtsA induction of the fusions identified in our screen. The MudJ fusions were transduced into strains containing pBAD or pRtsA and deletions of hilA or invF. As shown in Fig. 6, RtsA-dependent induction of sopA and sopB was significantly decreased, but not abolished, in the hilA deletion strain, whereas a deletion of invF completely blocked transcription of these fusions. This suggests that HilA regulates expression of these genes indirectly by activating expression of InvF, consistent with previous studies (2, 26, 67). Because RtsA can induce invF independently of HilA, some induction is still apparent in the hilA deletion strain.
FIG. 6.
Effect of invF and hilA mutations on RtsA induction of sopA, sopB, invC, and icgA fusions. The strains are ΔrtsAB7; contain pBAD30, pRtsA, pRtsB, or pRtsAB; and are ΔhilA::Cm or ΔinvF::Cm. The stains contain a lacZ transcriptional fusion to the gene specified. Overnight cultures were subcultured into LB (without salt)-ampicillin-0.2% l-arabinose and grown to an OD600 of ∼0.6 before assay of β-galactosidase. β-Galactosidase activity units are defined as (micromoles of ONP formed minute−1) × 103/(OD600 × milliliters of cell suspension) and are reported as means ± standard deviations, where n = 2. The strains used were plasmid-containing derivatives of JS306 through JS317. WT, wild type.
Expression of the invC fusion was analogous to that observed in the invF fusion described above; these genes are in an operon. Indeed, there was an apparent decrease in invC expression observed in the invF deletion, but we believe that this is due to polarity of the invF deletion construct on the invC fusion; InvF is not known to autoregulate (19, 44). Interestingly, RtsA induction of icgA (located on SPI4) was completely abolished by a mutation in hilA but was unaffected by the invF mutation (Fig. 6). This suggests that HilA regulates expression of icgA in an InvF-independent manner, but it is not known whether this regulation is direct or indirect.
RtsA induces expression of slrP independently of HilA and InvF.
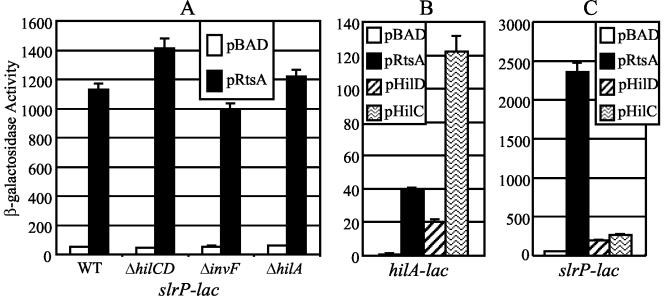
We wanted to determine whether RtsA could induce expression of other SPI1 effectors. Expression of slrP is induced in a HilA-independent manner (61). It is also known that SlrP is secreted primarily by the SPI1 TTSS and is required for colonization of Peyer's patches in the mouse small intestine (61, 82). Figure 7A shows that when RtsA is produced from the arabinose promoter under SPI1-repressing conditions, expression of slrP is induced 20- to 30-fold compared to the vector-only control. However, in contrast to other effectors examined, RtsA induction of slrP is independent of HilA, InvF, and HilD or HilC (Fig. 7A). This suggests that RtsA may induce expression of a subset of SPI1 effector proteins that are not directly controlled by HilA or InvF.
FIG. 7.
Effect of invF, hilA, and hilC-D mutations on RtsA induction of slrP (A) and ability of HilC, HilD, and RtsA to induce expression of hilA (B) and slrP (C). (A) The strains are ΔrtsAB7 and contain pBAD30 or pRtsA; ΔhilC-D::Cm, ΔhilA::Cm, or ΔinvF::Cm; and an slrP-lac fusion. (B and C) The strains contain a lac transcriptional fusion with hilA and slrP, respectively, and the chromosomal ΔrtsA ΔhilC-D::Cm mutations along with the plasmid specified. pHilD is pLS118, and pHilC is pLS119 (72). Overnight cultures were subcultured into LB (without salt)-ampicillin-0.2% l-arabinose and grown to an OD600 of ∼0.6 before assay of β-galactosidase. β-Galactosidase activity units are defined as (micromoles of ONP formed minute−1) × 103/(OD600 × milliliters of cell suspension) and are reported as means ± standard deviations, where n = 4. The strains used were plasmid-containing derivatives of JS318 through JS323. WT, wild type.
RtsA is a better inducer of slrP expression than HilC or HilD.
Our data suggest that HilC, HilD, and RtsA function in similar manners by binding to the same fragment of hilA to induce its expression. Therefore, we wanted to determine if HilC or HilD could also induce expression of slrP. To test this, we constructed strains containing ΔrtsA and ΔhilC-D::Cm mutations, either a hilA-lac or a slrP-lac fusion and pBAD, pRtsA, pLS118 (HilD), or pLS119 (HilC). Figure 7B shows that expression of hilA was induced ∼40-fold by RtsA, ∼20-fold by HilD, and ∼120-fold by HilC. These levels are consistent with previous data (72). Interestingly, RtsA induces expression of slrP 45-fold, while HilD and HilC are able to induce expression of slrP only four- and fivefold, respectively (Fig. 7B). Simplistically this suggests that RtsA acts as a better inducer of slrP expression than does either HilC or HilD, at least compared to their ability to induce hilA expression.
Expression of rtsA is induced under SPI1-inducing conditions.
It is clear that production of RtsA induces expression of the SPI1 invasion genes. We wanted to determine whether rtsA and rtsB were induced under the same conditions as the SPI1 invasion genes. We constructed rtsA-lac and rtsB-lac fusions in an otherwise wild-type background (JS324 and JS325, respectively) and then assayed the β-galactosidase activity in these strains after growth under SPI1-repressing and SPI1-inducing conditions. The β-galactosidase activity produced by the rtsA-lac fusion under SPI1-repressing conditions was 1.39 ± 0.08 U. Under SPI1-inducing conditions, the β-galactosidase activity increased to 21.65 ± 2.48 U. The β-galactosidase activity produced from the rtsB-lac fusion under SPI1-repressing conditions was 1.79 ± 0.44 U, versus 12.22 ± 0.86 U under SPI1-inducing conditions. Thus, the expression of rtsA and rtsB is significantly increased under SPI1-inducing conditions. Given that the rtsAB genes are separated by 11 bp and appear to be coordinately regulated, this suggests that they form an operon.
RtsB represses expression of the flagellin subunit gene fliC and regulatory genes flhDC.
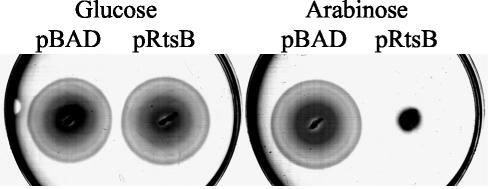
It appeared from our analysis of secreted proteins that strains producing RtsB had reduced levels of the flagellin subunit FliC/FljB in the culture supernatant (Fig. 1). To characterize this further, we tested the effect of RtsB production on motility by inoculating motility agar containing 0.2% glucose or 0.2% l-arabinose with strains containing either pRtsB or the pBAD vector alone. Both strains were motile in the presence of glucose. However, when grown in the presence of 0.2% l-arabinose, the strain containing pRtsB exhibited a significant motility defect compared to the control strain (Fig. 8).
FIG. 8.
Effect of RtsB production on motility. The strains are ΔrtsAB7 (JS250) and contain pBAD30 or pRtsB. Motility assays were performed in plates with 0.3% agar supplemented with either 0.2% glucose or 0.2% l-arabinose.
To analyze the effect of RtsB on flagellar gene expression, we introduced a fliC5050::MudJ or flhC5456::MudJ fusion into strains containing pBAD, pRtsA, pRtsB, and pRtsAB. We then assayed the β-galactosidase activity of the resulting strains after growth for 2.5 h in the presence of 0.2% l-arabinose. As shown in Fig. 2B, increased production of RtsB, but not RtsA, decreased expression of the fliC5050::MudJ and flhC5456::MudJ fusions approximately 5.5-fold. Thus, the decreased FliC/FljB observed in the culture supernatant is due to transcriptional repression of the flagellin genes by RtsB. Moreover, RtsB apparently acts as a repressor of flagellar expression by repressing expression of the master regulators of the flagellar operon, flhDC. We also examined the effect of loss of rtsA, rtsB, and rtsAB deletion mutations on both fliC-lac and flhC-lac expression; there was no significant decrease under the conditions tested (data not shown).
RtsB directly binds to the flhDC promoter region.
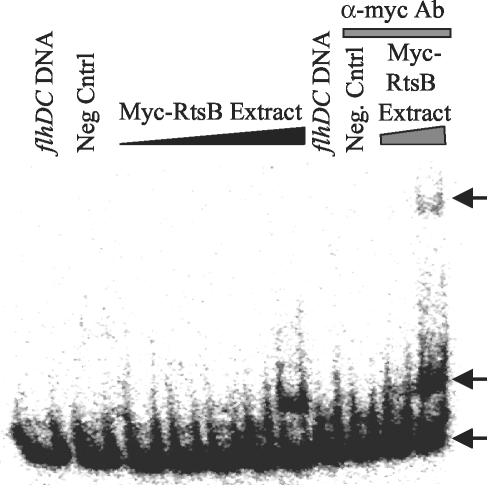
We wanted to determine whether repression of the flagellar master regulators was due to the direct binding of RtsB to the promoter region of flhDC. We performed gel shift assays with the flhDC promoter region and whole-cell extracts of E. coli strains (which otherwise lack rtsB) producing RtsB or Myc-RtsB. The Myc-RtsB construct represses expression of the flhDC-lac fusion (data not shown). Using whole-cell extracts of strains producing RtsB, we narrowed the region of the promoter that could be gel shifted to a 110-bp region corresponding to −4 to +106 relative to the start site of transcription (data not shown). Figure 9 shows that increasing concentrations of whole-cell extract from strains producing Myc-RtsB caused a significant shift in the migration of this flhDC DNA. The addition of anti-Myc antibody to the binding reactions caused a further shift in the migration of the flhDC DNA, showing that the gel shift is due to direct binding of RtsB. This suggests that RtsB decreases motility and represses expression of flagellar genes by acting as a repressor of flhDC.
FIG. 9.
Gel shift analysis of RtsB binding to the flhDC promoter region. Binding reaction mixtures contained ∼0.1 ng of 32P-labeled flhDC promoter region DNA corresponding to −4 to +106 bp from the start site of transcription. Increasing amounts of the Myc-RtsB whole-cell extract from 2 to 1,000 ng were included in the indicated reaction mixtures. The flhDC DNA lane has no extract. The negative control (Neg Cntrl) lane contains 1,000 ng of whole-cell extract of the vector control strain. One hundred nanograms of anti-Myc antibody (Ab) was added to the indicated reaction mixtures. The strain used was MG1655 with either pBAD or pCE82.
DISCUSSION
The ability of S. enterica serovar Typhimurium to sense its location within the host and respond by inducing and repressing expression of the appropriate virulence factors is critical to its survival. We have identified a regulatory operon on a Salmonella-specific island near tRNAPheU that encodes RtsA and RtsB. Sequence analysis suggests that RtsA is a member of the AraC/XylS family of transcriptional regulators and is related to HilC and HilD. This homology is most significant in the carboxy-terminal third of the proteins, which contains the putative DNA binding domain (RtsA versus HilC, 56% identity and 76% similarity; RtsA versus HilD, 60% identity and 75% similarity; HilC versus HilD, 58% identity and 72% similarity). However, the N-terminal domains, which presumably act to sense some environmental parameter, also show limited homology (RtsA versus HilC, 25% identity and 42% similarity; RtsA versus HilD, 21% identity and 46% similarity; HilC versus HilD, 24% identity and 50% similarity).
RtsA, HilC, and HilD apparently function in very similar fashion. All three proteins bind to approximately the same region of the hilA promoter. The results of our gel shift assays suggest that RtsA binds within the region from −189 to +19 from the start site of transcription. Much like HilC, RtsA appears to bind to at least two sites within this region as evidenced by multiple bands in the gel shift (64). HilC and HilD are known to independently bind to two regions of the hilA promoter, from approximately −231 to −179 and from −101 to −49, although they differ in their sequence requirements for recognition of the binding sites (64, 72). RtsA also induces expression of both hilC and hilD. HilC and HilD have been shown to bind both the hilC and hilD promoters, and either protein can activate expression of the hilC promoter in vitro (64). RtsA can induce expression of invF in a HilA-independent fashion. Both HilC and HilD also directly activate invF, and it has been suggested that they activate a promoter located upstream of the HilA-dependent promoter PinvF (2). The homology in the DNA binding domains of these proteins and the fact that they apparently activate the same set of promoters suggest that they may function by binding to similar DNA sites to directly activate transcription.
Although these three proteins are similar, they can each act independently. RtsA induces expression of hilA in the absence of both HilC and HilD. It has also been shown that both HilD and HilC can independently induce expression of a hilA-lac+ fusion in E. coli (72). RtsA is absent in this system. Why does serovar Typhimurium maintain three regulators of hilA that apparently perform the same function? We have three working hypotheses, which are not mutually exclusive: (i) RtsA, HilC, and HilD are active under different conditions or induce expression of hilA in response to different environmental cues. (ii) RtsA, HilC, and HilD differentially regulate expression of other genes independent of their effects on hilA expression. Indeed, it appears that RtsA, HilC and HilD are all capable of inducing expression of hilA, hilC, hilD, and invF but that RtsA is preferentially capable of inducing expression of slrP. This strongly supports a role for RtsA in expression of the SPI1 TTSS independent of its effects on hilA. (iii) RtsA, HilC, and HilD are required for signal amplification such that once one of these genes is turned on, expression of the others is also induced, resulting in greater induction of hilA. Regulation of hilA expression by RtsA, HilC, and HilD is an example of a feedforward loop. This type of regulatory cascade is common in yeast and presumably allows for increased sensitivity and/or tight temporal regulation (50). We believe that the inductions of RtsA, HilC, and HilD are self-reinforcing events leading to rapid and fully induced expression of hilA. Thus, this genetic switch is poised such that expression of the SPI1 TTSS is either essentially on or off.
The SPI4-carried gene icgA appears to encode a type 1 exported RTX pore-forming toxin or adhesin. SirA had previously been shown to induce expression of several SPI4 genes in a HilA-dependent manner, but the requirement for InvF in this induction was not determined (1). The previously identified fusions were within icgA, although at that time this region was incorrectly annotated (86). Here we show that RtsA induces expression of icgA in a HilA-dependent but InvF-independent manner. This suggests that HilA may directly regulate the expression of genes located outside SPI1 independently of InvF. It is not known what role IcgA plays during a serovar Typhimurium infection, but it is intriguing that a putative toxin or adhesin is coordinately regulated with the SPI1 invasion genes.
RtsB transcriptionally represses the flagellar operon by binding to and repressing expression of the flhDC promoter. Sequence analysis suggests that RtsB is a small protein (∼10 kDa), which contains a helix-turn-helix DNA binding motif that most closely resembles those in the LuxR/GerE family. However, RtsB does not appear to contain any distinct regulatory domain. RtsB binds directly to the flhDC promoter region within a 110-bp region from −4 to +106 from the start site of transcription (87). Ectopic production of RtsB in E. coli also blocks motility (data not shown), suggesting that RtsB can repress the E. coli flhDC operon. The E. coli and serovar Typhimurium flhDC promoters have the most significant homology immediately downstream of the −10 region (77, 87). Thus, RtsB binding may include the major transcriptional start site, and this binding could sterically hinder RNA polymerase, thus explaining how such a seemingly simple protein could act as a regulator. Regulation could be mediated by controlling the level of RtsB rather than by some conformational change in the protein in response to a particular signal. RtsB also induces expression of at least one other gene, yhgF. However, it is not clear whether RtsB-mediated induction is direct or indirect.
Although flagella are not essential for virulence of serovar Typhimurium in BALB/c mice (52), they affect interaction with the host at multiple levels (33, 41, 53, 73). Several studies have identified the class 3 flagellar protein FliZ as an activator of hilA expression, seemingly antagonistic to the role of RtsB (25, 42, 56). There are conflicting data about the role of the flagella and flagellar regulators and expression of hilA. A fliZ mutation decreases expression of hilA 2-fold (25, 42, 56), while a sirA mutation decreases expression of hilA 10-fold and induces expression of the flagella 100-fold (35). This suggests that the relationship between expression of the flagella and the SPI1 TTSS is complex and will require further study. The FliC/FljB flagellin proteins interact with Toll-like receptor 5 on the basolateral surface of epithelial cells to activate the NF-κB pathway in epithelial cells (31). This results in secretion of interleukin-8 and production of human β-defensin 2 (32, 80). FliC has also been shown to be a major CD4+ T-cell epitope in mice (60). However, strains that produce only FljB are attenuated during systemic infection in a mouse model (41).
Why does the cell want to upregulate expression of the SPI1 invasion genes while repressing expression of the flagellar genes? There are several possibilities. First, when serovar Typhimurium is invading epithelial cells, motility may be detrimental. The bacteria do not want to swim away from a host cell primed for invasion. Second, the flagellum is immunogenic and elicits a number of host responses (32, 60, 80). Therefore, when serovar Typhimurium is invading host tissue, it may be advantageous to stop producing the appendage. A problem with both of these arguments is that transcriptional repression of the flagellar genes would not be expected to immediately block motility or the immunological effects of preexisting flagella. Third, simultaneous production of SPI1 and the flagella could result in interference between the two TTSSs. Interference could manifest itself as misappropriate secretion of effectors or flagellin or more indirectly as increased periplasmic stress.
A number of genes have been shown to control expression of hilA. However, in many cases the mechanism of hilA regulation is unknown. Future experiments must address the regulation of rtsAB in order to place RtsA, RtsB, and the other regulators into regulatory pathways that control invasion and flagellar gene regulation. Of the known regulators of the SPI1 TTSS and flagellar genes, SirA is of particular interest, as it is known to induce expression of hilA while repressing expression of flhDC (35). At this time it is unclear how SirA regulates these two different TTSSs. RtsA and RtsB are induced by SPI1-inducing conditions. Thus, these environmental parameters are sensed upstream of RtsA in the regulatory scheme. To ultimately understand how RtsA affects expression of hilA and the SPI1 invasion genes during an infection, the regulation of rtsAB must be studied in detail.
Acknowledgments
This work was supported by grant 00-25 from the Roy J. Carver Charitable Trust.
We thank Kelly Hughes for the fliC5050::MudJ and flhC5456::MudJ fusions, Cathy Lee for plasmids pLS118 and pLS119, Anu Janakiraman for providing valuable strains, Jeff Gardner and Stan Maloy for helpful hints on performing the gel shift assays, Theresa Ho and Scott Minnich for valuable discussions, and the Slauch lab for productive comments. We also thank Jack Ikeda, Anne Thierauf, and Radha Krishnakumar for critically reading the manuscript.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. [DOI] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by CsrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 6.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 7.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 8.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 11.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Z. F., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 277:21624-21629. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077-14080. [DOI] [PubMed] [Google Scholar]
- 15.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 16.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 18.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 22.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichelberg, K., and J. E. Galan. 2000. The flagellar sigma factor FliA (σ28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect. Immun. 68:2735-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 27.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 28.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 29.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs, T. M., H. Deppisch, V. Scarlato, and R. Gross. 1996. A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol. 178:4445-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 32.Gewirtz, A. T., A. S. Rao, P. O. Simon, Jr., D. Merlin, D. Carnes, J. L. Madara, and A. S. Neish. 2000. Salmonella typhimurium induces epithelial IL-8 expression via Ca(2+)-mediated activation of the NF-kappaB pathway. J. Clin. Invest. 105:79-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gewirtz, A. T., P. O. Simon, Jr., C. K. Schmitt, L. J. Taylor, C. H. Hagedorn, A. D. O'Brien, A. S. Neish, and J. L. Madara. 2001. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Invest. 107:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillen, K. L., and K. T. Hughes. 1991. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J. Bacteriol. 173:6453-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen-Wester, I., and M. Hensel. 2002. Genome-based identification of chromosomal regions specific for Salmonella spp. Infect. Immun. 70:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartley, J. L., G. F. Temple, and M. A. Brasch. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho, T. D., and J. M. Slauch. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda, J. S., C. K. Schmitt, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, P. Adams, C. D. O'Connor, and A. D. O'Brien. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 69:3021-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 43.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 44.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 45.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimbrough, T. G., and S. I. Miller. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75-82. [DOI] [PubMed] [Google Scholar]
- 47.Ko, M., and C. Park. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komeda, Y., H. Suzuki, J. I. Ishidsu, and T. Iino. 1976. The role of cAMP in flagellation of Salmonella typhimurium. Mol. Gen. Genet. 142:289-298. [DOI] [PubMed] [Google Scholar]
- 49.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 89:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 51.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 52.Lockman, H. A., and R. Curtiss III. 1990. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect. Immun. 58:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lockman, H. A., and R. Curtiss III. 1992. Virulence of non-type 1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect. Immun. 60:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 55.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macnab R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 58.Maloy, S. R., V. J. Stewert, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 59.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 60.McSorley, S. J., B. T. Cookson, and M. K. Jenkins. 2000. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164:986-993. [DOI] [PubMed] [Google Scholar]
- 61.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller, S. I., and D. A. Pegues. 2000. Salmonella species, including Salmonella typhi, p. 2344-2363. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 63.Monack, D. M., D. Hersh, N. Ghori, D. Bouley, A. Zychlinsky, and S. Falkow. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J. Exp. Med. 192:249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 66.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 67.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 71.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 72.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 73.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silverman, M., and M. Simon. 1974. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J. Bacteriol. 120:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slauch, J. M., and A. Camilli. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 326:73-96. [DOI] [PubMed] [Google Scholar]
- 76.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takahashi, A., A. Wada, K. Ogushi, K. Maeda, T. Kawahara, K. Mawatari, H. Kurazono, J. Moss, T. Hirayama, and Y. Nakaya. 2001. Production of β-defensin-2 by human colonic epithelial cells induced by Salmonella enteritidis flagella filament structural protein. FEBS Lett. 508:484-488. [DOI] [PubMed] [Google Scholar]
- 81.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 84.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 85.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]
- 86.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yanagihara, S., S. Iyoda, K. Ohnishi, T. Iino, and K. Kutsukake. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105-111. [DOI] [PubMed] [Google Scholar]
- 88.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]