Abstract
In Bacillus subtilis expression of genes or operons encoding enzymes and other proteins involved in purine synthesis is affected by purine bases and nucleosides in the growth medium. The genes belonging to the PurR regulon (purR, purA, glyA, guaC, pbuO, pbuG, and the pur, yqhZ-folD, and xpt-pbuX operons) are controlled by the PurR repressor, which inhibits transcription initiation. Other genes are regulated by a less-well-described transcription termination mechanism that responds to the presence of hypoxanthine and guanine. The pur operon and the xpt-pbuX operon, which were studied here, are regulated by both mechanisms. We isolated two mutants resistant to 2-fluoroadenine in which the pur operon and the xpt-pbuX operon are expressed at increased levels in a PurR-independent manner. The mutations were caused by deletions that disrupted a potential transcription terminator structure located immediately upstream of the ydhL gene. The 5′ part of the ydhL leader region contained a 63-nucleotide (nt) sequence very similar to the 5′ ends of the leaders of the pur and xpt-pbuX operons. Transcripts of these regions may form a common tandem stem-loop secondary structure. Two additional genes with potential leader regions containing the 63-nt sequence are pbuG, encoding a hypoxanthine-guanine transporter, and yxjA, which was shown to encode a purine nucleoside transporter and is renamed nupG. Transcriptional lacZ fusions and mutations in the 63-nt sequence encoding the possible secondary structures provided evidence that expression of the pur and xpt-pbuX operons and expression of the ydhL, nupG, and pbuG genes are regulated by a common mechanism. The new pur regulon is designated the XptR regulon. Except for ydhL, the operons and genes were negatively regulated by hypoxanthine and guanine. ydhL was positively regulated. The derived amino acid sequence encoded by ydhL (now called pbuE) is similar to the amino acid sequences of metabolite efflux pumps. When overexpressed, PbuE lowers the sensitivity to purine analogs. Indirect evidence indicated that PbuE decreases the size of the internal pool of hypoxanthine. This explains why the hypoxanthine- and guanine-regulated genes are expressed at elevated levels in a mutant that overexpresses pbuE.
In Bacillus subtilis expression of the pur operon (purEKBCSQLFMNHD), encoding the enzymes which catalyze de novo synthesis of IMP, and expression of the xpt-pbuX operon, encoding xanthine phosphoribosyltransferase and a xanthine transporter, are subjected to dual regulation of transcription initiation and termination (3-5). The regulatory metabolites in the growth medium are purine bases and nucleosides. The mechanism controlling transcription initiation requires a DNA operator site, the PurR repressor, and phosphoribosylpyrophosphate (PRPP). PurR binds to the operator site and inhibits transcription. Uptake and metabolism of adenine from the medium result in a decrease in the size of the cellular PRPP pool, whereas guanosine and purine starvation has the opposite effect (26). When PRPP is abundant, PRPP binds to PurR and prevents the binding of PurR to DNA. The recognition site for the PurR protein has been found to be a tandem PurBox motif located at various positions both up- and downstream of the transcriptional start sites of the PurR regulon genes purR, purA, glyA, guaC, pbuO, and pbuG and the pur, yqhZ-folD, and xpt-pbuX operons (23, 27, 29).
A second transcription termination mechanism responds to hypoxanthine and guanine or their nucleoside derivatives. In the presence of purines transcription of the pur and xpt-pbuX operons terminates before RNA polymerase transcribes the first structural gene of each operon, resulting in a short transcript. In the absence of purines RNA polymerase transcribes the entire operon (3, 6). For the xpt-pbuX operon evidence has been obtained that the free bases hypoxanthine and guanine are the low-molecular-weight effectors (3). Analysis of the 5′ nucleotide sequence of a possible leader mRNA of both the pur and xpt-pbuX operons indicated that two mutually exclusive stem-loop structures can be formed. One structure forms a factor-independent transcription terminator. The 5′-proximal end of this terminator can also form an alternative secondary structure by base pairing with upstream nucleotides (3, 5). The genes encoding enzymes for pyrimidine nucleotide synthesis in B. subtilis are organized in a single operon, whose expression is also controlled by termination and antitermination. Within the pyr operon, which encodes the enzymes of the UMP biosynthetic pathway, there are three transcription terminators, each of which is preceded by another stem-loop structure, the antiterminator, whose formation prevents formation of the terminator. The PyrR protein plus UMP bind to pyr mRNA and disrupt the antiterminator, permitting the terminator to be formed (7, 13). The PyrR binding site is localized to a short stem-loop structure, the anti-antiterminator, which interferes with formation of the antiterminator structure when pyrimidines are abundant. The RNA-PyrR binding specificity has been studied in detail by introducing base changes in the RNA loop and amino acid changes in the PyrR protein (2, 21).
Mutants that overexpress pur genes have been selected in bacilli by selecting for resistance to toxic purine analogs (11, 20). The principle for this selection is to overcome the toxic effect either by overproducing purine compounds or by synthesizing increased amounts of purine biosynthetic enzymes or both. Such mutants have industrial importance for the production of purines and vitamins. Unfortunately, many of these mutants have not been genetically characterized.
Here we report characterization of a novel purine regulon in B. subtilis, which we designated the XptR regulon and which comprises the pur and xpt-pbuX operons and three genes, pbuG, pbuE (ydhL), and nupG (yxjA). Furthermore, we document that overexpression of the PbuE efflux pump results in increased expression of XptR regulon genes, most likely mediated by a decreased cellular concentration of hypoxanthine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains, plasmids, and DNA primers used are listed in Table 1. B. subtilis was grown in Spizizen minimal salt medium supplemented with 0.2% l-glutamate and 1 mg of thiamine per liter and with 0.4% glucose as the carbon source, as described previously (25). Amino acids, when required, were added to a final concentration of 40 mg/liter. For solubility reasons inosine and guanosine rather than hypoxanthine and guanine were used at a final concentration of 1 mM. Once inside a cell, both of these nucleosides are degraded to the corresponding free bases (18). For antibiotic resistance selection, antibiotics were used at the following concentrations: ampicillin, 100 mg/liter; neomycin, 5 mg/liter; erythromycin, 1 mg/liter; lincomycin, 25 mg/liter; and chloramphenicol, 6 mg/liter.
TABLE 1.
Bacterial strains, plasmids, and DNA primers used in this study
| Strain, plasmid, or primer | Genotype or characteristics | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | C. Anagnostopoulos |
| ED77 | trpC2 thr-5 hisA1 | QB-917, R. A. Dedonder |
| ED244 | trpC2 purM | H. H. Saxild and P. Nygaard |
| ED452 | trpC2 yxjA::pMutin4 (erm) | Y. Fujitaa |
| ED488 | trpC2 amyE::[xptT50A′-lacZ] | As LJ25, but containing the xptT50A mutation fused to lacZ |
| ED493 | trpC2 amyE::[yxjAA71G′-lacZ] | As LJ21, but containing the yxjAA71G mutation fused to lacZ |
| ED497 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] pbuG ydhL2 (FAr) | LJ25 spontaneously resistant to 1 μM FA and 0.2 mM 8-azaguanine |
| ED499 | trpC2 amyE::[pCATH1 ydhL′-lacZ (neo)] | 168 transformed with pCATH1, selecting for Neor |
| HH297 | trpC2 hpt::erm | 3 |
| HH441 | trpC2 thr-5 amyE::[pLJ24 ydhL′-lacZ (cat)] hpt::erm | LJ27 transformed with DNA from HH297, selecting for Err |
| HH442 | trpC2 thr-5 amyE::[pLJ24 ydhL′-lacZ (cat)] hpt::erm ydhL1 | LJ29 transformed with DNA from HH297, selecting for Err |
| BFA2255 | trpC2 yebB::pMutin4 (erm) | Micado databaseb |
| LJ4 | trpC2 amyE::[pLJ4 pbuG′-lacZ (cat)] | 168 transformed with pLJ4, selecting for Cmr |
| LJ6 | trpC2 amyE::[purEA85C′-lacZ] | As LJ20, but containing the purEA85C mutation fused to lacZ |
| LJ7 | trpC2 amyE::[purEC56A′-lacZ] | As LJ20, but containing the purEC56A mutation fused to lacZ |
| LJ8 | trpC2 amyE::[purE(ΔG41)′-lacZ] | As LJ20, but containing the purE(ΔG41) mutation fused to lacZ |
| LJ9 | trpC2 amyE::[purET82C′-lacZ] | As LJ20, but containing the purET82C mutation fused to lacZ |
| LJ10 | trpC2 amyE::[purET163A′-lacZ] | As LJ20, but containing the purET163A mutation fused to lacZ |
| LJ15 | trpC2 amyE::[pbuG(ΔT53-C54)′-lacZ] | As LJ4, but containing the pbuG(ΔT53-C54) mutation fused to lacZ |
| LJ17 | trpC2 amyE::[pbuGA27G′-lacZ] | As LJ4, but containing the pbuGA27G mutation fused to lacZ |
| LJ18 | trpC2 amyE::[pbuGA70T′-lacZ] | As LJ4, but containing the pbuGA70T mutation fused to lacZ |
| LJ20 | trpC2 amyE::[purEA85C′-lacZ] | 168 transformed with pLJ20, selecting for Cmr |
| LJ21 | trpC2 amyE::[pLJ21 yxjA′-lacZ (cat)] | 168 transformed with pLJ21, selecting for Cmr |
| LJ23 | trpC2 amyE::[pLJ23 ydhL′-lacZ (cat)] | 168 transformed with pLJ23, selecting for Cmr |
| LJ24 | trpC2 amyE::[pLJ24 ydhL′-lacZ (cat)] | 168 transformed with pLJ24, selecting for Cmr |
| LJ25 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] | 168 transformed with pLJ25, selecting for Neor |
| LJ26 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] ydhL1 (FAr) | LJ25 spontaneously resistant to 1 μM FA |
| LJ27 | trpC2 thr-5 ydhL1 (FAr) | ED77 transformed with LJ26, selecting for His+ and screened for FAr |
| LJ28 | trpC2 amyE::[pLJ21 yxjA′-lacZ (cat)] thr-5 ydhL1 | LJ27 transformed with pLJ21, selecting for Cmr |
| LJ29 | trpC2 amyE::[pLJ24 ydhL′-lacZ (cat)] thr-5 ydhL1 | LJ27 transformed with pLJ24, selecting for Cmr |
| LJ30 | trpC2 amyE::[pLJ4 pbuG′-lacZ (cat)] thr-5 ydhL1 | LJ27 transformed with pLJ4, selecting for Cmr |
| LJ32 | trpC2 ydhL::pLJ32 (erm) | 168 transformed with pLJ32, selecting for Err |
| LJ51 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] ydhL1 hpt::erm | LJ26 transformed with DNA from HH297, selecting for Err |
| LJ52 | trpC2 amyE::[pLJ52 ydhL1′-lacZ (cat)] | 168 transformed with pLJ52, selecting for Cmr |
| LJ53 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] hpt::erm | LJ25 transformed with DNA from HH297, selecting for Err |
| LJ54 | trpC2 amyE::[pLJ25 xpt′-lacZ (neo)] ydhL::pLJ42 (cat) | LJ25 transformed with pLJ42, selecting for Cmr |
| E. coli MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL(Strr) hsdR2(r− m−) mcrA mcrB | Laboratory stock |
| Plasmids | ||
| pMutin4 | Apr (E. coli), Err (B. subtilis); integrational vector for knockout mutations and formation of transcriptional lacZ fusions: IPTG-inducible Pspac promoter introduced to ensure expression of downstream genesc | 24 |
| PDG268cat/neo | Apr (E. coli), Cmr/Neor (B. subtilis); vector used for integration of transcriptional lacZ fusions into the amyE gene of B. subtilis | 18 |
| pBOE335 | Apr (E. coli), Cmr (B. subtilis); integrational vector, pUC19 containing the cat gene cloned into the KasI site | 18 |
| pLJ4 | pDG268cat digested with HindIII and BamHI and ligated to a 339-bp PCR fragment (primers Lars1 and Lars5) digested with the same enzymes | This study |
| pLJ20 | pDG268cat digested with HindIII and BamHI and ligated to a 312-bp PCR fragment (primers Y01 and Y03) digested with the same enzymes | This study |
| pLJ21 | pDG268cat digested with EcoRI and BamHI and ligated to a 319-bp PCR fragment (primers Lars8 and Lars9) digested with the same enzymes | This study |
| pLJ23 | pDG268cat digested with EcoRI and BamHI and ligated to a 213-bp PCR fragment (primers Lars11 and Lars12) digested with the same enzymes | This study |
| pLJ24 | pDG268cat digested with EcoRI and BamHI and ligated to a 369-bp PCR fragment (primers Lars10 and Lars12) digested with the same enzymes | This study |
| pLJ25 | pDG268neo digested with EcoRI and BamHI and ligated to a 262-bp PCR fragment (primers Y04 and Y05) digested with the same enzymes | This study/PICK> |
| pLJ32 | pMutin4 digested with EcoRI and BamHI and ligated to a 254-bp PCR fragment (primers Lars14 and Lars15) digested with the same enzymes | This study |
| pLJ42 | pBOE335 digested with EcoRI and BamHI and ligated to a 213-bp PCR fragment (primers Lars11 and Lars12) digested with the same enzymes | This study |
| pLJ52 | pDG268cat digested with EcoRI and BamHI and ligated to a 353-bp PCR fragment (primers Lars10 and Lars12) digested with the same enzymes | This study |
| pCATH1 | pDG268neo digested with EcoRI and BamHI and ligated to a 176-bp PCR fragment (primers Lars10 and CATH17) digested with the same enzymes | This study |
| Primersd | ||
| Y01 | 5′-GCCGGAATTCGTTCGATAATATCGTTGAC-3′ | This study |
| Y03 | 5′-GCGGGATCCTGATTCCTACTAGCGGC-3′ | This study |
| Y04 | 5′-GCCGGAATTCCAAAATTAAAGTTCGGG-3′ | This study |
| Y05 | 5′-GCGGGATCCTCCGTTTCAGTGTGCTTCC-3′ | This study |
| Lars1 | 5′-GCGGGATCCTGCGATAGCTGGTGCCC-3′ | This study |
| Lars5 | 5′-GCCGAAGCTTCGCGACTATTGTTCGC-3′ | This study |
| Lars8 | 5′-GCCGGAATTCGTGAAAATAGTTAAAAAGG-3′ | This study |
| Lars9 | 5′-GCGGGATCCCAATGAGACCGACAAGG-3′ | This study |
| Lars10 | 5′-GCCGGAATTCTCAACTGCTATCCCCC-3′ | This study |
| Lars11 | 5′-GCCGGAATTCACTCATCAACGGAAACG-3′ | This study |
| Lars12 | 5′-GCGGGATCCGTAGAAGCTGCAACAGG-3′ | This study |
| Lars14 | 5′-GCCGGAATTCAAGATTGAGCGGAAGCG-3′ | This study |
| Lars15 | 5′-GCGGGATCCATTCCGCGCGGCACGCC-3′ | This study |
| CATH17 | 5′-GCGGGATCCGTTTCCGTTCATGAGTG-3′ | This study |
| 5A-Ge | 5′-GTTTCTACCTAGTAACCGTAAAAAACTAGACTACAAG-3′ | This study |
| 3A-Ge | 5′-CTTGTAGTCTAGTTTTTTCACGGTTACTAGGTAGAAAC-3′ | This study |
IPTG, isopropyl-β-d-thiogalactopyranoside.
The underlined nucleotides indicate the positions of restriction sites.
Used to introduce the A71G mutation into pLJ21.
In vitro DNA manipulations and genetic techniques.
Isolation of DNA and basic molecular biology techniques were performed as previously described (22, 24, 25, 33).
Construction of transcriptional lacZ fusions.
Different promoter-containing PCR products were generated by using the primer combinations listed in Table 1. Random mutagenized sequences were generated by a modified PCR procedure (33). A Quick Change kit (Stratagene) was used to perform site-directed mutagenesis by using oligonucleotides with single nucleotide mismatches as PCR primers. The various DNA fragments were digested with restriction enzymes and ligated into pDG268cat, pDG268neo, or pMutin4 digested with the same enzymes and transformed into Escherichia coli MC1061 with selection for Apr. Plasmids extracted from E. coli were integrated into the B. subtilis chromosome as described previously (22, 28).
Enzyme assays and measurement of nucleobase and nucleoside uptake.
Cell extracts were made as described previously (26). Activities of β-galactosidase, purine phosphoribosyltransferases, and adenylosuccinate (sAMP) lyase were determined as described previously (3, 10, 26). All enzyme determinations were repeated at least three times. Enzyme activity was expressed in units; 1 U was equivalent to 1 nmol of product formed per min. The total protein content was determined by the Lowry method. Uptake of bases and nucleosides was performed as described by Saxild and Nygaard (25).
Bioinformatic tools.
Searches for specific nucleotide sequences in the B. subtilis genome sequence were performed by using the integrated BLAST search feature of the SubtiList web server (http://genolist.pasteur.fr/SubtiList/).
RESULTS
Isolation and characterization of a mutation affecting xpt-pbuX and pur operon expression.
A transcriptional xpt-lacZ fusion strain (LJ25) was constructed and used in a white-blue screening procedure for isolation of mutations affecting hypoxanthine- and guanine-controlled gene expression. The PCR-generated DNA fragment fused to lacZ contained the entire untranslated xpt leader sequence, the xpt promoter region, and a deleted PurR binding site. The PurR binding site upstream of the xpt promoter was deleted to ensure that only defects in the hypoxanthine-guanine control mechanism were obtained by the screening procedure (23). Strain LJ25 formed blue colonies on minimal medium agar plates and white colonies on minimal medium agar plates containing hypoxanthine and guanine. Approximately 107 cells were plated on a minimal medium agar plate containing 1 μM 2-fluoroadenine (FA). Forty colonies appeared after 24 h of incubation at 37°C. To become toxic, FA must be phosphoribosylated to the nucleoside monophosphate level. Therefore, mutants defective in adenine phosphoribosyltransferase (encoded by apt) are frequently obtained by this selection procedure (25). Alternatively, overexpression of purine biosynthetic enzymes might also give rise to resistance to toxic purine analogs (11). We were interested in isolating mutants of the latter type. We therefore streaked the 40 spontaneously FAr clones on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing minimal medium agarplates with or without hypoxanthine plus guanine. One clone (LJ26) was light blue on the purine-containing plate, while the rest of the colonies showed wild-type-like purine control. LJ25 (wild type) and LJ26 (FAr) were grown in liquid minimal media containing and not containing purines and assayed for β-galactosidase (xpt′-lacZ), xanthine phosphoribosyltransferase (XPRTase) (xpt), sAMP lyase (pur operon, purB), and adenine phosphoribosyltransferase (APRTase) (apt) activities. LJ26 had the same APRTase activity as strain LJ25, whereas the levels of β-galactosidase, XPRTase, and sAMP lyase were greater in LJ26 cells grown in the presence of hypoxanthine and guanine. In purine-free medium, the enzyme levels of LJ26 were slightly higher than those of the wild type. The FAr-conferring mutation clearly had a trans-acting effect since it affected gene expression at three different loci on the chromosome (Table 2). The FAr mutation was moved to another strain by congression. This strain, LJ27, showed the same elevated XPRTase and sAMP lyase levels in purine-containing medium as the donor strain, strain LJ26 (data not shown).
TABLE 2.
Expression of purB (sAMP lyase), xpt (XPRTase), apt (APRTase), and an xpt-lacZ transcriptional fusion integrated into the amyE locus in mutants resistant to FAa
| Strain | Relevant genotype and characteristics | Purines added | Enzyme activities (U/mg of protein)
|
|||
|---|---|---|---|---|---|---|
| β-Galactosidase | XPRTase | sAMP lyase | APRTase | |||
| LJ25 | amyE::xpt′-lac | − | 288 | 21 | 226 | 19 |
| + | 7 | 3 | 24 | ND | ||
| LJ26 | amyE::xpt′-lacZ FAr | − | 310 | 35 | 304 | 22 |
| + | 154 | 7 | 229 | ND | ||
The values are means from three experiments. Cells were grown in glucose minimal medium with and without purines (hypoxanthine and guanine) and analyzed as described in Materials and Methods. The variations were less than 25%. ND, not determined.
Generalized transduction with phage AR9 (25) was used to locate the FAr mutation on the chromosome. The FAr phenotype was found to be closely linked to ydhL located between 53 and 54 degrees on the B. subtilis genome (12). ydhL and its upstream regulatory region were amplified from strain LJ27 by PCR, and the nucleotide sequence was determined. It was found that the chromosome of LJ27 had suffered a 16-nucleotide (nt) deletion located just upstream of the ydhL translational start signal. The deleted sequence included one of the stems of a potential transcription termination stem-loop structure (Fig. 1). The mutation was designated ydhL1. A second ydhL mutant that was resistant to FA was isolated by a procedure similar to that used for LJ26. Strain ED497 had the same phenotypes for xpt and pur operon expression as strain LJ26 (data not shown). The nucleotide sequence of the ydhL leader region was determined, and a 52-bp deletion of the ydhL leader terminator was observed (Fig. 1). The ydhL regulatory region was amplified from LJ25 and LJ27 and was used to construct transcriptional lacZ fusions that were integrated into the amyE locus of wild-type strain 168. Low levels of ydhL expression were observed in strains containing the wild-type ydhL sequence fused to lacZ (strains LJ24 and LJ32), whereas strongly elevated expression from the ydhL1 regulatory region was observed in strain LJ52 (Table 3).
FIG. 1.
Nucleotide sequence of the ydhL regulatory region. The discrepancy between this sequence and the official B. subtilis genome sequence is indicated by an asterisk above the inserted C nucleotide. The sequence corresponds to the cloned sequence in front of lacZ in LJ24. LJ23 contains the same downstream lacZ fusion point as LJ24, and the bent arrow pointing to the right indicates the 5′ end of the cloned sequence. ED499 contains the same 5′ sequence as LJ24, and the 3′ lacZ fusion point is indicated by the bent arrow pointing to the left. The underlined nucleotides indicate the deleted segments of the ydhL1 and ydhL2 mutations. The arrows arranged in pairs (arrows 1 and 2; arrows 3 and 4; and arrows 5 and 6) above the sequence indicate palindromic sequences capable of forming various stem-loop structures when they are transcribed to RNA. The stem-loops composed of sequences 1 and 2 and of sequences 3 and 4 comprise the conserved 63-nt sequence found in front of XptR regulon genes. Sequences 5 and 6 constitute a transcription terminator structure. RBS, ribosome binding site.
TABLE 3.
Expression of ydhL′-lacZ transcriptional fusions as affected by purines and YdhLa
| Strain | Relevant genotype and lacZ fusionb | YdhL status | β-Galactosidase activity (U/mg of protein)
|
|
|---|---|---|---|---|
| Without purines | With purines | |||
| LJ24 | amyE::ydhL′-lacZ (nt 1 to 370) | Wild-type level | 0.7 | 1.5 |
| LJ32 | ydhL::(pLJ32 ydhL′-lacZ) | YdhL disruptant | 1.4 | 20 |
| LJ23 | amyE::ydhL′-lacZ (nt 157 to 370) | Wild-type level | 0.4 | 0.4 |
| ED499 | amyE::ydhL′-lacZ (nt 1 to 174) | Wild-type level | 6.7 | 6.0 |
| LJ52 | amyE::ydhL1′-lacZ (nt 1 to 370 with 16-nt ydhL1 deletion) | Wild-type level | 3,540 | 3,630 |
| LJ29 | amyE::ydhL′-lacZ ydhL1 (nt 1 to 370) | Overexpression | 0.6 | 0.5 |
Characterization of the ydhL regulatory region.
From the nucleotide sequences determined as described above it became clear that the previously published genome sequence (12) most likely contained a sequencing error after position 625985, which in our sequences is followed by a C residue. This residue is absent in the previously published genome sequence. The insertion of an extra C changed the original ydhL reading frame and resulted in extension of its 5′ end to an ATG start codon located immediately downstream of a potential ribosome binding site (Fig. 1). Deletion of 156 nt of the 5′ end of the ydhL fragment fused to lacZ in strain LJ24 resulted in a complete loss of ydhL-lacZ expression (strain LJ23) (Fig. 1 and Table 3). Deletion of 197 nt of the 3′ end of the LJ24 ydhL fragment resulted in low ydhL-lacZ expression (strain ED499) (Fig. 1 and Table 3). This located the ydhL promoter (probably the −35 region) between the fusion points defined by the fusions in strains LJ23 and ED499 and, most importantly, to a position upstream of the ydhL1 deletion mutation. The position of the promoter region relative to the ydhL translational start site indicates that ydhL is preceded by an untranslated leader sequence that is at least 176 nt long, which encodes a transcription termination structure together with other potential secondary structures (Fig. 1).
Regulation of ydhL expression.
The expression of ydhL in strain LJ24 was induced twofold in response to addition of purines (Table 3). The ydhL′-lacZ fusion of strain LJ24 was transformed into strain LJ27 (ydhL1), resulting in strain LJ29 (amyE::ydhL′-lacZ ydhL1). In LJ29, which was presumed to overproduce YdhL, hypoxanthine and guanine could not induce expression of the ydhL-lacZ fusion (Table 3). ydhL-lacZ expression was also studied in a ydhL-negative background. The mutation was introduced by integrating plasmid pMutin4 into ydhL as previously described (28). Intragenic insertion of pMutin plasmids results in gene disruption and in the formation of a transcriptional lacZ fusion to the upstream regulatory sequence. Strain LJ32 contained pMutin4 inserted into ydhL; in this strain YdhL deficiency significantly stimulated purine induction of the ydhL promoter (Table 3). It appeared that high levels of YdhL repressed ydhL expression and that ydhL expression was activated in the presence of hypoxanthine and guanine. The latter hypothesis is in contrast to the regulation of expression of the pur and xpt-pbuX operons, which are repressed by hypoxanthine and guanine.
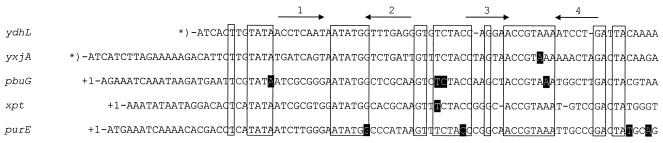
Similar untranslated leader sequences are found upstream of ydhL, the xpt operon, the pur operon, pbuG, and yxjA.
We observed that the ydhL leader sequence, which is composed of a possible transcription terminator located downstream of several other potential secondary structures, resembles the leader sequences in front of the pur and xpt-pbuX operons. Alignment of the leader sequences revealed a highly conserved nucleotide sequence in the 5′ ends of all three leaders (Fig. 2). This 63-nt sequence was used as the query sequence in a B. subtilis genome search for other genes with potential leader sequences similar to the ydhL, pur, and xpt leaders. We found two additional genes, pbuG and yxjA, which have leader sequences with a potential transcription terminator and which are capable of forming other upstream secondary structures. An alignment of the 5′ ends of all five transcriptional units is shown in Fig. 2. The five sequences can be folded into similar secondary structures composed of two consecutive stem-loop structures. The nucleotides defining the loops and the sequences flanking the stems are nearly 100% conserved in all five sequences, while the stem sequences are subject to greater variation. To analyze expression of pbuG and yxjA during growth with and without hypoxanthine and guanine, the β-galactosidase activities were determined in strains with pMutin4 integrated into pbuG (BFA2255) and yxjA (ED452). The enzyme activity was reduced in both strains five- to eightfold by hypoxanthine and guanine (Table 4). To test whether the ydhL1 mutation affected pbuG and yxjA expression, pbuG′- and yxjA′-lacZ transcriptional fusion strains and the corresponding ydhL1 derivatives were constructed in strains with intact pbuG and yxjA genes. Both pbuG expression and yxjA expression were repressed 16- to 20-fold in the presence of hypoxanthine and guanine. This repression was largely lost in a ydhL1 genetic background (Table 4). Based on the data in Tables 2, 3, and 4 we concluded that ydhL, xpt-pbuX, the pur operon, pbuG, and yxjA constitute a novel regulon, which is regulated by a putative hypoxanthine- and guanine-controlled transcription termination-antitermination mechanism.
FIG. 2.
Alignment of the nucleotide sequences containing a conserved 63-nt sequence found in the untranslated leader mRNA of five transcriptional units in B. subtilis. Identical nucleotides in all five sequences are enclosed in boxes. The arrows above the ydhL sequence indicate the positions of palindromic sequences found at the same positions relative to the identical nucleotides in all five mRNA leader regions. The highlighted nucleotides indicate the positions of single-base-pair substitutions that result in derepressed expression of the gene located downstream. Asterisks indicate that the exact +1 transcription start sites for ydhL and yxjA have not been established.
TABLE 4.
Effects of purines and the ydhL1 mutation on expression of genes having a leader sequence similar to the ydhL genea
| Strain | Relevant genotype | β-Galactosidase activity (U/mg of protein)
|
|
|---|---|---|---|
| Without purines | With purines | ||
| ED452 | yxjA::pMutin4 | 43 | 8 |
| BFA2255 | pbuG::pMutin4 | 141 | 18 |
| LJ21 | amyE::yxjA′-lacZ | 126 | 8 |
| LJ28 | amyE::yxjA′-lacZ ydhL1 | 96 | 54 |
| LJ4 | amyE::pbuG′-lacZ | 1,071 | 51 |
| LJ30 | amyE::pbuG′-lacZ ydhL1 | 775 | 503 |
Cells were grown and analyzed as described in Table 2, footnote a.
Mutagenic analysis of the upstream conserved leader region.
In order to verify the importance of the conserved sequences in the putative leader region, random mutagenesis was performed on the upstream regulatory sequences of xpt, purE, and pbuG. PCR fragments containing the leader and the promoter region were amplified under mutagenic conditions and fused to lacZ in pDG268 plasmid derivatives. The resulting plasmids were integrated into the amyE locus. Transformants that were not repressed by purines and therefore formed blue colonies on X-Gal plates containing purines were isolated and sequenced. The β-galactosidase levels in cell extracts of transformants that had only single base replacements or deletions are shown in Table 5. In addition, a mutation in the yxjA leader region in the second loop structure was introduced by site-directed mutagenesis (strain ED493). All 10 mutants listed in Table 5 showed high β-galactosidase activity in the presence of purines. Most clones were mutated in one of the highly conserved nucleotides, and the mutations mapped in one of the two putative loop structures between sequence 1 and sequence 2 and between sequence 3 and sequence 4 (Fig. 2), between the two stems, upstream of the 5′-proximal stem, or downstream of the 3′-proximal stem. A total of 44 derepressed mutants with mutations in the pur, xpt, and pbuG leaders were sequenced and assayed for β-galactosidase activity in the absence or presence of purines. Even though the majority of the mutants had more than one mutation, they all had at least one mutation in one of the regions mentioned above or in the terminator sequence. This clearly confirmed the importance of the conserved nucleotides in the loops and the sequences flanking the stems for hypoxanthine and guanine control of gene expression.
TABLE 5.
Mutational analysis of the proposed leader in the purE, pbuG, xpt, and yxjA regulatory region: effect of purines on expression of transcriptional fusionsa
| Strain | Relevant genotype | Mutationb | β-Galactosidase activity (U/mg of protein)
|
|
|---|---|---|---|---|
| Without purines | With purines | |||
| LJ20 | amyE::purE′-lacZ | None | 753 | 79 |
| LJ6 | amyE::purE′-lacZ | A85C | 1,008 | 957 |
| LJ7 | amyE::purE′-lacZ | C56A | 728 | 853 |
| LJ8 | amyE::purE′-lacZ | ΔG41 | 623 | 636 |
| LJ9 | amyE::purE′-lacZ | T82C | 954 | 836 |
| LJ10 | amyE::purE′-lacZ | T163A | 1,121 | 633 |
| LJ4 | amyE::pbuG′-lacZ | None | 1,071 | 51 |
| LJ15 | amyE::pbuG′-lacZ | Δ(T53-C54) | 477 | 466 |
| LJ17 | amyE::pbuG′-lacZ | A27G | 438 | 348 |
| LJ18 | amyE::pbuG′-lacZ | A70T | 428 | 565 |
| LJ25 | amyE::xpt′-lacZ | None | 288 | 7 |
| ED488 | amyE::xpt′-lacZ | T50A | 516 | 182 |
| LJ21 | amyE::yxjA′-lacZ | None | 126 | 8 |
| ED493 | amyE::yxjA′-lacZ | A71G | 110 | 63 |
Function of the pbuG and yxjA genes.
pbuG encodes a high-affinity hypoxanthine/guanine permease (23, 25), and yxjA encodes a putative nucleoside transport protein with 32% amino acid sequence identity to the B. subtilis NupC pyrimidine nucleoside transporter (22). By determining uptake of nucleosides in wild-type strain 168 and yxjA mutant strain ED452, we found that uptake of uridine was the same in the two organisms, whereas the levels of inosine and guanosine uptake in the yxjA mutant were 20% of the wild-type levels (data not shown). Because of the defect in purine nucleoside uptake in the yjxA mutant, we suggest that yxjA should be renamed nupG (nucleoside permease guanosine).
Function of the ydhL gene.
A program used to predict transmembrane helices of proteins (http://www.cbs.dtu.dk/services/-TMHMM/) indicated that there are 12 transmembrane segments in YdhL. The derived amino acid sequence of YdhL exhibits 32 to 38% identity to the amino acid sequences of putative chloramphenicol resistance proteins from B. subtilis (ybcL) (accession number A69746) and Pseudomonas aeruginosa (MFS transporter) (F883233). These findings prompted us to determine the susceptibility of ydhL mutants to various antibiotics, including chloramphenicol. We found that neither ydhL-defective mutant LJ32 nor YdhL-overproducing mutant LJ26 was more or less susceptible to chloramphenicol, kanamycin, neomycin, erythromycin, lincomycin, novobiocin, nalidixic acid, or tetracycline than wild-type cells (data not shown). The susceptibilities to FA and two other toxic purine analogs, 8-azaguanine and 8-azaxanthine, were also tested (Table 6). Interestingly, the decreased susceptibility to FA, which was the selection principle used for isolation of ydhL1 mutant strain LJ26, was paralleled by increased susceptibility to 8-azaxanthine. Uptake of adenine and hypoxanthine was found to be reduced in the ydhL1 mutant compared with the uptake in the wild type and an ydhL disruptant (Table 6). Our working hypothesis is that ydhL encodes an efflux system, which when overexpressed extrudes purine analogs and lowers purine base uptake. To test whether YdhL is involved in excretion of the purine bases contained in nucleic acids, two types of experiments were performed. In one approach direct determination of purine excretion was analyzed. In the other approach, possible effects related to the size of the intracellular pool of purine bases were assessed. Wild-type cells (strain LJ25) and ydhL mutant cells (strains LJ26 ydhL1 and LJ54 ydhL) were used. Cross-feeding experiments on solid media showed that the three strains supported weak growth of a purine-requiring mutant (strain ED244 purM) to the same extent (data not shown). The same result was obtained when strain ED244 was incubated in cell-free filtrates of minimal salt growth medium in which the same strains (LJ25, LJ26, and LJ54) had been grown to an optical density at 436 nm of 3. In these experiments a purine concentration of 1 μM arising from the cells growing in the medium would have been detected.
TABLE 6.
Phenotypes of ydhL mutants; susceptibilities to purine analogs and purine base transporta
| Strain | Relevant genotype | MIC (μg/ml) of:
|
Purine transport (U/mg [dry wt])
|
|||
|---|---|---|---|---|---|---|
| FA | azaG | azaX | Adenine | Hypoxanthine | ||
| LJ25 | Wild type | 0.5 | 25 | 12 | 5.3 | 2.6 |
| LJ26 | ydhL1 | 15 | 25 | 6 | 1.9 | 1.1 |
| LJ32 | ydhL | 0.5 | 25 | 12 | 4.8 | 2.9 |
MICs were determined by a twofold agar dilution technique with minimal salts medium by streaking bacteria on plates. azaG, 8-azaguanine; azaX, 8-azaxanthine. Purine base transport was determined as described in Materials and Methods.
It has been reported previously that expression of the xpt-pbuX operon is repressed in an hpt mutant (3). In such a mutant hypoxanthine cannot be salvaged and hence accumulates intracellularly. This creates a situation similar to the situation that occurs when hypoxanthine is abundant in the growth medium. The effects of the hpt and ydhL1 mutations on expression of xpt′-lacZ and ydhL1′-lacZ were examined. It must be remembered that xpt expression is repressed by hypoxanthine and guanine, while ydhL expression is induced. Expression of the xpt-pbuX operon was repressed and ydhL gene expression was induced twofold in the hpt mutant (Table 7). This was expected because of the increased pool of hypoxanthine and guanine. In the hpt ydhL1 double mutant the effect of the hpt mutation was reduced (Table 7). The most plausible explanation for this is that the size of the endogenous hypoxanthine-guanine pool is decreased by increased hypoxanthine-guanine efflux due to a hyperactive YdhL protein. We therefore propose a new designation for ydhL, pbuE, in which the E stands for efflux.
TABLE 7.
Effects of the hpt and ydhL1 mutations on expression of xpt-lacZ and ydhL-lacZ transcriptional fusionsa
| Strain | Relevant genotype | β-Galactosidase activity (U/mg of protein)
|
|
|---|---|---|---|
| Without purines | With purines | ||
| LJ25 | amyE::xpt′-lacZ | 288 | 7 |
| LJ53 | amyE::xpt′-lacZ hpt | 7 | 1.4 |
| LJ51 | amyE::xpt′-lacZ hpt ydhL1 | 128 | 9 |
| LJ24 | amyE::ydhL′-lacZ | 0.7 | 1.5 |
| HH441 | amyE::ydhL′-lacZ hpt | 1.3 | 3.7 |
| HH442 | amyE::ydhL′-lacZ hpt ydhL1 | 1.3 | 1.8 |
Cells were grown and analyzed as described in Table 2, footnote a.
DISCUSSION
We identified a second pur regulon in B. subtilis controlling expression of the pur operon, the xpt-pbuX operon, and the pbuE (ydhL), nupG (yxjA), and pbuG genes. This regulon, which we designated the XptR regulon, is different from the PurR regulon. However, some genes (the pur operon, the xpt-pbuX operon, and pbuG) belong to both regulons. When B. subtilis is grown in minimal salt medium, both the PurR regulon and the XptR regulon, except for the pbuE gene, are expressed. In other words, the enzymes required for de novo purine synthesis and transporters of purine bases and nucleosides are synthesized. When purine compounds are present in the growth medium, they are readily taken up and used for purine nucleotide synthesis. At the same time purines have an effect on expression of the pur genes, either directly or by affecting the PRPP pool size. The PurR regulon responds to changes in the PRPP pool. A small cellular PRPP pool facilitates the binding of PurR to the tandem PurBoxes, which results in inhibition of transcription (27). When the PRPP pool is large, PRPP binds to PurR and prevents its binding to the tandem PurBoxes, thus stimulating transcription. The XptR regulon, except for the pbuE gene, is negatively regulated by the free bases hypoxanthine and guanine, which act synergistically (3). Adenine and hypoxanthine and their nucleoside derivatives are the best purine sources for B. subtilis, as judged from the growth of purine auxotrophs on these compounds and from incorporation studies with wild-type cells (17, 18). When present, these compounds serve as sole purine sources, and de novo purine synthesis is shut down. Guanine, guanine nucleosides, and xanthine, on the other hand, are less well used, and in their presence a considerable portion of the purine nucleotides synthesized is formed via the de novo pathway.
Uptake and metabolism of purine compounds present in the growth medium also have an effect on the cellular nucleotide pool, and this affects the activity of key enzymes of the de novo purine pathway (26). De novo IMP synthesis is controlled by feedback inhibition of the first enzyme of the pathway, glutamine PRPP amidotransferase. The strongest single inhibitor is AMP, but GMP, ADP, and GDP also inhibit the enzyme synergistically (15). The branching from IMP to form AMP and GMP is feedback controlled at the first enzyme of the branch by the corresponding end products (9, 26, 32).
An increase in the size of the PRPP pool is seen when cells are starved for purines and when guanine and guanosine are abundant (26). A reduction in the PRPP pool size is seen when adenine and adenosine are present, most likely because of an increase in the size of the adenylate nucleotide pool, which results in ADP inhibition of PRPP synthetase activity (1). During purine starvation a large PRPP pool stimulates the synthesis of enzymes involved in de novo purine synthesis (increased expression of the pur operon and purA, encoding sAMP synthase), and there is increased capacity for synthesis of the precursor molecules glycine and one-carbon units by increased expression of folD and glyA (23). Furthermore, the capacity to convert GMP to IMP is increased as a result of an increased level of GMP reductase encoded by guaC (23). Due to increased expression of pbuG, pbuO, and xpt-pbuX during purine starvation, the capacity for uptake of purine bases is also increased. When an increase in the PRPP pool size is caused by guanine and guanosine in the medium, both the PurR and XptR control mechanisms operate. Of the genes belonging to the PurR regulon, expression of guaC, purA, glyA, purR, and folD is induced, whereas expression of the pur operon, pbuG, and xpt-pbuX is reduced. Under these conditions genes encoding enzymes that have special importance for the utilization of guanine as the sole purine source (namely, guaC and purA) are induced. At the same time the demand for the de novo pathway is reduced. A decrease in pur operon expression is caused by guanine, which also inhibits transcription of pbuG, nupG, and the xpt-pbuX operon. The pbuE gene is induced and may be involved in excretion of excess purine bases. When the PRPP pool is small because of metabolism of adenine and adenosine from the growth medium, expression of the PurR regulon is reduced due to the small PRPP pool. This makes sense because under these conditions there is a reduced need for enzymes encoded by the pur operon, including purine transporters, GMP reductase (guaC), and sAMP synthase (purA). The deamination of adenine to hypoxanthine (19) ensures synthesis of IMP, which is further converted to GMP by IMP dehydrogenase and GMP synthetase, enzymes that are not part of the pur regulons. Hypoxanthine affects only expression of the XptR regulon by reducing the synthesis of purine biosynthetic enzymes and transporters of purine bases and nucleosides. In agreement with the conclusions described above, the most dramatic reduction in expression of both regulons is seen when mixtures of purines, like adenine and guanosine, are combined (18). Altogether, we have now a more or less complete picture of how regulation of purine synthesis on the genetic level accounts for the metabolic data. The physiological significance of the control of expression of the two pur regulons by two overlapping control mechanisms is that it ensures balanced synthesis of purine nucleotides by affecting the levels of purine biosynthetic enzymes and transporters. This control is mediated via the availability and composition of purines in the growth medium.
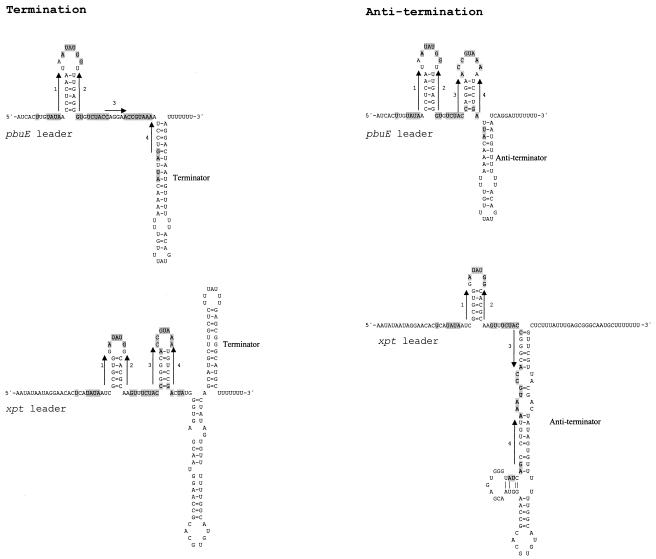
As indicated above and shown in Fig. 2, all of the XptR regulon transcriptional units have leader sequences capable of forming alternative secondary structures. The leader sequence of pbuE (ydhL) was analyzed by using standard RNA fold software (mfold v. 3.1; M. Zuker; http://www.bioinfo.rpi.edu/-applications/mfold/-old/rna), and the results are shown in Fig. 3. A secondary structure containing the terminator stem-loop was predicted by the software to be the most stable form (−31 kcal) of the transcript. Software parameters were modulated to show other possible foldings of the leader sequence. The second most stable structure was the form containing the conserved 63-nt sequence folded into a tandem stem-loop structure, followed downstream by the antiterminator stem-loop (Fig. 3). The stability of the latter form was calculated to be −21 kcal, indicating that this form is less stable than the form containing the terminator stem-loop. Since pbuE expression is induced in the presence of hypoxanthine and guanine, it is tempting to suggest that formation of the tandem stem-loop structure (resulting in antitermination) is stabilized under these conditions. A similar analysis was performed with the xpt-pbuX operon leader sequence. In contrast to expression of pbuE, expression of the xpt-pbuX operon and the rest of the XptR regulon transcriptional units was repressed in the presence of hypoxanthine and guanine. Therefore, it must be expected that the presence of hypoxanthine and guanine favors a secondary structure containing a terminator stem-loop. In the case of the xpt-pbuX leader sequence, the software predicted that the structure containing the terminator stem-loop would be the most stable structure (−56 kcal) (Fig. 3). Modulation of the software parameters resulted in folding of an alternative structure in which the sequence of the second stem-loop of the tandem stem-loop structure participated in formation of the antiterminator stem-loop (Fig. 3). Now, the problem is that the predicted stability of the antiterminator-containing structure is lower (−20 kcal) than the predicted stability of the terminator-containing structure. This contradicts the model proposed for pbuE, in which the presence of hypoxanthine and guanine stabilizes formation of the tandem stem-loop structure. One important reservation must be noted when computer-generated RNA secondary structures are analyzed. The software is generally designed to give the most stable form of a given RNA sequence. When we loaded the program with the full-length leader sequences, the software did not take into account that mRNA folding is a dynamic process in which the RNA folds immediately after it has been synthesized. It is very unlikely that a nonfolded full-length leader mRNA molecule is present in the cell. Therefore, the folding of the nascent leader mRNA may be influenced by regulatory signals that eventually lock the conformation in an energetically less favorable structure. In all XptR regulon leader mRNA sequences the sequence of the tandem stem-loop structure is synthesized first. Therefore, the folding of this sequence must influence whether the downstream antiterminator or terminator stem-loops are formed in response to the absence or presence of hypoxanthine and guanine. Therefore, detailed in vitro RNA folding studies have to be performed in order to propose a model for this time- and corepressor-dependent process.
FIG. 3.
Possible secondary structures of the ydhL and xpt leader mRNA. Highlighted nucleotides indicate the nucleotides enclosed in boxes in Fig. 2. The arrows indicate the palindromic sequences responsible for formation of the tandem stem-loop structure characteristic of the XptR regulon genes.
What is the role of the characteristic tandem stem-loop structure in purine-regulated expression of the XptR regulon transcription units? The obvious answer is that it functions as a cis-acting element in the control mechanism. From what has been reported about control of B. subtilis pyr operon expression and most recently about control of expression of thiamine and riboflavin biosynthetic genes and other genes in E. coli and B. subtilis, two possible roles may be ascribed to this structure. One model for control of the XptR regulon is a model similar to that for control of pyr operon expression (21). pyr operon expression is controlled by the RNA binding protein PyrR, which, when UMP is abundant in the cell, binds to the PyrR binding site, which is a stem-loop structure formed at three different positions in the pyr operon leader sequence. Binding of PyrR-UMP to this structure stimulates the formation of terminator structures located downstream. Several genetic and biochemical strategies have been used in our laboratory to obtain purine regulatory mutants of B. subtilis with mutations affecting the XptR regulon or to purify a trans-acting protein factor from B. subtilis cell extracts. None of these attempts have been successful, indicating that the XptR regulon may not be controlled by a trans-acting regulatory protein. This leads to a second possible model, which resembles the model for control of expression of thiamine and riboflavin biosynthetic genes in E. coli and B. subtilis (16, 30, 31) and the T and S box transcription termination control systems which control amino acid biosynthetic genes in B. subtilis (8, 14). In B. subtilis these biosynthetic genes are controlled by direct binding of the effector molecule (thiamine pyrophosphate in the case of thiamine genes, flavin mononucleotide in the case of riboflavin genes, uncharged tRNA in the case of T box genes, and S-adenosylmethionine in the case of S box genes) to secondary structures of leader mRNA sequences found in the corresponding transcripts. In favor of this model two facts may be taken in to account. One fact is that a trans-acting protein that controls the XptR regulon has not been found. The second fact is that the pbuE-encoded efflux pump of B. subtilis appears to be responsible for adjustment of the cellular hypoxanthine and guanine pools. The fact that pbuE expression is induced by an elevated hypoxanthine-guanine pool may indicate that B. subtilis has a highly regulated mechanism that ensures an optimal balance between purine nucleotide demand and the cellular biosynthetic capacity. The fact that regulation of the XptR regulon is focused on adjusting the concentration of hypoxanthine and guanine suggests that these molecules may be directly involved in the regulatory mechanism. We are currently attempting to test the latter possibility by in vitro transcription analysis.
Acknowledgments
We thank Jenny Steno Christensen and Kirsten Hansen for excellent technical assistance.
This work was supported by Danish Natural Science Research Council grant 21-02-0492 and by the Saxild Family Foundation.
REFERENCES
- 1.Arnvig, K., B. Hove-Jensen, and R. L. Switzer. 1990. Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur. J. Biochem. 192:195-200. [DOI] [PubMed] [Google Scholar]
- 2.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christiansen, L. C., S. Schou, P. Nygaard, and H. H. Saxild. 1997. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 179:2540-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide biosynthesis. J. Biol. Chem. 262:8274-8287. [PubMed] [Google Scholar]
- 5.Ebbole, D. J., and H. Zalkin. 1988. Characterization of the Bacillus subtilis pur operon: new insights into gene regulation, p. 51-55. In J. A. Hoch (ed.), Genetics and biotechnology of bacilli, vol. 2. Academic Press Inc., New York, N.Y.
- 6.Ebbole, D. J., and H. Zalkin. 1988. Detection of pur operon-attenuated mRNA and accumulated degradation intermediates in Bacillus subtilis. J. Biol. Chem. 263:10894-10902. [PubMed] [Google Scholar]
- 7.Ghim, S.-Y., and R. L. Switzer. 1996. Characterization of cis-acting mutations in the first attenuator region of the Bacillus subtilis pyr operon that are defective in pyrimidine-mediated regulation of expression. J. Bacteriol. 178:2351-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy, F. J., W. C. Winkler, and T. M. Henkin. 2002. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA 99:11121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii, K., and I. Shiio. 1970. Regulation of purine ribonucleotide synthesis by end product inhibition. III. Effect of purine nucleotides on succino-AMP synthetase of Bacillus subtilis. J. Biochem. 68:171-176. [DOI] [PubMed] [Google Scholar]
- 10.Jochimsen, B., P. Nygaard, and T. Vestergaard. 1975. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Mol. Gen. Genet. 143:85-91. [DOI] [PubMed] [Google Scholar]
- 11.Kuninaka, A. 1986. Nucleic acids, nucleotides, and related compounds, p. 71-114. In H. J. Rehm and G. Reed (ed.), Bio/technology. VCH Verlagsgesellschaft, Weinheim, Germany.
- 12.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Lu, Y., R. J. Turner, and R. L. Switzer. 1996. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl. Acad. Sci. USA 93:14462-14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel, B. A., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer, E., and R. L. Switzer. 1979. Regulation of n-glutamine phosphoribosylpyrophosphate amidotransferase activity by end products. J. Biol. Chem. 254:5397-5402. [PubMed] [Google Scholar]
- 16.Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA. A mechanism to control transcription in bacteria. Cell 111:747-756. [DOI] [PubMed] [Google Scholar]
- 17.Nygaard, P. 1983. Utilization of preformed purine bases and nucleosides, p. 27-93. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, London, United Kingdom.
- 18.Nygaard, P. 1993. Purine and pyrimidine salvage pathways, p. 359-378. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 19.Nygaard, P., P. Duckert, and H. H. Saxild. 1996. Role of adenine deaminase in purine salvage and nitrogen metabolism and characterization of the ade gene in Bacillus subtilis. J. Bacteriol. 178:846-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins, J. B., A. Sloma, T. Hermann, K. Theriault, E. Zachgo, T. Erdenberger, N. Hannett, N. P. Chatterjee, V. Williams II, G. A. Rufo, Jr., R. Hatch, and J. Pero. 1999. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol.Bio/technol. 22:8-18. [Google Scholar]
- 21.Savacool, H. K., and R. L. Switzer. 2002. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J. Bacteriol. 184:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxild, H. H., L. N. Andersen, and K. Hammer. 1996. dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J. Bacteriol. 178:424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saxild, H. H., K. Brunstedt, K. I. Nielsen, H. Jarmer, and P. Nygaard. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxild, H. H., J. H. Jacobsen, and P. Nygaard. 1995. Functional analysis of the Bacillus subtilis purT gene encoding formate-dependent glycinamide ribonucleotide transformylase. Microbiology 141:2211-2218. [DOI] [PubMed] [Google Scholar]
- 25.Saxild, H. H., and P. Nygaard. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 169:2977-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxild, H. H., and P. Nygaard. 1991. Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing nucleotide pools. J. Gen. Microbiol. 137:2387-2394. [DOI] [PubMed] [Google Scholar]
- 27.Shin, B. S., A. Stein, and H. Zalkin. 1997. Interaction of Bacillus subtilis purine repressor with DNA. J. Bacteriol. 179:7394-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagner, V., E. Dervyn, and D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 29.Weng, M., P. L. Nagy, and H. Zalkin. 1995. Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. USA 92:7455-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler, W., A. Nahvi, and R. R. Breaker. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952-959. [DOI] [PubMed] [Google Scholar]
- 31.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99:15908.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, T. W., and K. G. Scrimgeour. 1973. Properties of inosinic acid dehydrogenase from Bacillis subtilis. II. Kinetic properties. Can. J. Biochem. 51:1391-1398. [DOI] [PubMed] [Google Scholar]
- 33.Zeng, X., and H. H. Saxild. 1999. Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 181:1719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]