Abstract
The InvE protein positively regulates the expression of virulence genes ipaBCD in Shigella sonnei. The InvE has significant homology with ParB of plasmid P1, which is known as a plasmid partitioning factor with DNA binding ability. Although the DNA binding activity of InvE has been predicted, it is not known whether the DNA binding activity is necessary for type III secretion system-associated gene expression. In this study, we determined the transcription start site of the icsB-ipaBCD operon (ipa operon) and constructed a series of deletions of the icsB promoter region in the Escherichia coli K-12 background. The deletion study revealed that an 86-bp region upstream of the icsB transcription start site was essential for expression of the ipa operon, where the ParB binding motif (ParB BoxA-like sequence) was observed. Purified glutathione S-transferase-InvE fusion protein bound directly to the −93 to −54 region (designating the icsB transcription start site as nucleotide +1) containing the ParB BoxA-like sequence. These results indicated that InvE bound directly to the promoter region.
The gram-negative bacterial genus Shigella includes the causative agent of bacillary dysentery in humans. The essential steps for Shigella virulence are invasion of epithelial cells, intracellular multiplication, and spread of bacteria into adjacent cells (16). The invasion ability of shigellae is dependent upon a 31-kb region of the 230-kb virulence plasmid. The common 31-kb region is essential for virulence. This region includes genes for invasins, molecular chaperones, motility, regulation, and a specialized type III secretion system. A similar plasmid is also found in enteroinvasive Escherichia coli strains (10, 11, 14, 20, 28, 29).
The 31-kb region contains two divergently transcribed loci that encode more than 30 genes. One locus includes the ipaBCD genes for effector molecules and the icsB and ipgC genes, and the other locus includes a cluster of about 20 genes for Mxi and Spa proteins, which are components of the type III secretion system and function in surface presentation and secretion of the IpaBCD effector proteins (1, 2, 3, 31, 36). Ipa effector proteins act as triggers for entry into epithelial cells and are required for escape from phagosomes (14, 20). The IpaB protein also has a lysin-like, cytotoxin-like function, by which it lyses vacuole membranes and induces apoptosis of macrophages (38). The IcsB and IpgC proteins are encoded upstream of the ipa genes. IcsB forms protrusions at the surface of infected cells, and IpgC is a molecular chaperone that associates with IpaB and IpaC in the bacterial cytoplasm. IpgC stabilizes IpaB and IpaC and prevents their premature association (21). The expression of the icsB-ipa operon region is positively controlled by a regulator cascade composed of InvE (VirB) and VirF, which are encoded on the virulence plasmid (8, 12, 24, 30).
The primary regulator VirF is an AraC-like transcription factor that activates transcriptional expression of the secondary regulatory gene invE (26). InvE is required for transcription of the ipa, mxi, and spa operons but a mechanism of activation has not been clarified. When the bacteria encounter a 37°C environment, VirF activates transcription of both icsA and invE, initiating the cascade of virulence gene expression (19, 37).
InvE protein has been identified as an essential factor for the expression of the virulence genes of Shigella sonnei (37). InvE has no homology to conventional transcriptional factors. However, InvE has significant homology to the ParB and SopB proteins, which are required for plasmid partitioning and maintenance of plasmid copy number of the P1 and P7 plasmids and F plasmid, respectively. But there are some differences in function between ParB and InvE. ParB is involved in plasmid partitioning, while InvE is probably not, because InvE does not complement a parB mutation (H. Watanabe, unpublished data). ParB autorepresses parAB transcription (15), while InvE activates the expression of invasion genes. The ParB dimer binds specifically to the parS sequence of plasmid P1. A helix-turn-helix (HTH) region of ParB recognizes the BoxA motifs of the parS site (5, 35).
The predicted secondary structure of InvE contains two motifs found in transcriptional factors: HTH, and a putative leucine zipper. Beloin et al. reported that the oligomerization of InvE requires the leucine zipper domain and that the ability of InvE to activate virulence gene expression depends on the presence of the HTH motif (4). However, it is not clear how InvE regulates virulence gene expression.
In this study, we focused on identification of the cis element required for activation of expression of the ipa genes by InvE. First, we determined the transcription start site of icsB, which is the leadoff gene of the ipa operon. Then, we confirmed that a glutathione S-transferase (GST)-InvE fusion protein bound specifically to the region around the transcription start site of icsB. Based on these findings, we concluded that the region upstream of the ipa operon is essential for IpaB expression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli K-12 CC118 (18) was the host for pSS120 and its derivatives. E. coli MC1061 (34) was the host for overproduction of the GST-InvE fusion protein. Strain HW1273 is an ampicillin-resistant E. coli K-12 strain carrying S. sonnei virulence plasmid pHW1273 (pSS120::Tn1) which was derived from transconjugation of E. coli CC118 with S. sonnei HW383 (37). Plasmid pHW749, containing virF and invE genes, was used as the source of these activators (37). Plasmid pJK1142 is a derivative of F plasmid-replicon vector pJK282 carrying a 31-kb region spanning from a part of ipaB to the mxi-spa genes of S. sonnei plasmid (14). Plasmid pHW735 is a derivative of pJK1142 which has a Tn3-lac insertion in ipaB, which was used for determination of the transcription start site of icsB (37). In this study, all the experiments were performed in E. coli. We are assuming that the transcriptional signals are well conserved between S. sonnei and E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli K-12 | ||
| MC1061 | hsdR hsdM araD139 Δ(ara-leu)7679 Δlac(IPOZYA) galU galK rpsL | 34 |
| CCI18 | ΔphoA20 galE galK thi rpsE rpoB recA1 argE(Am) ΔlacX74 | 18 |
| HW1273 | CC118 harboring pHW1273 | 14 |
| Plasmids | ||
| pHW735 | pJK282 + BamHI fragment carryng most of ipaB::Tn3-lac of pHB76, Δ(invE virF)Kmr | 37 |
| pHW749 | Both invE and virF cloned into pHSG 415, Cmr | 37 |
| pHW1273 | Large virulence plasmid pSS120 of S. sonnei HW383 carrying Tn1, Apr | 14 |
| pKD46::Cm | λ Red helper plasmid:oriR101 repA101(Ts)P-araB-gam-bet-exo Cmr | This study |
| pKD13 | Kmr gene with FRT sequence | 7 |
| pCP20 | FLP helper plasmid; pSC101 replicon (Ts) bla+cat+ Flp(λRp) c1857 Apr Cmr | 7 |
| pTT0606 | pHW1273 with region upstream of icsB promote deleted from −1740 to −136 (using primers H2 and H5 for disruption; see Table 2) | This study |
| pTT0818 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −41 (using primers H2 and H6 for disruption; see Table 2) | This study |
| pTT1405 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −74 (using the primers H2 and H10 for disruption; see Table 2) | This study |
| pTT1608 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −84 (using primers H2 and H11 for disruption; see Table 2) | This study |
| pTT1804 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −94 (using primers H2 and H12 for disruption; see Table 2) | This study |
| pTT2002 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −104 (using primers H2 and H13 for disruption; see Table 2) | This study |
| pTT2201 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −85 (using primers H2 and H13-1 for disruption; see Table 2) | This study |
| pTT2202 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −86 (using primers H2 and H13-2 for disruption; see Table 2) | This study |
| pTT2203 | pHW1273 with region upstream of icsB promoter deleted from −1740 to −87 (using primers H2 and H13-3 for disruption; see Table 2) | This study |
| pGEX-5X-3 | Expression vector, tacP lacIqSj26 Apr | 33 |
| pQW2001 | pGEX-5X-3 + a fragment encoding full-length of InvE protein, Apr | This study |
Media and buffers.
Bacteria were grown according to standard methods in Luria-Bertani (LB) broth (23) or on 1.5% Bacto agar plates, both containing appropriate antibiotics. Antibiotic concentrations were as follows: kanamycin, 40 μg/ml; chloramphenicol, 25 μg/ml; and ampicillin, 40 μg/ml. YENEB (0.75% Bacto yeast extract, 0.8% Bacto nutrient broth) was used for preparation of competent cells (32). During the course of protein purification, a lysis buffer (1% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride in phosphate-buffered saline [0.8% NaCl, 0.02% KCl, 0.3 mM Na2HPO4, and 0.15 mM KH2PO4]) was used for the bacterial suspension step. For dilution of protein samples, we used an elution buffer consisting of 50 mM Tris-HCl (pH 7.8) containing 10 mM reduced glutathione.
Preparation and manipulation of DNA.
Preparation and manipulation of DNA were carried out essentially as described by Maniatis et al. (17).
RNA preparation and primer extension.
Total cellular RNA was extracted from E. coli K-12 MC1061 harboring pHW735 and pHW735 or pHW749 in order to study transcription of icsB. RNA was prepared essentially as described by Chomczynski and Sacchi (6). Bacterial cultures were grown with shaking at 37°C in 40 ml of LB medium overnight. RNA samples from each culture were prepared with Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions, and the RNA was quantified by measuring the absorbance at 260 nm (A260). The RNA sample was dissolved in H2O and stored at −70°C.
For the primer extension assay, we used a synthetic primer (5′-GAGGATCATACTTTATTAACTC-3′, complementary to nucleotides 126 to 147 in Fig. 1) that had been 5′-end labeled with [γ-32P]ATP (specific activity, >3,000 Ci/mmol; Amersham United Kingdom) with T4 polynucleotide kinase (Megalabel; Takara, Tokyo, Japan). Total RNA (20 μg) was annealed to 1.0 pmol of 5′-end-labeled primer at 80°C for 2 min and then at 42°C for 45 min in 50 μl of buffer containing 100 mM Tris-HCl (pH 7.5), 100 mM KCl, and 20 mM MgCl2. Then, the four deoxynucleoside triphosphates, dithiothreitol, and reverse transcriptase from avian sarcoma-related virus (Takara, Tokyo, Japan) were added to the solution at final concentrations of 2 mM each, 1 mM, and 20 U per 50 μl, respectively. The reaction was continued for 1 h at 42°C. The synthesized DNA was extracted with phenol-chloroform, precipitated with ethanol, dissolved in DNA sequencing solution (27), and analyzed by electrophoresis on an 8% polyacrylamide gel containing 8.3 M urea, followed by autoradiography. As a standard, the above 32P-labeled primer was annealed to alkaline-denatured DNA from pHW735, and a dideoxy chain termination sequencing reaction was amplified with Klenow enzyme, as described elsewhere (27). The samples were loaded on the gel with the synthesized DNA described above.
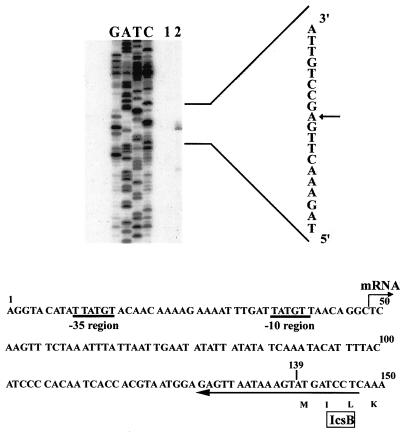
FIG. 1.
Primer extension mapping of the 5′ end of transcript for icsB and alignment of 5′-end nucleotide sequence around translation initiation site of icsB. Total cellular RNAs were extracted from MC1061 harboring pHW735 (lane 1) and MC1061 harboring pHW735 and pHW749 (lane 2) grown in LB medium and annealed to the 32P-labeled primer (5′-GAGGATCATACTTTATTAACTC-3′). DNA was synthesized by the reaction of reverse transcriptase. Lanes G, A, T, and C, standard sequence ladder synthesized by using the same primer and pHW735 DNA as the template. This indicates the sequence of the coding (non-RNA-like) strand, a part of which is shown in vertically ordered characters on the right. The arrow indicates the detected 5′ end of icsB mRNA. Below the gel, the alignment of the 5′-end sequence of the noncoding (RNA-like) strand around the translation initiation site of icsB is shown. The nucleotide sequence is complementary to the S. sonnei pSS120 DNA sequence reported by Arakawa et al. (GenBank accession no. D50601). The hooked arrow at T-49 shows the 5′ end of the icsB transcript on the noncoding strand. The −10 and −35 regions of a possible promoter are indicated by thick underlines. The horizontal arrow denotes the position and direction of the primer used in this experiment. The translation initiation site of icsB is A-139 in this figure, and the deduced amino acid sequence starting at this site is also indicated in the one-letter code.
Construction of promoter deletion mutants by a PCR-based gene disruption method.
The icsB promoter deletion mutants of pHW1273 (designated pTT0606, pTT0818, pTT1405, pTT1608, pTT1804, pTT2002, pTT2201, pTT2202, and pTT2203) were constructed by the PCR-based gene disruption method reported by Datsenko and Wanner (7).
For construction of the deletion series, the PCR-based gene disruption method with λ Red recombinase (7) was used with some modifications. Because strain HW1273 is ampicillin resistant, the bla gene of plasmid pKD46 (7) was exchanged with cat by an in vitro transposase reaction with the GPS-LS kit (New England Biolabs). The resultant plasmid was designated pKD46::Cm.
Strain HW1273 with λ Red helper plasmid (pKD46::Cm) was grown in 5-ml YENB cultures with chloramphenicol and 10 mM l-arabinose at 30°C to an optical density at 600 nm of 0.6 and was then made electrocompetent by concentrating the bacteria 100-fold and washing three times with ice-cold distilled water (7). Next, linear DNA for the construction of deletion mutants was synthesized by PCR. PCR primers (Table 2.) were about 70-bp single-stranded DNAs corresponding to the sites which flank the upstream regions of the icsB promoter to be deleted and the kanamycin resistance gene of pKD13. Plasmid pKD13 was used as a template DNA for PCR to introduce a kanamycin resistance cassette with the FLP recognition target (FRT) sequence at the both ends of the kanamycin resistance gene. PCR products were purified and suspended in distilled water. Transformation was performed by electroporation with 50 μl of competent cells (HW1273 carrying pKD46::Cm) prepared as described above and 100 ng of the linear PCR product. Shocked cells were added to 1 ml of SOC (2% Bacto tryptone, 0.5% Bacto yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgSO4, 20 mM glucose), incubated overnight at room temperature, and then spread onto agar to select kanamycin-resistant transformants, which should result from recombination between pHW1273 and the linear PCR product, and subsequent deletions of the upstream of icsB promoter region. After primary selection, mutants were maintained on medium without an antibiotic. They were colony purified once nonselectively at 42°C and then tested for chloramphenicol sensitivity to confirm the loss of the helper plasmid pKD46::Cm, which is temperature sensitive for replication.
TABLE 2.
Primers designed for gene disruptiona
| Primer | Sequence (5′ → 3′) |
|---|---|
| H2 | (−1790)-ATACCCAAGGCTCGGCAAATAACATCTGCTAAATCTTCCATATATTCCT(−1741)CATTCCGGGGATCCGTCGACC |
| H5 | (−84)-CCCACAAGTTAAAGTGTCTGATATATCAGGCTCGGAGTGTTATAGAAAA(−135)AGTGTAGGCTGGAGCTGCTTC |
| H6 | (+14)-ATTTAGAAACTTGAGCCTGTTAACATAATCAAATTTTCTTTTGTTGTACATAAT(−40)GTGTAGGCTGGAGCTGCTTC |
| H10 | (−24)-CTTTTGTTGTACATAATATGTACCTCGTGAGCATATGTAGTGCTCGTTTC(−73)GTGTAGGCTGGAGCTGCTTC |
| H11 | (−34)-ACATAATATGTACCTCGTGAGCATATGTAGTGCTCGTTTCATCATGAAA(−83)TGTGTAGGCTGGAGCTGCTTC |
| H12 | (−44)-TACCTCGTGAGCATATGTAGTGCTCGTTTCATCATGAAATCCCACAAGT(−93)TGTGTAGGCTGGAGCTGCTTC |
| H13 | (−54)-GCATATGTAGTGCTCGTTTCATCATGAAATCCCACAAGTTAAAGTGTCT(−103)GGTGTAGGCTGGAGCTGCTTC |
| H13-1 | (−35)-CATAATATGTACCTCGTGAGCATATGTAGTGCTCGTTTCATCATGAAAT(−84)CGTGTAGGCTGGAGCTGCTTC |
| H13-2 | (−36)-ATAATATGTACCTCGTGAGCATATGTAGTGCTCGTTTCATCATGAAATC(−85)CGTGTAGGCTGGAGCTGCTTC |
| H13-3 | (−37)-TAATATGTACCTCGTGAGCATATGTAGTGCTCGTTTCATCATGAAATCCC(−86)GTGTAGGCTGGAGCTGCTTC |
Underlined sequences correspond to the flanking regions of the kanamycin resistance gene of pKD13 (7). Dotted sequences correspond to the sites of the icsB promoter which flank regions to be deleted. The numbers indicate the position relative to the icsB transcription start site at +1.
For elimination of the kanamycin resistance genes of the recombinants, kanamycin-resistant mutants were transformed with pCP20, a chloramphenicol resistance plasmid that exhibits temperature-sensitive replication and thermal induction of FLP (flipase) synthesis, which causes recombination between FRT sequences. Chloramphenicol-resistant transformants were selected at 30°C, after which a few were colony purified once nonselectively at 42°C and then tested for loss of all antibiotic resistances. The majority lost the FRT-flanked kanamycin resistance gene on pHW1273 and the FLP helper plasmid pCP20 simultaneously. The resultant plasmids were derivatives of pHW1273 with deletions of icsB upstream regions.
SDS-PAGE and immunoblotting analysis.
Whole-cell extracts of HW1273 and its derivatives (20 μl) were subjected to electrophoresis under denaturing conditions. Proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) and then electrophoretically transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 5% skim milk in phosphate-buffered saline containing 1% Tween 20 and then incubated with a 1:25,000 dilution of anti-IpaB serum (13). Membranes were incubated with secondary horseradish peroxidase-conjugated anti-mouse immunoglobulin antibodies at a dilution of 1:20,000 and developed with the ECL kit (Amersham, London, United Kingdom). The signals were detected by exposure to X-ray film (Fuji, Tokyo, Japan) at room temperature.
Overproduction and purification of GST-InvE fusion protein.
Plasmid pGEX-5X-3 (Amersham, London, United Kingdom) was used for expression and purification of InvE protein as a GST fusion protein (33). A DNA fragment precisely corresponding to the reading frame of invE, starting at the initiation codon and ending at the termination codon, was synthesized by PCR with the primers 5′-CGGGATCCCCATGGTGGATTTGTGC-3′ and 5′-GGCGAATTCCATCAGTGTTCGATGT-3′ with BamHI and EcoRI linkers at the respective regions. This fragment was then digested with BamHI and EcoRI and ligated into the corresponding sites of pGEX-5X-3. The resultant plasmid was designated pQW2001. MC1061 harboring pQW2001 was cultured to an optical density at 600 nm of approximately 0.5. After addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM to induce expression of the cloned fusion gene, culturing proceeded until an optical density at 600 nm of 1.0 was reached. Then the cells were harvested by centrifugation, suspended in the lysis buffer described above, frozen at −20°C, and thawed. After 3 min of sonication, soluble crude extract was obtained by centrifugation.
A 10-ml sample of this extract was absorbed into a 0.4-ml glutathione-Sepharose 4B gel (Pharmacia) previously equilibrated with phosphate-buffered saline. The gel was washed extensively with the above buffer, and bound proteins were then eluted twice with 0.2 ml of the elution buffer described above at 4°C for 1 h. Protein concentration was determined with a protein assay kit (Bio-Rad). The protein was stored at −70°C.
Gel mobility shift assay.
A DNA fragment carrying the regulatory region upstream of the icsB-ipa operon was used as the probe. The double-stranded DNAs used for probes were amplified by PCR or made from complementary synthetic oligonucleotides (ESPEC Oligo Service Co., Tokyo, Japan) and labeled with digoxigenin (DIG)-11-ddUTP (Roche Diagnostics) with 1 U of terminal deoxynucleotidyl transferase at 37°C for 30 min in a solution of 200 mM potassium cacodylate, 25 mM Tris-HCl (pH 6.6), 0.25 mg of bovine serum albumin per ml, and 5 mM CoCl2. After the labeling reaction, the probes were precipitated with ethanol, suspended in H2O, and stored at −20°C. Eight femtomoles of each DIG-labeled probe was incubated with the GST-InvE sample at 25°C for 30 min in a final volume of 20 μl containing 20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% (wt/vol) Tween 20, 30 mM KCl, 50 ng of poly(dI-dC) per μl, and 5 ng of poly-l-lysine per μl. After the addition of 5 μl of 0.25× Tris-borate-EDTA (TBE) containing 40% glycerol, incubated samples were run on a 6% polyacrylamide gel (79:1) in 0.25× TBE, followed by electroblotting onto a nylon membrane (Hybond-N+; Amersham, London, United Kingdom) and fixation by UV cross-linking. Detection of DNA fragments by anti-DIG Fab fragment-alkaline phosphatase conjugate (Roche Diagnostics) and substrate CSPD (Tropix Inc.) was performed as directed by the manual from the manufacturer (Roche Diagnostics). The signals were recorded by exposure to X-ray film (Fuji, Tokyo, Japan) at room temperature.
RESULTS
Determination of transcription start site and transcriptional regulation of icsB mRNA.
In a previous study, we reported that both InvE and VirF are required for expression of the ipaBCD genes encoded on the large plasmid of S. sonnei and suggested that the icsB-ipgABC-ipaBCD genes constitute an operon (ipa operon) (37). In this study, we analyzed the predicted location of the ipa operon promoter, upstream of icsB, the leadoff gene of the ipa operon.
First, to precisely confirm the location of the promoter element, we determined the transcription initiation site of the ipa operon. We analyzed the icsB transcript by primer extension mapping in order to determine the transcription start site of icsB. The template RNAs were extracted from cultures of MC1061/pHW735 and MC1061/pHW735/pHW749. Both strains were grown in LB at 37°C, and RNAs prepared from the cultures were used for the experiment with a primer complementary to the icsB mRNA at nucleotides 126 to 147 (Fig. 1). Figure 1 (lane 1) shows the result of mapping. Comparison of the band with a DNA sequencing ladder as a standard showed that the 5′ end of the mRNA corresponded to nucleotide T-49, 90 bp upstream of the translation initiation codon of icsB (Fig. 1, lane 2). Upstream of nucleotide T-49, the sequences TTATGT (−35 region) and TATGTT (−10 region) were found at nucleotides 10 to 15 and 36 to 41, respectively (Fig. 1), and the distance between these two sequences was 20 bp.
It should be pointed out that the promoter of the ipa operon has a −35 sequence and that the spacing between the −35 and −10 regions is also large (20 bp). These features perhaps justify the requirement for activators. This suggests that the extended band detected in this experiment corresponded to the transcription initiation site of icsB. The intensity of the band in the primer extension reaction with RNAs from the strain harboring pHW735 (neither virF or invE) was much lower than that of RNA from the strain harboring pHW735/pHW749 (containing both virF and invE), although the same amount of RNA was used for the reactions (compare lanes 1 and 2 in Fig. 1). This is consistent with the observation that expression of icsB is dependent on the presence of both virF and invE at the transcriptional level.
Construction of promoter deletion series for ipa operon.
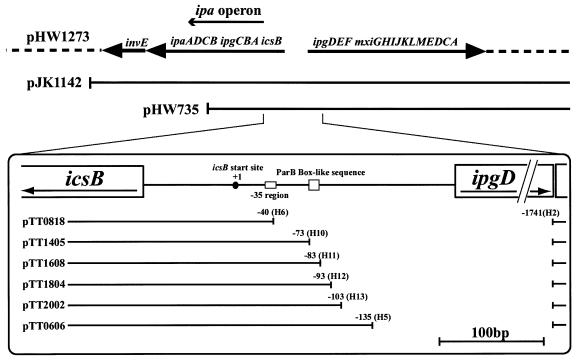
Sasakawa et al. reported that there is promoter activity within the 1.9-kb HindIII fragment containing the translation start site of icsB (31). However, the 1.9-kb HindIII sequence and VirF-InvE activators were not enough for full InvE-dependent activation. Then, in order to determine the essential cis-activating region upstream of the icsB gene, we used the large virulence plasmid pHW1273, which contains the whole region required for full expression of the ipa operon.
In order to define the promoter region that is necessary for ipa operon expression, we constructed mutants of the virulence plasmid of S. sonnei, pHW1273 to create a deletion series with λ Red recombinase as described in Materials and Methods. Strains carrying the parent plasmid pHW1273 and the deletion derivatives pTT0606, pTT0818, pTT1405, pTT1608, pTT1804, and pTT2002 (Fig. 2) in E. coli strain CC118 were designated HW1273, TT0606, TT0818, TT1405, TT1608, TT1804, and TT2002, respectively. For each strain, three independent transformants were chosen for PCR verification and sequencing. We confirmed the presence of the intact ipaB gene in all transformants by PCR. In each mutant, the deleted region was replaced by the nucleotides reported by Datsenko and Wanner (7).
FIG. 2.
Map around ipa operon of pHW1273. The line and dotted line indicate the genomic region of the plasmid. Deletion mutants of the pTT series were constructed as described in Materials and Methods by the use of the primers described in Table 2. Nucleotide +1 is the icsB transcription start site.
An 86-bp region upstream of the transcription start site of icsB is necessary for IpaB expression.
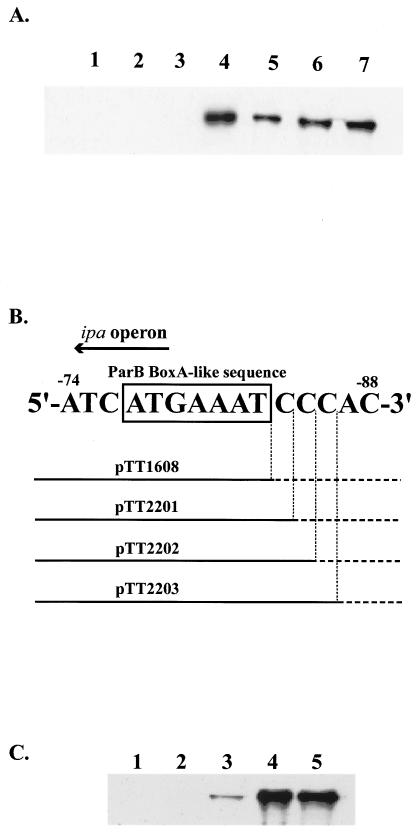
In order to determine the effect of each deletion on IpaB expression, we used immunoblotting analysis with antiserum specific to IpaB. HW1273 harboring pHW1273 expressed IpaB protein, whereas TT0818, in which the region from −1740 to −41 (icsB transcription start site designated +1) was deleted and which lacked the putative promoter region of the ipa operon, did not express IpaB (Fig. 3A, lanes 7 and 1). In Fig. 3A, IpaB expression by HW1273, which contains the entire upstream region of icsB, was considered maximum expression. Deletion of the region from −1740 to −136 (TT0606) retained IpaB expression at the same level as HW1273 (Fig. 3A, lane 6). These results showed that an important cis-acting region for IpaB expression was located between −136 and −41.
FIG. 3.
Effect of ipa operon promoter deletions on IpaB expression. (A) Whole-cell proteins of E. coli CC118 harboring the parent plasmid pHW1273 (lane 7) or its derivatives pTT0818 (lane 1), pTT1405 (lane 2), pTT1608 (lane 3), pTT1804 (lane 4), pTT2002 (lane 5), and pTT0606 (lane 6) were separated in a 12% polyacrylamide gel, immunoblotted, and detected with antiserum specific to IpaB. (B) Schematic diagram of pTT1608, pTT2201, pTT2202, and pTT2203 around the ParB BoxA-like sequence. At the top of this figure, the solid arrow indicates the direction of ipa operon transcription. The DNA sequence indicates the −88 to −74 region (icsB transcription start site as nucleotide +1), and the box indicates the position of the ParB BoxA-like sequence. Under the DNA sequence, thick horizontal lines indicate the predicted promoter region of the ipa operon in the derivatives, and horizontal dotted lines indicate the deleted regions. (C) Whole-cell proteins of E. coli CC118 harboring the parent plasmid pHW1273 (lane 5) or its derivatives pTT1608 (lane 1), pTT2201 (lane 2), pTT2202 (lane 3), and pTT2203 (lane 4) were separated in a 12% polyacrylamide gel, immunoblotted, and detected with antiserum specific to IpaB.
Mutant plasmids with further sequential deletions at 10-bp intervals between −136 and −41 were constructed with specially designed primers as described above. pTT1405, pTT1608, pTT1804, and pTT2002 lacked the nucleotides from −1740 to −74, −84, −94, and −104, respectively (Tables 1 and 2, Fig. 1). Deletion from −1740 to −84 (TT1608) reduced IpaB expression (Fig. 3A, lane 3). However, deletion from −1740 to −94 (TT1804) retained IpaB expression at almost the same level as HW1273 (Fig. 3A, lane 4).
Next, we constructed sequential deletion mutants of the 10-bp stretch between −94 and −84 with the above method. The mutant plasmids pTT2201, pTT2202, and pTT2203 lacked the nucleotides from −1740 to −85, −86, and −87, respectively (Fig. 3B). These deletion derivatives were used to transform E. coli strain CC118, creating strains designated TT2201, TT2202, and TT2203, respectively. Deletion from −1740 to −85 (TT2201) caused reduction of IpaB expression to an extent similar to that of TT1608 (Fig. 3C, lane 2). Deletion from −1740 to −86 (TT2202) resulted in IpaB expression at a very low level (Fig. 3C, lane 3). Deletion from −1740 to −87 (TT2203) resulted in IpaB expression at the same level as in HW1273 (Fig. 3C, lane 4). Based on these findings, we hypothesized that the intact promoter of the ipa operon is located in the 86-bp region upstream of the transcription start site of icsB. At the 3′ end of the region, we found the BoxA-like sequence 5′-ATGAAAT-3′ (−83 to −77), which is known as a repeated motif of the ParB binding site in the parS region of plasmid P1 (9). We named this sequence a ParB BoxA-like sequence. In conclusion, the ParB BoxA-like sequence and a further 3 bp (−86 to −84) were required for the full expression of IpaB. Next we examined whether InvE binding to the region was direct or not.
Direct binding of InvE to icsB upstream region.
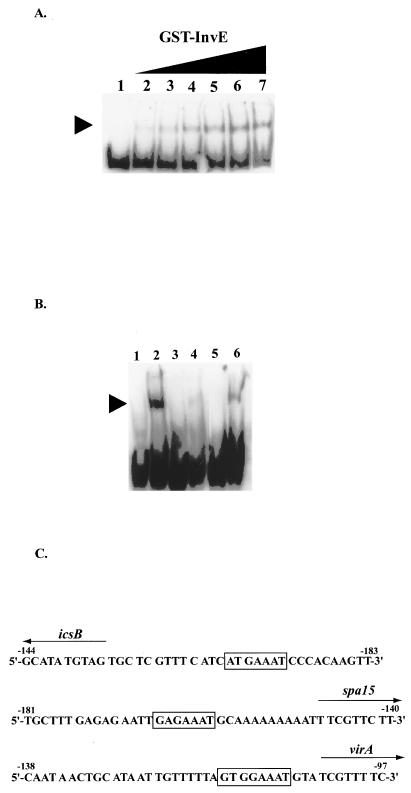
In order to clarify the mechanism by which InvE activates ipa operon expression, we determined whether the activation pathway is direct. First, we investigated the ability of InvE to bind to the DNA fragment corresponding to the icsB upstream region by gel shift assay. For this purpose, we purified GST-InvE as described in Materials and Methods. As shown in Fig. 4A, the specific PCR-amplified 150-bp DNA fragment, probe Q (from −143 to +7, with the icsB transcription start site designated nucleotide +1), was shifted upon addition of GST-InvE. The increased band shift was in proportion to the GST-InvE concentration. When the concentration of protein added to the reaction mixture was in the range of 0.3 to 9.6 μM, only one shifted band was observed, suggesting that one DNA-protein complex was formed. Also, there was no retarded band for GST protein, the negative control (data not shown). Therefore, we searched for an InvE binding site(s) within this 150-bp region.
FIG. 4.
Binding of GST-InvE to the putative ipa promoter region. (A) Eight femtomoles of probe Q (from −143 to + 7, with the icsB transcription start site designated nucleotide + 1) was synthesized by PCR, and the 3′ terminus was labeled with DIG-11-ddUTP. The binding reaction mixtures are described in Materials and Methods and were incubated at 25°C for 30 min. The amounts of GST-InvE were 0, 0.3, 0.6, 1.2, 2.4, 4.8, and 9.6 μM (lanes 1 to 7, respectively) in a final reaction volume of 20 μl. To the left of the gel, the arrowhead indicates the position of protein-DNA complexes containing GST-InvE and probe Q. (B) Each probe was synthesized by the manufacturer (ESPEC Oligo Service Co., Tokyo, Japan); probe I consisted of −93 to −54 (lanes 1 and 2), probe II consisted of the same sequence as probe I with an alteration of the ParB BoxA-like sequence from 5′-ATGAAAT-3′ to 5′-CGTCCCG-3′ (lanes 3 and 4), and probe III consisted of the same sequence as probe I with an alteration of the −63 to −54 sequence from 5′-GCATATGTAG-3′ to 5′-TACGCGTGCT-3′ (lanes 5 and 6). The icsB transcription start site is designated nucleotide + 1. Eight femtomoles of the probes at the 3′ termini were labeled with DIG-11-ddUTP. Probes were incubated at 25°C for 30 min in the reaction mixture described in Materials and Methods in the absence ofGST-InvE (lanes 1, 3, and 5) or the presence of 3.0 μM GST-InvE sample (lanes 2, 4, and 6) in a final reaction volume of 20 μl. To the left of the gel, the arrowhead indicates the position of a protein-DNA complex containing GST-InvE and probe I or III. (C) Comparison of the putative InvE binding regions in S. sonnei. Beloin et al. revealed that InvE (VirB) binds to the regions upstream of these genes (icsB, spa-15, and virA) specifically (4). The numbers indicate the positions of these regions (translation start site of each gene as nucleotide +1). The boxes indicate the position of the ParB BoxA-like sequence. The solid arrows indicate the direction of each gene transcription.
Based on the data from deletion mutants described above, 40-bp fragments (probe I, from −93 to −54; probe II, the same as probe I with altered sequences between −83 and −77; and probe III, the same as probe I with altered sequences between −63 and −54) were used as the target DNA in gel mobility shift experiments. The results of the assays with the GST-InvE sample and probes I, II, and III are shown in Fig. 4B. Probes I and III were clearly shifted when incubated with GST-InvE at a final concentration of 3.0 μM (Fig. 4B, lanes 2 and 6), but probe II with the altered ParB BoxA-like region sequences was shifted little under the same condition (Fig. 4B, lane 4). We also observed that the ParB BoxA-like sequence was conserved in the putative InvE binding regions upstream of the spa-15 and virA genes, which are regulated by InvE (Fig. 4C). These results indicate that the ParB BoxA-like sequence is necessary for InvE binding.
DISCUSSION
InvE plays a central role in controlling expression of the virulence genes in Shigella spp., but little is known about the mechanism of transcriptional regulation. A recent study indicated that InvE has a DNA-binding function and interacts specifically with the putative promoter of the ipa operon (4). However, it has not been shown genetically whether the InvE binding site is essential for activation of the ipa operon in vivo, and the minimal cis region in the ipa promoter has not been determined. In this study, we used virulence plasmid pHW1273 for determination of the cis region essential for InvE-dependent activation.
First, we determined the transcription start site of ics-ipa. The transcription start site was located 90 bp upstream of the icsB translation initiation site, and sequences characteristic of E. coli promoters for σ70 were found upstream of the transcriptional start site. The transcription of the mRNA was dependent upon InvE. These results suggest the presence of a cis region necessary for binding of InvE in addition to a conventional σ70 promoter.
To determine the cis region that is essential for ipa expression, we constructed a series of promoter deletions of virulence plasmid pHW1273 with a PCR-based gene disruption method and analyzed the IpaB expression of the mutants with anti-IpaB antibody. The minimal region essential for full expression of the ipaB gene contained a 7-nucleotide sequence. The sequence (5′-ATGAAAT-3′) was homologous to a BoxA motif in the parS site of plasmid P1, which is required for specific DNA binding of the ParB protein (25). Here we designated a ParB BoxA-like sequence. This finding is in good agreement with the fact that InvE protein has significant homology with ParB protein, especially the DNA binding domain (37). The deletion mutants lacking the ParB BoxA-like sequence and two nucleotides in its vicinity lost the expression of IpaB, while strains maintaining the intact ParB BoxA-like sequence with three cytosine nucleotides showed full expression of IpaB. These results indicated that the ParB BoxA-like sequence and three cytosine residues in the vicinity were essential for the expression of the ipa operon. A study with Shigella flexneri showed binding of VirB (InvE) to various promoter regions (4). These regions in S. sonnei also contain potential copies of the 7-bp ParB BoxA-like sequence [5′-(A/G)(A/T)G(G)AAAT-3′] (Fig. 4C). This would strengthen the prediction about the binding specificity of VirB (InvE). These results led us to examine direct binding of InvE protein to the promoter region by gel shift assay.
GST-InvE protein bound to a region between nucleotides −93 and −54 (DNA probe I in Fig. 4B) upstream of the icsB transcription start site at +1. This region contained the ParB BoxA-like sequence. When the sequences of the ParB BoxA-like region were altered, the ability of GST-InvE to bind to the probe was reduced (Fig. 4B, lane 4). This result again supports the hypothesis that InvE binding requires the ParB BoxA-like sequence in the ipa promoter region.
The identity of InvE and ParB is 42.8%, especially high in the predicted DNA binding motif (37). The ParB HTH region recognizes the BoxA sequence on the parS site (35). An HTH motif of InvE may also bind to the ParB BoxA-like sequence on the ipa promoter. However, nobody has shown whether ParB alone is sufficient for binding to the BoxA sequence, which seems to be the case with InvE. Further work is required to determine the possible cofactors that interact with InvE, which is competent for transcriptional activation of ipa genes. The present study provides a useful starting point for further study of transcriptional regulation by InvE.
Acknowledgments
This work was supported by grants from the Ministry of Health, Labor and Welfare and the Ministry of Education, Science and Technology.
REFERENCES
- 1.Adler, B., C. Sasakawa, N. Okada, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol. Microbiol. 7:59-1768. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, G. P., A. E. Hromockyj, C. Coker, and A. T. Maurelli. 1991. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect. Immun. 59:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333-15344. [DOI] [PubMed] [Google Scholar]
- 5.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 with PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677-684. [DOI] [PubMed] [Google Scholar]
- 9.Funnell, B. E., and L. Gagnier. 1993. The P1 plasmid partition complex at parS. II. Analysis of ParB protein binding activity and specificity. J. Biol. Chem. 268:3616-3624. [PubMed] [Google Scholar]
- 10.Hale, T. L., P. J. Sansonetti, P. A. Schad, S. Austin, and S. B. Formal. 1983. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect. Immun. 40:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hale, T. L., E. V. Oaks, and S. B. Formal. 1985. Identification and antigenic characterization of virulence-associated, plasmid-coded protein of Shigella spp. and enteroinvasive Escherichia coli. Infect. Immun. 50:620-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale, T. L. 1991. Genetic basis of virulence in Shigella species. Microbiol. Rev. 55:206-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, K., T. Nakajima, T. Sasaki, and H. Watanabe. 1991. Localization of IpaB protein in Escherichia coli K-12 MC1061 strain carrying Shigella sonnei form I plasmid pSS120. Microbiol. Immunol. 35:335-341. [DOI] [PubMed] [Google Scholar]
- 14.Kato, J., K. Ito, A. Nakamura, and H. Watanabe. 1989. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect. Immun. 57:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong, S. M., C. C. Yeo, and C. L. Poh. 2001. Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site parS. Mol. Microbiol. 40:621-633. [DOI] [PubMed] [Google Scholar]
- 16.Makino, S., C. Sasakawa, K. Kamata, and M. Yoshikawa. 1986. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in Shigella flexneri 2a. Cell 46:551-555. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurelli, A., T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurelli, A., T., B. Baundry, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequence involved in invasion HeLa cells by Shigella flexneri 2a. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menard, R., P. J. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partition of IpaB and IpaC invasins of Shigella flexneri. Cell 79:515-535. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Parsot, C. 1994. Shigella flexneri: genetics of entry and intercellular dissemination in epithelial cells. Curr. Top. Microbiol. Immunol. 192:217-241. [DOI] [PubMed] [Google Scholar]
- 25.Radnedge, L., M. A. Davis, and S. J. Austin. 1996. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 15:1155-1162. [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai, T., C. Sasakawa, and M. Yoshikawa. 1988. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30kDa virF protein. Mol. Microbiol. 2:589-597. [DOI] [PubMed] [Google Scholar]
- 27.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasakawa, C., K. Kamata, T. Tobe, T. Suzuki, and M. Yoshikawa. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasakawa, C., J. M. Buysse, and H. Watanabe. 1992. The large virulence plasmid of Shigella. Curr. Top. Microbiol. Immunol. 180:21-44. [DOI] [PubMed] [Google Scholar]
- 31.Sasakawa, C., K. Komatsu, T. Tobe, T. Suzuki, and M. Yoshikawa. 1993. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J. Bacteriol. 175:2334-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma, R. C., and R. T. Schimke. 1996. Preparation of electrocompetent E. coli with salt-free growth medium. BioTechniques 20:42-44. [DOI] [PubMed] [Google Scholar]
- 33.Smith, D. B., and K. S. Johnson. 1988. Single step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 34.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3-lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surtees, J. A., and B. E. Funnell. 2001. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276:12385-12394. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesan, M., J. M. Buysse, and D. J. Kopecko. 1988. Characterization of invasion plasmid antigen (ipaBCD) gene from Shigella flexneri: DNA sequence analysis and control of gene expression. Proc. Natl. Acad. Sci. USA 85:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe, H., E. Arakawa, K.-I. Ito, J.-I. Kato, and A. Nakamura. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of InvE with ParB of plasmid P1. J. Bacteriol. 172:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zychlinsky, A., B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti. 1994. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol. 11:619-627. [DOI] [PubMed] [Google Scholar]