Abstract
The integron-borne blaVEB-1 gene encodes an extended-spectrum β-lactamase. This gene was associated mostly with IS1999 and rarely with an additional IS2000 element in Pseudomonas aeruginosa isolates from Thailand, whereas IS1999 was only very rarely associated with blaVEB-1 in Enterobacteriaceae. Expression experiments and promoter study identified promoter sequences in IS1999 that increased the expression of VEB-1 in P. aeruginosa.
Antibiotic resistance in many gram-negative clinical isolates is due to resistance genes that are captured and expressed in class 1 integrons. These integrons possess two conserved regions located on each side of integrated gene cassettes (14). The 5′ conserved segment (5′-CS) includes a gene encoding an integrase, intI1, the cassette integration site, attI1, and the promoter Pant, sometimes associated with a second promoter P2, which are responsible for the expression of gene cassettes (6, 14). The 3′ conserved segment (3′-CS) includes, along with another open reading frame of unknown function, the disinfectant (qacEΔ1) and the sulfonamide (sul1) resistance determinants (14). The expression of the inserted gene cassettes depends not only on sequences of promoters Pant/P2 but also on the gene cassette position relative to the 5′-CS (2).
The integron-borne blaVEB-1 gene encodes the extended-spectrum β-lactamase VEB-1 (Vietnamese extended-spectrum β-lactamase) found initially in an Escherichia coli clinical isolate from Vietnam (13). Subsequently, the veb-1 gene cassette was identified in two Pseudomonas aeruginosa clinical isolates from Thailand (9, 18). In those strains, blaVEB-1 was associated with either one (IS1999) or two (IS1999/IS2000) insertion sequences (IS) that were inserted upstream of blaVEB-1 in the integron-specific recombination site, attI1. Bacterial IS may bring promoters located in or near their inverted-repeat sequences (IR) that are capable of modulating the expression of neighboring antibiotic resistance genes (7).
Previous studies performed with ceftazidime-resistant Enterobacteriaceae and P. aeruginosa strains isolated in 1999 from the Siriraj Hospital, Bangkok, Thailand (3, 4), resulted in reports that out of 37 enterobacterial isolates, 18 were blaVEB-1 positive (10 E. coli, 4 Enterobacter cloacae, 1 Enterobacter sakazakii, and 3 Klebsiella pneumoniae) and 19 were blaVEB-1 negative. Out of 33 ceftazidime-resistant P. aeruginosa isolates, 31 were blaVEB-1 positive. blaVEB-1 was mostly plasmid located in Enterobacteriaceae, whereas this gene was mostly chromosome encoded in P. aeruginosa (3, 4). Moreover, spreading of blaVEB-1-containing P. aeruginosa strains was detected in several unrelated isolates carrying different integrons of various sizes and structures (3, 4).
Distribution of IS1999 and IS2000 in blaVEB-1-positive isolates.
The distribution of IS1999, an IS10-like element, and IS2000, which belongs to the IS5 family (7, 9), was investigated using the same blaVEB-1-positive isolates (Table 1). Dot blot hybridizations were performed using whole-cell DNAs (12, 15) of the 18 Enterobacteriaceae- and the 31 P. aeruginosa-positive isolates (Table 1). Using ECL nonradioactive labeling and detection kits (Amersham Biosciences, Orsay, France), hybridizations were performed under high-stringency conditions. The probes consisted of PCR-generated fragments internal to IS1999 and IS2000 (15) (primer sequences are available upon request).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or isolate(s) or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains or isolates | ||
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG | Life Technologies, Eragny, France |
| P. aeruginosa | ||
| KG2505 | P. aeruginosa reference strain that does not express the naturally chromosome-encoded AmpC β-lactamase and is deficient for the multidrug efflux system MexAB-OprM mexA::res-ΩSm ampC::res-ΩSm of PAO1; Smr | 11 |
| Isolate JES | Clinical isolates from a French patient hospitalized in Thailand; blaVEB-1 positive; Cazr | 9 |
| Isolates 1 to 31 | Clinical isolates from the Siriraj Hospital, Bangkok, Thailand; blaVEB-1 positive; Cazr | 3 |
| E. coli | ||
| Isolates 1 to 10 | Clinical isolates from the Siriraj Hospital, Bangkok, Thailand; blaVEB-1 positive; Cazr | 4 |
| E. cloacae | ||
| Isolates 11 to 14 | Clinical isolates from the Siriraj Hospital, Bangkok, Thailand; blaVEB-1 positive; Cazr | 4 |
| E. sakazakii | ||
| Isolate 15 | Clinical isolates from the Siriraj Hospital, Bangkok, Thailand; blaVEB-1 positive; Cazr | 4 |
| K. pneumoniae | ||
| Isolates 16 to 18 | Clinical isolates from the Siriraj Hospital, Bangkok, Thailand; blaVEB-1 positive; Cazr | 4 |
| Plasmids | ||
| pBK-CMV phagemid | Neor, Kanr | Stratagene, La Jolla, Calif. |
| pCR-Blunt II-TOPO | Kanr | Invitrogen, Cergy Pontoise, France |
| pBBR1MCS.3 | Low-copy-number broad-host-range cloning vector; Tetr | 5 |
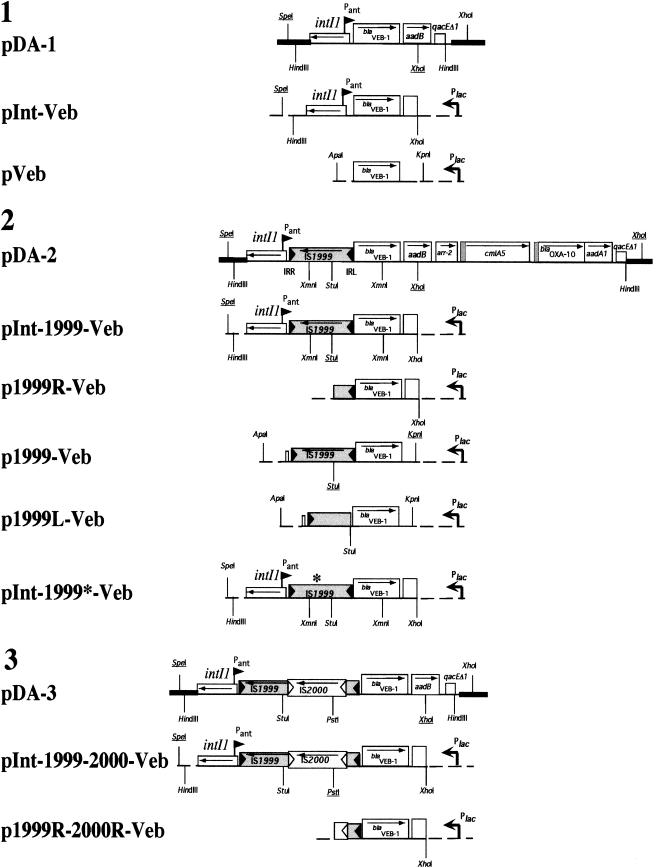
| pDA-1 | pBK-CMV recombinant plasmid containing genomic HindIII fragment from P. aeruginosa 14, including blaVEB-1 | This work |
| pDA-2 | pBK-CMV recombinant plasmid containing genomic HindIII fragment from P. aeruginosa 1, including blaVEB-1 and IS1999 | This work |
| pDA-3 | pBK-CMV recombinant plasmid containing genomic HindIII fragment from P. aeruginosa JES, including blaVEB-1 and IS1999/IS2000 | This work |
| pInt-Veb | A 2.4-kb SpeI-XhoI fragment from plasmid pDA-1, containing | This work |
| blaVEB-1 inserted in the SpeI-XhoI site of pBBR1MCS.3 | ||
| pInt-1999-Veb | A 3.7-kb SpeI-XhoI fragment from pDA-2, containing blaVEB-1 and IS1999 inserted in the SpeI-XhoI site of pBBR1MCS.3 | This work |
| pInt-1999-2000-Veb | A 4.9-kb SpeI-XhoI fragment from pDA-3, containing blaVEB-1 and IS1999/IS2000 inserted in the SpeI-XhoI site of pBBR1MCS.3 | This work |
| pTOP1 | A 1.1-kb fragment resulting from PCR amplifications using VEBCAS-A/VEBCAS-B primers and genomic DNA of P. aeruginosa 1 as template; this fragment, including the veb-1 gene cassette, was inserted in the cloning site of pCR-BluntII-TOPO | This work |
| pTOP2 | A 2.5-kb fragment resulting from PCR amplifications using 5′-CS/VEBCAS-B primers and genomic DNA of P. aeruginosa 1 as the template; this fragment, including the veb-1 gene cassette and IS1999, was inserted in the cloning site of pCR-BluntII-TOPO | This work |
| pTOP3 | A 1.1-kb fragment resulting from PCR amplifications using VEBCAS-AStu/VEBCAS-B primers and genomic DNA of P. aeruginosa 1 as the template; this fragment, including a StuI restriction site in the veb-1 gene cassette upstream of blaVEB-1, was inserted in the cloning site of pCR-BluntII-TOPO | This work |
| pTOP4 | A 2.2-kb fragment resulting from PCR amplifications using MUTIS1999Stop/VEBCAS-B primers and genomic DNA of P. aeruginosa 1 as the template; this fragment includes a site-directed mutation generating after transcription a stopcodon within the IS1999 transposase sequence; this fragment, inserted in the cloning site of pCR-BluntII-TOPO, includes the two XmnI sites that are present in IS1999 and blaVEB-1 sequences and that are located on each side of the mutation | This work |
| pVeb | A 1.1-kb KpnI-ApaI fragment from pTOP1, containing the veb-1 gene cassette inserted in the KpnI-ApaI site of pBBR1MCS.3 | This work |
| p1999R-Veb | pInt-1999-Veb from which a 1.9-kb SpeI-StuI fragment has been deleted that includes intII and the left end of IS1999 | This work |
| p1999-Veb | A 2.6-kb KpnI-ApaI fragment from pTOP1, containing the veb-1 gene cassette and IS1999, inserted in the KpnI-ApaI site of pBBR1MCS.3 | This work |
| p1999L-Veb | A 2.1-kb KpnI-StuI fragment from pTOP3, containing the veb-1 gene cassette, inserted in the KpnI-StuI site of p1999-Veb, containing the left end of IS1999 | This work |
| pInt-1999*-Veb | A 1.5-kb XmnI fragment from pTOP4, containing parts of the mutated IS1999 and blaVEB-1 sequences inserted in the XmnI site of pInt-1999-Veb | This work |
| p1999R-2000R-Veb | pInt-1999-2000-Veb from which a 1.8 kb SpeI-PstI fragment has been deleted that includes intII and the right ends of IS1999/IS2000 | This work |
Superscript “r,” resistance. Abbreviations: Caz, ceftazidime; Kan, kanamycin; Neo, neomycin; Sm, streptomycin; Tet, tetracycline.
Out of 18 blaVEB-1-positive enterobacterial isolates, none possessed IS2000 and only one K. pneumoniae isolate carried IS1999. In contrast, IS1999 was found in 28 out of 31 (90%) blaVEB-1-containing P. aeruginosa isolates and 2 out of these 28 isolates had an additional IS2000 inserted within the IS1999 sequence.
Thus, the frequent association mostly of IS1999, which is sometimes associated with IS2000, with the blaVEB-1 gene in P. aeruginosa and its absence from Enterobacteriaceae, especially from E. coli, led us to study the contribution of these IS on β-lactamase expression in both bacterial species.
Influence of IS1999/IS2000 on blaVEB-1 expression.
Using three blaVEB-1-positive P. aeruginosa isolates as templates (P. aeruginosa 14, 1, and JES), recombinant plasmids were constructed containing blaVEB-1 without any IS (pDA-1), with IS1999 (pDA-2), and with IS1999 and IS2000 (pDA-3) (Fig. 1 and Table 1). Plasmids were constructed by standard recombinant techniques (15). The low-copy-number and broad-host-range cloning vector pBBR1MCS.3 that replicates in E. coli and in P. aeruginosa (5) was used for subcloning experiments, generating recombinant plasmids pInt-Veb, pInt-1999-Veb, and pInt-1999-2000-Veb (Fig. 1 and Table 1). Recombinant plasmids were introduced by electroporation into E. coli DH10B (12) and P. aeruginosa KG2505 (11, 17). P. aeruginosa KG2505 does not express the naturally chromosome-encoded AmpC β-lactamase and is deficient for the multidrug efflux system MexAB-OprM (11).
FIG.1.
Schematic map of the constructs used in this study. Constructs 1 (pDA-1, pInt-Veb, and pVeb), 2 (pDA-2, pInt-1999-Veb, pInt-1999*-Veb, p1999R-Veb, p1999-Veb, and p1999L-Veb), and 3 (pDA-3, pInt-1999-2000-Veb, and p1999R-2000R-Veb) were cloned from genomic DNAs of P. aeruginosa clinical isolates 14, 1, and JES, respectively. The blaVEB-1 gene was inserted in opposite orientation to Plac, thus removing any contribution of promoter Plac in β-lactamase expression. The stop codon resulting from a site-directed mutagenesis experiment is shown by an asterisk (construct pInt-1999*-Veb). Restriction sites that were used at each cloning step are underlined. The coding regions are shown as boxes, with an arrow indicating the orientation of their transcription. The IR of IS1999 and IS2000 are shown by filled and empty triangles, respectively. IRL and IRR of IS1999 are indicated for pDA-2. The broken arrows indicate the promoter Plac. Thin dashed lines indicate ligation in the multiple-cloning site of the shuttle vector pBBR1MCS.3.
Sequencing was performed using laboratory-designed primers on an ABI PRISM 3100 automated sequencer (Applied Biosystems, Les Ullis, France). Sequence analysis of inserts of all the recombinant plasmids revealed the presence of the two previously characterized promoters: (i) the weak Pant promoter and (ii) the P2 promoter that is likely nonfunctional (as previously shown) due to its reduced spacing between the −10 and −35 promoter sequences (14 bp instead of 17 bp) (2, 6).
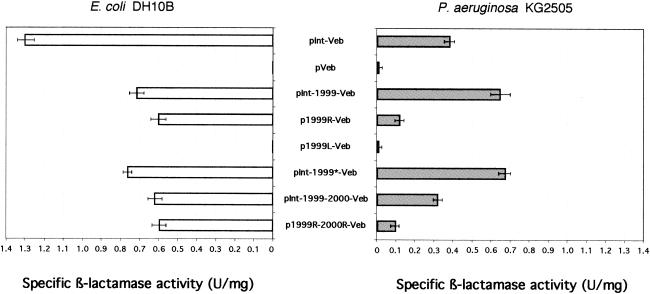
Using β-lactamase extracts from cultures of E. coli DH10B and P. aeruginosa KG2505 harboring recombinant plasmids (Fig. 2) prepared as previously described (10), blaVEB-1 gene expression was then investigated by measuring the specific β-lactamase activities for cefepime (a β-lactam antibiotic hydrolyzed specifically by VEB-1). Total protein contents and the initial rate of cefepime hydrolysis were determined as previously described (13).
FIG. 2.
Comparative study (using cefepime as the substrate) of the specific activity of β-lactamase VEB-1 from cultures of E. coli DH10B (left panel) and P. aeruginosa KG2505 (right panel) strains harboring recombinant plasmids. Error bars represent standard deviations calculated from five independent cultures. No measurable activity was detected for E. coli DH10B(pVEB) and for E. coli DH10B(p1999L-VEB).
To determine whether IS1999 and IS2000 carry active promoter sequences located in their right ends, Pant was deleted from pInt-Veb, pInt-1999-Veb, and pInt-1999-2000-Veb, generating recombinant plasmids pVeb, p1999R-Veb, and p1999R-2000R-Veb, respectively (Fig. 1 and Table 1).
In E. coli, the highest β-lactamase activity was measured for E. coli DH10B(pInt-Veb), which had blaVEB-1 located just downstream of the promoter Pant of the class 1 integron (Fig. 1 and 2). Insertion of IS1999 (pInt-1999-Veb), which moved blaVEB-1 away from Pant, decreased blaVEB-1 expression (50% decrease). After the removal of the promoter Pant (p1999R-Veb and p1999R-2000R-Veb), significant activity (45%) was still measured, suggesting the presence of an IS1999-located promoter. Similar specific activities were obtained for E. coli DH10B(pInt-1999-Veb), (p1999R-Veb), (pInt-1999-2000-Veb), and (p1999R-2000R-Veb), suggesting that Pant made only a minor contribution to the overall β-lactamase expression. Thus, the activity measured in E. coli DH10B(pInt-1999-Veb) and (pInt-1999-2000-Veb) is mostly due to the presence of an IS1999-located promoter (and possibly to the presence of IS2000).
Determination of specific activity of P. aeruginosa KG2505 cultures showed that surprisingly, the highest activity was measured for P. aeruginosa KG2505(pInt-1999-Veb) [a 60% increase compared to the activity of cultures of P. aeruginosa KG2505pInt-Veb)] (Fig. 2). However, insertion of IS2000 (pInt-1999-2000-Veb) or deletion of the Pant promoter along with the left end(s) of IS1999 and/or IS2000 (p1999R-Veb and p1999R-2000R-Veb) led to a decrease in β-lactamase activity. Thus, the right end of IS1999, which includes the left IR (IRL), most likely carried a functional outward-directed promoter capable of driving blaVEB-1 transcription in P. aeruginosa.
To determine whether the left end of IS1999 carried additional promoter sequences, plasmid p1999L-Veb was constructed. This plasmid contained two-thirds of the IS1999 sequence, including the right IR (IRR) located upstream of blaVEB-1 (Fig. 1). Strains harboring p1999L-Veb plasmid had very low levels of β-lactamase activity similar to those observed for strains harboring pVeb, suggesting the absence of functional promoter sequences in the left end of IS1999.
To determine whether the functional transposase of IS1999 can influence blaVEB-1 expression, the plasmid pInt-1999*-Veb, which contains an interrupted open reading frame encoding the IS1999 transposase, was generated. No significant modification of blaVEB-1 expression was measured for strains harboring pInt-1999*-Veb compared to those harboring pInt-1999-Veb (Fig. 2). Thus, the transposase had no or little effect on β-lactamase expression in E. coli and P. aeruginosa.
Mapping of promoter Pout of IS1999.
The precise location of the right-end-located promoter of IS1999 was determined with a primer extension system-AMV reverse transcriptase kit (Promega, Charbonnières, France). Total RNAs were extracted with a Qiagen RNeasy Maxi kit (Qiagen, Courtaboeuf, France). cDNAs were synthesized using 32P end-labeled primers Vebprom and Vebprom 1999, which annealed to the left end of veb-1 gene cassette and to the right end of IS1999. Using a Sequenase version 2 DNA sequencing kit (Amersham Biosciences), manual sequencing was performed with the same primers. Sequencing and extension products were separated on an 8% polyacrylamide gel and were visualized by autoradiography after overnight exposure at −80°C.
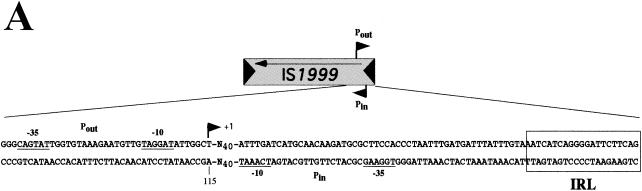
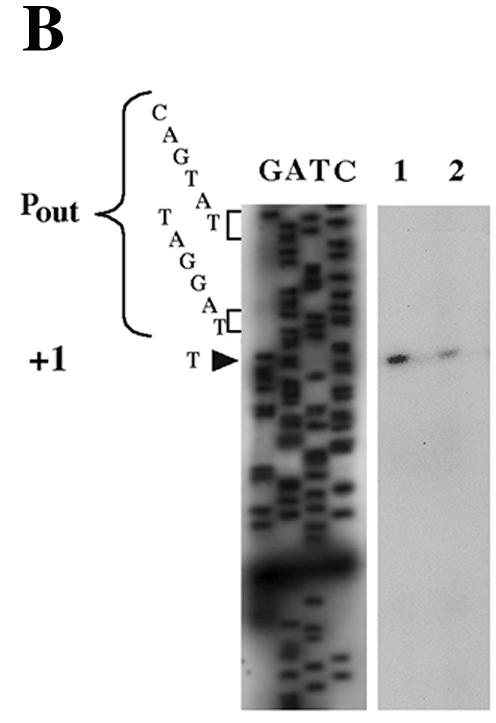
Primer extension experiments (performed with Vebprom primer and RNAs from E. coli DH10B and P. aeruginosa KG2505 strains containing recombinant plasmids pInt-1999-Veb and pInt-1999-2000-Veb) generated a cDNA starting at a thymidine at nucleotide position 115 (Fig. 3). Analysis of the sequence located upstream of bp 115 revealed a putative −35 promoter region (CAGTAT) separated by 17 bp from a −10 region (TAGGAT) (Fig. 3). This promoter was located close to the IRL of IS1999 at a position similar to that of the promoter Pout (−35 [CAGAAT] and −10 [TAAAAT]) identified in the related IS element, IS10 (16). Using the primer Vebprom 1999, which annealed upstream of Pout, no extension product was identified, indicating that no promoter sequence was located further inside of IS1999 or in IS2000.
FIG. 3.
(A) Structure of IS1999. The IS1999 IR are shown by filled triangles. The arrows indicate the orientation of transcription. The outward-directed promoter, Pout, and the promoter of the transposase gene, Pin, are indicated by broken arrows. The −10 and −35 regions for Pin and Pout are underlined. Nucleotide position 115 (according to the sequence GenBank AF133697) in IS1999 corresponds to the transcription start of Pout. The IRL sequence is boxed. (B) Mapping of transcription initiation. Primer Vebprom was extended using RNAs from cultures of E. coli DH10B(pInt-1999-Veb) (lane 1) or P. aeruginosa KG2505(pInt-1999-Veb) (lane 2) as the templates. Equal volumes (2 μl) of the extension product obtained from P. aeruginosa and E. coli were loaded onto the gel. Size markers were from sequencing reactions generated from pInt-1999-Veb DNA primed with Vebprom. G-, A-, T-, and C-specific lanes are indicated. The nucleotide sequences on the left side correspond to that of the complementary strand, which was deduced from the sequencing reaction. The −10 and −35 promoter sequences of Pout regions are shown, and the +1 transcriptional initiation site is indicated by an arrowhead. Similar results were obtained for RNA extracted from E. coli DH10B and P. aeruginosa KG2505 harboring pInt-1999-2000-Veb recombinant plasmid (data not shown).
Conclusions.
This work identified a functional outward-directed promoter, Pout, of IS1999 that was capable of driving blaVEB-1 expression in E. coli and in P. aeruginosa. The level of expression obtained from Pant or Pout was about fourfold higher in E. coli than in P. aeruginosa. Furthermore, our results suggest that an association between IS1999 and the Pant promoter enhances blaVEB-1 expression by about 60% in P. aeruginosa but not in E. coli. An increase in β-lactamase expression may change bacteria from being susceptible to being intermediate or even resistant. This is the case for P. aeruginosa harboring plasmid pInt-1999-Veb, for which certain β-lactams, such as piperacillin and cefepime, that are extensively prescribed in hospitals display a twofold increase in MICs (data not shown). Thus, in a hospital environment, where bacteria may be under constant antibiotic pressure, IS1999 (by enhancing the blaVEB-1 gene expression) might bring a selective advantage to P. aeruginosa, at least when it is present on a low-copy-number plasmid (pBBR1MCS). Future experiments will be directed towards determination of blaVEB-1 gene expression in its native environment, i.e., the chromosome of P. aeruginosa isolates, to see whether our plasmid-mediated system mimics the chromosomal situation.
An increase of blaVEB-1 expression in P. aeruginosa KG2505 (pInt-1999-Veb) might be the result of a cooperative effect between Pout and Pant promoters. Indeed, the maximum amount of expression was obtained only when both promoters were present. In E. coli(pInt-1999-Veb), however, both promoters were present and still the expression decreased compared to that seen with E. coli(pInt-Veb). These data may reflect major differences in transcriptional properties between P. aeruginosa and E. coli. The role of IS2000 in the blaVEB-1-containing integron remains unclear, but decrease of blaVEB-1 expression after IS2000 insertion into IS1999 argues against its role in β-lactamase expression.
Most of the genes inserted in class 1 integrons are expressed from a common promoter region (Pant/P2). In a few cases, however, other promoters of the expression of gene cassettes have been reported (1, 8). This work identified for the first time an IS-located promoter capable of driving expression of downstream-located gene cassettes in an integron structure.
Acknowledgments
We thank N. Gotoh for providing P. aeruginosa strain KG2505.
This work was funded by a grant from the Ministère de la Recherche (grant UPRES-EA 3539), Université Paris XI, Paris, France.
REFERENCES
- 1.Bissonnette, L., S. Champetier, J.-P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 4.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop 2nd, and K. M. Peterson. 1994. PBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 6.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 7.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naas, T., Y. Mikami, T. Imae, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 10.Naas, T., W. Sougakoff, A. Casetta, and P. Nordmann. 1998. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto, K., N. Gotoh, and T. Nishino. 2001. Pseudomonas aeruginosa reveals high intrinsic resistance to penem antibiotics: penem resistance mechanisms and their interplay. Antimicrob. Agents Chemother. 45:1964-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile elements. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Simons, R. W., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1983. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell 34:673-682. [DOI] [PubMed] [Google Scholar]
- 17.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]