Abstract
The Rgg family of transcription regulators is widely distributed among gram-positive bacteria; however, how the members of this family control transcription is poorly understood. In the pathogen Streptococcus pyogenes, the Rgg family member RopB is required for transcription of the gene that encodes the secreted SpeB cysteine protease. Expression of the protease follows distinct kinetics that involves control of transcription in response to the growth phase. In this study, the contribution of RopB to growth phase control was examined. The gene encoding the protease (speB) and ropB are transcribed divergently from a 940-bp intergenic region. Primer extension analyses, in conjunction with reporter fusion studies, revealed that the major region controlling the transcription of both speB and ropB is adjacent to ropB and that the promoters for the two genes likely overlap. Furthermore, it was found that RopB is a DNA-binding protein that specifically binds to sequences in this control region. The interrelationship between ropB and speB expression was further reflected in the observation that transcription of ropB itself is subject to growth phase control. However, while expression of ropB from a promoter expressed during the early logarithmic phase of growth could complement a ropB deletion mutant, ectopic expression of ropB did not uncouple the expression of speB from its growth phase signal. These data implicate other factors in growth phase control and suggest that regulation of ropB expression itself is not the central mechanism of control.
The pathogenesis of a bacterial infection is a multistage process characterized by adaptive responses on the part of both the pathogen and the host. For the pathogen, transmission to a new host, specific interaction with tissues at the initial site of infection, and growth in the initial host compartment, often followed by dissemination to other host tissues, all represent stages that require the expression of distinct subsets of the pathogen's genome. The host's adaptive response involves sensing of the pathogen and then modification of the site of infection to produce a harsh environment that is not conducive to microbial growth. Successful pathogens have evolved sensory programs to recognize the host's response in order to mount an appropriate adaptive response. Thus, it is not surprising that the expression of most bacterial virulence genes is highly regulated and that mutations in these genes generally result in an attenuated ability to cause infection.
Virulence regulatory genes have been most thoroughly studied among the gram-negative bacteria, where it has become apparent that many are members of several large conserved families of proteins that modulate transcription. Each of these families is characterized by a common structure and mechanism of action that have been adapted to individual regulatory programs. An understanding of the general paradigm by which a particular family functions often provides crucial insight into how any individual member functions in the regulation of virulence. Much less is known about virulence regulatory networks in gram-positive bacteria. However, the recent influx of genomic information has revealed large conserved families of regulators that are unique to the gram-positive pathogens. One example is the family of regulators that share homology with the prototype member rgg of the oral bacterium Streptococcus gordonii (38). Members of the rgg-like family have been identified in the genomes of S. pyogenes (8, 27), S. pneumoniae (40), S. agalactiae (17), S. mutans (32), S. oralis (15), S. sanguis (42), S. equi (M. N. Neely and M. Caparon, unpublished data), and Listeria monocytogenes (16). Some genomes, including those of S. pyogenes (13, 27) and S. gordonii (25, 41), contain multiple family members, and rgg-like genes have also been found in nonpathogens as well, including Lactococcus lactis (35) and Lactobacillus sakei (33). How the members of this extensive family function to regulate gene expression is not understood. For example, while the Rgg protein of S. gordonii has been shown to associate with DNA (43), it has not been clearly established that any member of this extensive family of proteins binds specifically to DNA to regulate transcription.
In S. pyogenes, one of the major secreted proteins during the transition from the logarithmic to the stationary phase of growth in vitro is the SpeB cysteine protease. This organism is the cause of many different suppurative and nonsuppurative diseases, including pharyngitis (strep throat), cellulitis, necrotizing fasciitis, and rheumatic fever (2). The contribution of SpeB to the pathogenesis of any of these diseases is not well understood (1, 3, 19, 23, 24, 26, 39). However, it is clear that expression of the gene that encodes the protease (speB) is regulated by the rgg family member ropB (also known as rgg) (8, 27). Consistent with other rgg family members, ropB is located adjacent to speB in the S. pyogenes chromosome (27), transcription of speB has an absolute dependence on ropB, and speB encodes a protein that is secreted from the bacterial cell. Proteomic profiling has revealed that ropB regulates the expression of several other proteins that are preferentially secreted from cells in the stationary phase of growth, including acting as a repressor of the mitogenic factor DNase (11). Transcription profiling has suggested that ropB may have a much greater influence over expression of the proteome via its ability to influence the expression of a number of unrelated virulence regulatory programs (10). The importance of ropB is further illustrated by the fact that a ropB mutant has a reduced ability to cause disease in an animal model of streptococcal myositis (29).
Similar to other rgg family members, little is known about how ropB functions to regulate the growth phase pattern of transcription of speB or any of its other target genes. In the present study, we characterized the intergenic region between ropB and speB to identify elements involved in transcriptional control and showed that RopB binds to DNA in the vicinity of the major speB promoter. This study also revealed that transcription of ropB itself is regulated in a growth phase-dependent manner that resembles that of speB. However, ectopic expression of ropB during the logarithmic phase of growth does not alter the expression pattern of speB. Thus, control of ropB transcription does not represent the major determinant for control of the speB regulatory program. These data suggest that additional factors must interact with ropB in order to control secreted protease production in S. pyogenes.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Streptococcus experiments were done with derivatives of S. pyogenes HSC5 (20) cultured as described previously (20). When appropriate, antibiotics were added at the following concentrations: kanamycin at 25 μg/ml for Escherichia coli and 500 μg/ml for S. pyogenes, erythromycin at 750 μg/ml for E. coli and 1 μg/ml for S. pyogenes, chloramphenicol at 15 μg/ml for E. coli and 3 μg/ml for S. pyogenes, and ampicillin at 50 μg/ml for E. coli.
Northern blot analysis.
RNA samples were prepared from cultures harvested at the onset of stationary phase (approximately 6 h) as described elsewhere (27), and 10-μg samples were subjected to Northern blot analysis as described previously (27). The probe used for the speB transcript consisted of a 550-bp DNA fragment that was amplified by PCR from HSC5 chromosomal DNA with primers speB1850for and speB2400rev (Table 1) as described elsewhere (27). Relative levels of the speB transcripts were determined by comparison to a probe that hybridized to the transcript for recA, similar to that described previously (27).
TABLE 1.
Primers used in this study
| Name | Sequencea |
|---|---|
| speB-20primerext | CTGCATTTGCTTTTAACGCTAC |
| speB-600primerext | GTTTGTCAATGCTTTTATTGAC |
| ropB intergene | GGAAGACTGTCTGACAGAGCGG |
| ropB-stop-BamHI | CGCGGATCCCCTCAGGACAGTTTATGTTTAATGG |
| speB-3′-fwd | GGCGATTTCAGAATTGATGGCTG |
| speB-3′-rev | GCCACCAGTACCAAGAGCTGAAGGG |
| glnQ-5′ | GGAATTGATGCCAAATTCTACGAAGGTG |
| glnQ-3′ | GATAACACGGTTAGCTACTTGGCGAGC |
| pSPE-speB+ 180EcoRI | ATATCTTCTGGAATTCGTCAACCTGCTTTG |
| pSPE10-speB-1000BamHI | GAATGCCTAATGGATCCAACGGTTTCACAA |
| pSPE11-speB-800BamHI | GTCATATGTGGATCCTTTCTAATC |
| pSPE12-speB-700BamHI | GCAGCTATGAAGGATCCATAAGGTTAAAAGGAAG |
| pSPE13-speB-400BamHI | GGTGTTGTGGATCCTATCACGATC |
| pSPE14-speB-250BamHI | GATGATAAGTGGATCCACTCATAGCGTC |
| pSPE15-speB-20BamHI | GTACCGTTAAAAGGATCCGCAGTAG |
| pSPE17-700ropB-XhoI | CCTTATGGCCTCGAGATAGCTGC |
| pSPE17-inverse-250speB-XhoI | GATGAGAATACTCGAGTTGGGTTGTC |
| pROP-ropB + 100EcoRI | GTTTTTTGAATTCCTAATCTATTCAACGG |
| pROP10-speB360BamHI | CTTCTGCGCTTCGGGATCCTGCTTTG |
| pROP11ropB1600BamHI | CGATGTCGCGGATCCCATGAATGGTAATAG |
| pROP12-ropB1400BamHI | GCTAGGCAGACGGATCCGCAAGATCAGC |
| pROP13-ropB1200BamHI | CCATTAGTTGACTCGTAGGATCCTCCCTGG |
| pROP14-ropB1000BamHI | CCTTTTAACCTTATGGATCCATCATAGCTGC |
| pROP15-ropB900BamHI | CATATGACAGTGGATCCAACTATCGCATCTG |
| 5′-ropB-IFD | AAAACTGCAGGGAGGTCGTGACCAAAAAGGCGGC |
| 3′-ropB-IFD | AAAACTGCAGGGAGGTCGTGACCAAAAAGGCGGC |
| ropB-5′-rev-IFD | TCCCCCGGGTTCACCAATTTCCATATGTCAAGC |
| ropB-3′-fwd-IFD | TCCCCCGGGATTAAACATAAACTGTCCTGAGGC |
| rofA-5′-BglII | GGAAGATCTCATATATGAGCTTACCACG |
| rofA-3′-BamHI | CGCGGATCCAGTTCCTCACAATAATGG |
| ropB-5′-RBS-BamHI | CGCGGATCCGACATCAACTAGGAAGGC |
| ropB-stop-PstI | AAAACTGCAGCCTCAGGACAGTTTATGTTTAATGG |
| ropB-5′-SmaI | TCCCCCGGGATGGAAATTGGTGAAACCG |
| 5′-Upstream-pro | CAAGCCTTCCTAGTTGATGTC |
| 3′-Upstream-pro | GTCAATGCTTTTATTGACTTCTC |
| 5′-Downstream-pro | GAGAACGGTCCCATATCATC |
| speBpro-20rev | TCCCCCGGGCTGCATTTGCTTTTAACGGTAC |
Underlined sequences denote restriction site incorporated into primer.
Primer extension analysis.
The 5′ ends of the speB transcript(s) was determined with (i) avian myeloblastosis virus reverse transcriptase (Promega) in accordance with the manufacturer's directions and (ii) RNA samples prepared as described above. The primers used for extension included speB-20primerext and speB-600primerext, which were end labeled with 32P as previously described (27). Extended products were visualized by autoradiography following electrophoresis through a 6% polyacrylamide-urea sequencing gel, and the 5′ ends were determined by comparison to a sequencing reaction generated with a modified T7 DNA polymerase (Sequenase 2.0; Amersham), the same end-labeled primer and a plasmid template that contained a DNA segment that encompassed the entire speB-ropB intergenic region.
Transcript analysis by RT-PCR.
S. pyogenes cultures were grown in C medium; aliquots were harvested at 3, 5, 7, and 24 h; and total RNA was isolated as described above. First-strand cDNA synthesis was performed with SuperScript II reverse transcriptase (Invitrogen) with 5 μg of RNA, 250 ng of random hexamers as primers (Invitrogen), 0.5 mM deoxynucleoside triphosphate mix, 10× reverse transcription (RT) buffer, 5 mM MgCl2, and 10 mM dithiothreitol in accordance with the manufacturer's directions. With 2 μl of the cDNA reaction mixture, specific transcripts were detected with primer pairs that amplified an internal segment of the target cDNAs, including ropB (ropB-intergene and ropB-stop-BamHI), speB (speB-3′-fwd and speB-3′-rev), and the housekeeping gene glnQ (glnQ-5′ and glnQ-3′). To ensure that no contaminating DNA was present in the RNA samples and that all cDNA synthesis was derived from RNA templates, PCR amplification was performed on RNA samples in the absence of the first-strand cDNA synthesis reaction.
Analysis of promoter structure with reporter fusions.
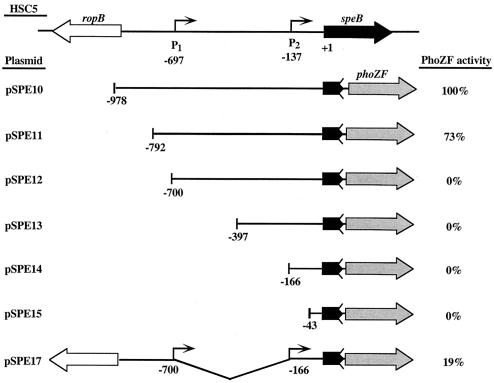
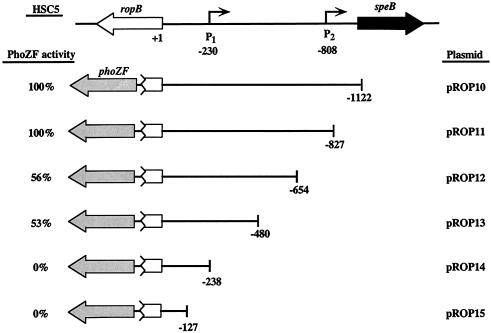
Reporter fusions were constructed with a gene encoding an alkaline phosphatase that has been modified for secretion (PhoZF) (18). Regions of interest were amplified from chromosomal DNA templates by PCR and inserted between the EcoRI and BamHI sites of the phoZF-containing E. coli-streptococcal shuttle plasmid pAGB5 (18). All amplifications of the speB promoter fragments used primer speB+180EcoRI and one of the following primers (the 5′ end of the resulting fragment is in parentheses): speB-1000BamHI (full length), speB-800BamHI (−792), speB-700BamHI (−700), speB-400BamHI (−397), speB-250BamHI (−166), and speB-20BamHI (−43). The plasmids containing these fragments were designated pSPE10 to pSPE15, respectively (see Fig. 2). An internal deletion was constructed by inverse PCR with plasmid pSPE10 (see Fig. 2) and primers 700ropB-XhoI and inverse-250speB-XhoI. The resulting plasmid (pSPE17) contains a deletion between bp −700 and −166 (see Fig. 2). All amplifications of ropB promoter fragments used primer ropB+100EcoRI and one of the following primers (the 5′ end of the resulting fragment is in parentheses): speB360BamHI (full length), ropB1600BamHI (−827), ropB1400BamHI (−654), ropB1200BamHI(−480), ropB1000BamHI (−238), and ropB900BamHI (−127). The plasmids containing these fragments were designated pROP10 to pROP15, respectively (see Fig. 3). Transformation of HSC5 with pSPE10 to pSPE17 and pROP10 to pROP15 by electroporation produced strains that were designated SPE100 to SPE107 and ROP100 to ROP105, respectively. For analysis of reporter activity, strains were cultured as described above and secreted alkaline phosphatase activity was determined after 9 h of growth as explained elsewhere (18).
FIG. 2.
Analysis of the speB promoter. The importance of sequences in the speB-ropB region for transcription of the speB promoter were evaluated with fusions to the reporter gene phoZF. The alkaline phosphatase activities produced by various truncations of the intergenic regions are shown relative to the activity produced from the full-length intergenic region (pSPE10). One hundred percent activity for pSPE10 was 125 U, calculated as enzyme units per unit of optical density at 600 nm as described by Brinkman and Beckwith (6). The number at the left end of each truncation indicates the 5′-terminal nucleotide relative to the first nucleotide of the speB start codon. Bent arrows represent 5′ ends of the speB transcripts identified by primer extension. The zigzag line after speB in each fusion construct indicates that transcription and not translation fusions to phoZF were constructed. Construct pSPE17 has an internal deletion that is shown by the connecting line below the line illustrating the intergenic sequence.
FIG. 3.
Analysis of the ropB promoter. The importance of sequences in the speB-ropB region for transcription of the ropB promoter was evaluated as described in the legend to Fig. 2. The number at the right end of each truncation indicates the 5′-terminal nucleotide relative to the first nucleotide of the start codon for ropB. Bent arrows represent 5′ ends of the speB transcripts and are numbered relative to the start codon of ropB. Alkaline phosphatase activity is shown as a percentage of that produced by the full-length intergenic region (pROP10). One hundred percent activity for pROP10 was 32 U, calculated as enzyme units per unit of optical density at 600 nm as described by Brinkman and Beckwith (6). All other designations are the same as those described for Fig. 2.
Construction of a ropB deletion mutant.
An in-frame deletion in ropB was constructed with primers 5′-ropB-IFD and 3′-ropB-IFD to amplify a 1,594-bp fragment containing the entire HSC5 ropB open reading frame. The fragment was inserted into a standard vector, and an in-frame deletion of 807 bp was made by inverse PCR with the primers ropB-5′-rev-IFD and ropB-3′-fwd-IFD. The resulting mutant allele (ropBΔ6-274) was inserted between the PstI and BamHI sites of the shuttle vector pJRS233 (31) to create pDLR3, which was used to replace the wild-type ropB allele in HSC5 by a previously described method (34). The chromosomal structure of the resulting mutant, MNN100, was confirmed by PCR.
Construction of a ropB expression plasmid.
A streptococcal expression vector was created with a previously described S. pyogenes promoter to express the ropB transcript as follows. The rofA promoter (18) was amplified by PCR with HSC5 chromosomal DNA and primers rofA-5′-BglII and rofA-3′-BamHI, and the resulting fragment was inserted into the BamHI site of pLZ12 (30). The wild-type allele of ropB was amplified by PCR with primers ropB-5′-RBS-BamHI and ropB-stop-PstI, and the restriction sites introduced into the primers were used to insert the resulting fragment between the BamHI and PstI sites downstream of the rofA promoter segment, creating pMNN23. The sequence of the inserted fragments was confirmed, and then pMNN23 was used to transform the wild type (HSC5) and the ropB mutant (MNN100) for the analyses described later in the text.
Protease activity assays.
Analysis of SpeB proteolytic activity was determined by measurement of zones of clearance on skim milk agar medium following growth at 37°C in an anaerobic environment or in cell-free supernatants with the substrate fluorescein isothiocyanate-casein as described previously (21). Uninoculated C medium was used to determine background values, and proteolytic activity is presented as relative to the activity of wild-type strain HSC5 at 20 h. Cell numbers were adjusted to that of HSC5 for each time point tested. For selected samples, inclusion of the cysteine protease inhibitor E64 (Sigma) (27) confirmed that typically >95% of the proteolytic activity was due to SpeB.
Construction of MBP-RopB.
A fusion protein joining maltose-binding protein (MBP) and RopB was constructed as follows. The ropB open reading frame was amplified by PCR from HSC5 genomic DNA with primers RopB-5′-SmaI and ropB-stop-PstI. The resulting fragment was digested with SmaI and PstI (sites in primers) and inserted between the XmnI and PstI sites of pMAL-c2 (New England Biolabs), which produced an in-frame fusion of the entire ropB coding sequence to the 3′ end of the malE gene encoded on the vector. The fidelity of the construct was confirmed by DNA sequencing, and the resulting plasmid was designated pMAL-ropB. Cell lysis and protein purification by amylose affinity chromatography were performed in accordance with the manufacturer's (New England Biolabs) specifications. Purified protein was concentrated with Centricon YM-10 centrifugal filter units (Millipore), and concentrations were determined by the bicinchoninic acid method with bovine serum albumin as the standard (37). Electrophoresis through sodium dodecyl sulfate-polyacrylamide gels and staining with Coomassie blue confirmed the expected size of MBP-RopB (∼74 kDa) and that preparations were typically >95% pure.
DNA-binding studies.
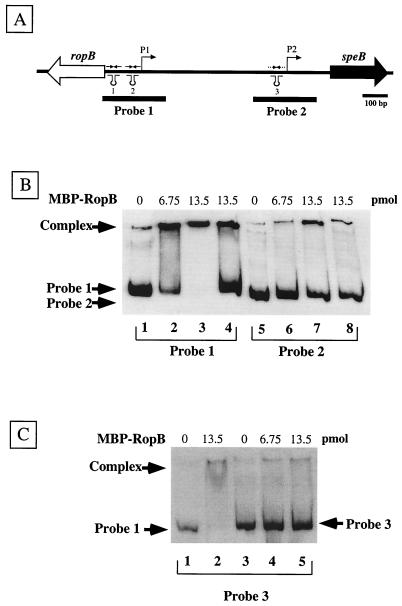
Probes for DNA-binding studies were generated by PCR amplification from HSC5 chromosomal DNA and biotinylated with a commercial DNA biotinylation kit (DNADetector; KPL). Probe 1 is 345 bp long and was synthesized with primers 5′-Upstream-pro and 3′-Upstream-pro. Probe 2 is 319 bp long and was synthesized with primers 5′-Downstream-pro and SpeBpro-20rev. The chromosomal regions corresponding to the probes are shown in Fig. 4A. Electrophoretic mobility shift assays were performed under the following conditions. A constant amount of labeled and gel-purified probe (5 ng) was mixed with increasing amounts of purified MBP-RopB protein (6.75 to 13.5 pmol) in reaction mixtures containing 20 mM HEPES (pH 7.2), 1 mM EDTA, 10 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 5% glycerol, and 500 ng of poly(dI-dC) in a total volume of 20 μl. A 20-fold excess of unlabeled probe was added to selected reaction mixtures. Following incubation at room temperature for 45 min, samples were run on a nondenaturing Tris-borate-EDTA 10% polyacrylamide gel and then transferred to a nylon membrane (Biodyne B; Pall) by alkaline transfer in accordance with the manufacturer's (KPL) directions. Chemiluminescent detection of biotinylated DNA on membranes was accomplished with a commercial reagent (DNADetector; KPL), followed by exposure to X-ray film.
FIG. 4.
RopB binds to sequences in the vicinity of the speB P1 promoter. (A) The DNA contained on two labeled probes encompassing sequences in the vicinity of the P1 and P2 promoters is shown by the solid bars under the chromosome. (B) The ability of MBP-RopB to bind to the probes was evaluated in an electrophoretic mobility shift assay. Lanes 1 to 4 contain 5 ng of probe 1 incubated with no protein (lane 1) or with 6.75 and 13.5 pmol of MBP-RopB (lanes 2 and 3, respectively). Lanes 5 to 8 contain 5 ng of probe 2 incubated with no protein (lane 5) or with 6.75 and 13.5 pmol of MBP-RopB (lanes 6 and 7, respectively). A 20-fold excess of unlabeled probe 1 or 2 was added to reaction mixtures with 13.5 pmol of MBP-RopB analyzed in lanes 4 and 8, respectively. (C) Ability of MBP-RopB to bind to an unrelated fragment outside of the speB-ropB intergenic region. Lanes 1 and 2 contain 5 ng of probe 1 with no protein (lane 1) and 13.5 pmol of MBP-RopB (lane 2). Lanes 3 to 5 contain 5 ng of probe 3 incubated with no protein (lane 3) or 6.75 and 13.5 pmol of MBP-RopB (lanes 4 and 5, respectively).
RESULTS
Analysis of the speB promoter region.
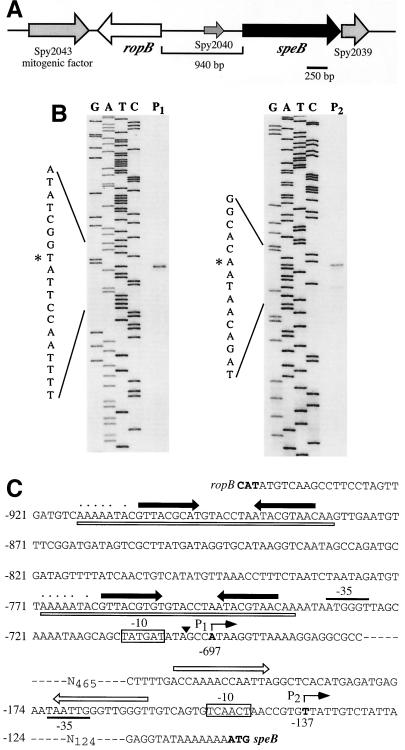
The genes encoding SpeB and RopB are located adjacent to one another on the streptococcal chromosome but transcribed in opposite directions (Fig. 1A). An interesting feature of this locus is a long 941-bp intergenic region between speB and ropB that does not contain any long open reading frame. The region does contain numerous small open reading frames of 56 or fewer codons, and most annotations of the genomic information available for several S. pyogenes strains recognize only a single small open reading frame of 56 residues (13) (e.g., Spy2040 of strain SF370). The importance of any feature of the intergenic region is unknown. Previous data demonstrated that two distinct transcripts could be detected by a speB-specific probe in a Northern blot analysis of RNA isolated from S. pyogenes HSC5 (27). To further define the regions involved in control of speB expression, the ends of these two transcripts were identified by primer extension with primers that annealed to various sequences within the intergenic region. Extension products that terminated at two distinct nucleotides were consistently obtained (the products obtained by extension of two representative primers are shown in Fig. 1B). These transcript ends were located 578 bp apart at positions −697 and −137 relative to the first nucleotide of the initiation codon of the speB open reading frame. The loci corresponding to the distal and proximal transcript ends were designated P1 and P2, respectively (Fig. 1C). Regions similar to the consensus −10 element of a σ70 promoter were located upstream of P1 and P2, although regions similar to a consensus −35 sequence are not present in either promoter (Fig. 1C). The P1 promoter was preceded by two copies of a long nearly identical direct repeat (identical at 33 of 35 positions), and each direct repeat contained an internal 9-bp inverted repeat (Fig. 1C). An inverted repeat identical at 12 of 14 positions lies directly upstream of the P2 promoter (Fig. 1C).
FIG. 1.
Identification of the 5′ ends of the speB transcripts. (A) Organization of the ropB-speB region on the streptococcal chromosome. Gene designations are based on the genome sequence of S. pyogenes SF370 (18). (B) Primer extension analyses to identify the 5′ ends of the two transcripts previously determined for speB. The primer extension products obtained for the longer (P1) and shorter (P2) transcripts are shown. The sequencing ladder used to determine the terminal nucleotides of each transcript (denoted by an asterisk) are shown at the left of the extension products. (C) Features of the P1 and P2 promoters. The initiation nucleotides of the P1 and P2 promoters are in bold and marked by the bent arrow above the sequence. The numbers below the nucleotides and at the left of the sequence indicate nucleotide positions relative to the first nucleotide of the speB start codon. The closed arrows above the sequence show the locations of an inverted repeat present in two copies near P1, and the open boxes beneath the sequence indicate a large direct repeat. Nucleotides that encode a poly(U) tract adjacent to the inverted repeats on the strand opposite to the one shown are indicated by the dotted line above the sequence. The poly(U) tract and repeats have the characteristics of a rho-independent terminator for transcripts encoded on the opposite strand.
Sequences required for speB transcription.
The importance of the sequence features that flank each transcript end was analyzed through the construction of fusions between the speB promoter regions and a reporter gene. This analysis was done with a gene that encodes a modified alkaline phosphatase (PhoZF) that was previously used for analysis of the divergently transcribed rofA and prtF promoters of S. pyogenes (18). The initial construct resulted from the introduction of the entire intergenic region in a position to transcribe phoZF in the same direction as speB transcription in the vector pABG5 (18) (pSPE10, Fig. 2). Following introduction into S. pyogenes HSC5, the pattern of alkaline phosphatase expression paralleled the pattern of expression of SpeB cysteine protease activity and characteristically initiated late in log phase, just prior to the entry of the culture into the stationary phase of growth (data not shown). Truncations of the intergenic region were then analyzed after 9 h of growth, in comparison to the full-length region, to determine the minimal required sequence for transcription of speB. A truncation that terminated 95 bp upstream of P1 retained high levels of activity, although they were reduced about 25% relative to the activity of the full-length construct (compare pSPE11 to pSPE10, Fig. 2). In contrast, a truncation that removed the putative −10 and −35 regions of P1 resulted in complete loss of transcriptional activity, despite the fact that P2 was unaltered (pSPE12, Fig. 2). Additional P2-containing constructs also demonstrated no activity (pSPE13 and pSPE14, Fig. 2), as did a construct that lacked both P1 and P2 (pSPE15, Fig. 2). The observed drop from high to background levels of activity between truncations at −792 and −700 suggested that the intervening 92 bp contain sequences essential for expression of speB. An internal deletion of the region between P1 and P2 (−700 to −166 bp) reduced activity even though the region upstream of P1 remained intact (pSPE17, Fig. 2). Taken together, these data suggest (i) that the entire extensive intergenic region is important in cis for transcription of speB; (ii) that P1 represents the principal promoter of speB and is controlled by elements, which may include the direct repeats, in sequences upstream of the −35 region; (iii) that P2 is not an autonomous promoter; and (iv) that important cis-acting control regions for speB transcription are located adjacent to the ropB open reading frame.
Identification of the regions required for ropB transcription.
The proximity of speB control regions to ropB suggests that the speB and ropB promoters are oriented such that they oppose each other. However, despite the use of numerous primers, all attempts to obtain an extension product to define the end of the ropB transcript were unsuccessful, possibly because of secondary structure generated by interactions between the extensive repetitive sequences in this region. Thus, to define the region that controls transcription of ropB and to determine the relative distance between the ropB and speB promoters, the region upstream of ropB was analyzed with fusions to the phoZF reporter (Fig. 3). When introduced so that the promoter for ropB would direct transcription of phoZF, the entire intergenic region and a truncation to approximately the location of P2 produced equivalent activities (pROP10 and pROP11, Fig. 3). However, truncation to −654 bp relative to the start of ropB reduced activity to about half of maximum (pROP12, Fig. 3) and this level was unaffected by the removal of an additional 174 bp (pROP13, Fig. 3). All further truncations reduced activity to background values (pROP14 and pROP15, Fig. 3). These data indicate (i) that, similar to that of speB, transcriptional control of ropB requires extensive sequences in cis that extend up to 800 bp upstream of ropB; (ii) that elements essential for control of ropB reside in the vicinity of P1 between 238 and 480 bp upstream of ropB; and (iii) that it is highly likely that the ropB and speB P1 promoters oppose each other.
RopB binds specific sequences in the ropB-speB intergenic region.
Numerous members of the Rgg family of regulators have been shown to be essential activators of transcription of their target genes. However, these regulators lack homology to any other known class of transcription activator, and how they function to regulate transcription is unknown. For example, it has not been shown whether these regulatory proteins have DNA binding activity. In order to determine if RopB can bind DNA, the protein was overexpressed and purified from E. coli as a fusion to MBP (MBP-RopB; see Materials and Methods). The ability of MBP-RopB to bind to the speB-ropB intergenic region was examined with an electrophoretic mobility shift assay. Reactions between a 345-bp probe that included the sequences flanking P1 that were shown above to be critical for expression of speB (probe 1, Fig. 4A) and MBP-RopB resulted in decreased mobility of the probe compared to the mobility of the probe alone, and the amount of DNA in the shifted band was dependent on the concentration of MBP-RopB in the reaction mixture (compare lane 1 with lanes 2 and 3, Fig. 4B). In contrast, a second probe derived from sequences that included P2 (probe 2, Fig. 4A) showed little to no decreased mobility when MBP-RopB was added to the reaction mixture (compare lane 5 to lanes 6 and 7, Fig. 4B). The addition of an excess of unlabeled probe 1 to a reaction mixture containing labeled probe 1 and MBP-RopB resulted in a decreased amount of labeled DNA in the shifted complex, demonstrating that binding of MBP-RopB is specific (compare lanes 3 and 4, Fig. 4B). Additional controls for specificity included that an unrelated MBP fusion protein did not bind probe 1 or 2 (data not shown) and a probe made from a different region of the streptococcal chromosome did not to bind MBP-RopB (probe 3, Fig. 4C).
Expression kinetics of ropB and speB.
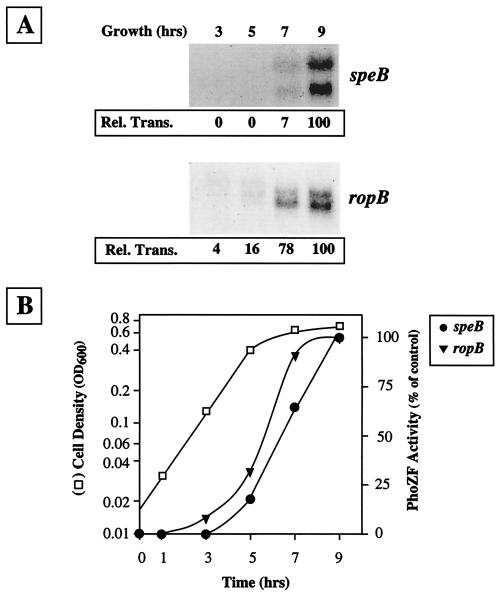
As noted above, speB is regulated in a striking growth phase pattern and transcriptional expression is dependent on ropB. Since the major control region for expression of speB, including the P1 promoter and RopB binding sites, are located adjacent to ropB and the control regions for ropB, it was of interest to determine whether expression of ropB is constitutive or shows a distinct kinetic pattern. When a Northern blot analysis was used to directly compare the kinetics of expression of speB and ropB, it was found that ropB was expressed in a growth phase pattern that resembled that of speB (Fig. 5A). Similar to speB, a ropB-specific probe hybridized to two distinct bands, which were only detected at later, rather than earlier, time points during culture growth. However, expression of ropB appeared to initiate at an earlier time point than that of speB (compare ropB and speB at 7 h, Fig. 5A). The kinetics of ropB expression was confirmed with phoZF fusions containing the entire intergenic region in speB- and ropB-specific orientations. Consistent with data from the Northern blot analyses, expression of ropB promoter-dependent PhoZF activity initiated in late log phase in a pattern similar to that of speB promoter-dependent activity (Fig. 5B).
FIG. 5.
Kinetics of ropB and speB expression. (A) Northern blot analysis of total RNA harvested at the indicated time points during growth. Transcripts for the two genes were visualized with either ropB-specific or speB-specific probes. The value for relative transcription (Rel. Trans.) listed under each lane was determined by comparison to the same RNA sample hybridized with a probe for the recA transcript. (B) Expression kinetics evaluated with reporter fusions. Activity of the PhoZF reporter in HSC5 strains containing a fusion of the speB and ropB promoters contained in the full-length intergenic region (pSPE10 and pROP10, respectively) was determined relative to the cell density of the culture as shown. Activity is presented relative to the maximal activity obtained in early stationary-phase cultures and is normalized for the density of the culture. OD600, optical density at 600 nm.
Role of ropB in growth phase control.
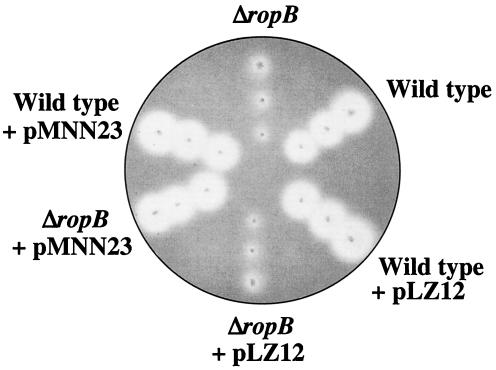
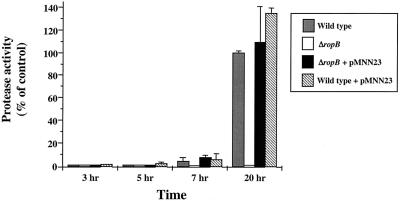
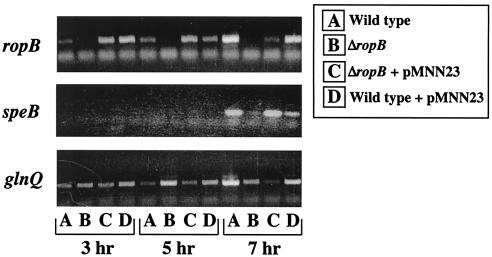
Since ropB also demonstrates growth phase expression kinetics, one model proposes that the signal for expression of speB is channeled directly via ropB. In this model, the signal acts exclusively to initiate transcription of ropB. Once RopB is present, it acts as the sole factor required to trigger expression of speB. The model predicts that the major event required for speB expression is the time point at which ropB is transcribed. To test this model, a vector was established for ectopic expression of ropB from a promoter that is expressed in the early, rather than the late, log phase of growth. The promoter for rofA has this characteristic and has the additional advantage that it has been shown to be highly active when plasmid encoded under the growth conditions used in these experiments for expression of speB (14, 18). A plasmid was constructed that contains ropB under the control of the rofA promoter (pMNN23) and was introduced into both the wild type (HSC5) and a mutant (MNN100) that contains an in-frame deletion in ropB. The deletion resulted in removal of the region encoding 271 of the 281 amino acids in the internal region of the ropB polypeptide but left the ropB-speB intergenic promoter region intact. Consistent with previous data (8, 27), mutant MNN100 made little proteolytic activity on protease indicator plates compared to HSC5 (ΔropB versus wild type, Fig. 6). Introduction of a plasmid alone had no effect on the protease expression profiles of the mutant or HSC5 (wild type + pLZ12 and ΔropB + pLZ12, Fig. 6). However, when the plasmid contained ropB under the control of the rofA promoter, the protease activity of the complemented mutant strain was indistinguishable from that of HSC5 (compare ΔropB + pMNN23 to wild type and wild type + pMNN23, Fig. 6). These data show that ectopic expression of ropB can complement a chromosomal deletion of ropB. However, the protease indicator plates measure the accumulation of protease over time and are not sensitive for kinetics. Analysis of SpeB protease activity in cell-free supernatants at different time points revealed that ectopic expression of ropB had no influence on the pattern of expression of speB. Both the mutant and wild-type strains complemented with pMNN23 expressed protease activity in a pattern similar to that of the wild-type strain alone (Fig. 7). Since the protease zymogen must be activated following its secretion from the cell, it was possible that protease activity did not accurately measure expression because activating conditions are only generated following the log phase of growth. Analysis of transcript levels by RT-PCR demonstrated that this was not the case. Although not quantitative for absolute message levels, no transcripts for speB could be detected in wild-type and complemented wild-type and mutant strains at early time points during growth (3 and 5 h for speB, Fig. 8). This was despite the fact that transcripts of ropB could be detected at the earliest point analyzed (3 h, Fig. 8). No transcripts for speB or ropB were detected in the ropB mutant at any time point (Fig. 8), and samples subjected to PCR without prior treatment with reverse transcriptase failed to show a band at any time point to indicate that contamination by DNA did not influence the results (data not shown). Finally, RT-PCR with primers to detect the housekeeping gene glnQ demonstrated that amplifiable RNA was present in each sample (Fig. 8). Because shifting of the timing of ropB expression did not influence the kinetics of speB transcription, these data do not support growth phase control of ropB as the event required for expression of speB.
FIG. 6.
Complementation of RopB deletion demonstrated by protease expression. Derivatives of S. pyogenes strain HSC5 (wild type) were grown on protease indicator plates overnight at 37°C in an anaerobic environment. The strains evaluated included MNN100 (ΔropB), HSC5 with the vector alone (wild type + pLZ12), MNN100 with the vector alone (Δrop + pLZ12), MNN100 with a ropB-complementing plasmid (Δrop + pMNN23), and HSC5 with a ropB-complementing plasmid (wild type + pMNN23). Clear zones around colonies indicate protease activity.
FIG. 7.
Ectopic expression of ropB does not alter the expression kinetics of speB. The chimeric plasmid pMNN23 contains ropB under the control of a promoter expressed during the early logarithmic phase of growth. Levels of protease activity produced in various strains at the indicated time points were measured with the substrate fluorescein isothiocyanate-casein and are reported relative to the activity of the wild-type strain at 20 h. The strains analyzed included HSC5 (wild type), MNN100 (ΔropB), MNN100 plus ectopically expressed ropB (ΔropB + pMNN23), and HSC5 plus ectopically expressed ropB (wild type + pMNN23). The data presented represent the mean and standard error of the mean derived from seven independent experiments.
FIG. 8.
Confirmation of ectopic expression of ropB. The presence of mRNA for ropB (top), speB (middle), and the housekeeping gene glnQ (bottom) was determined for cultures at the time points indicated by nonquantitative RT-PCR. The strains evaluated were identical to those listed in Fig. 7. The results shown are representative of at least three different RT-PCR assays from three separate RNA isolations.
DISCUSSION
The SpeB cysteine protease of S. pyogenes is remarkable for its multiple layers of regulation. Not only are the secretion and processing of the protease polypeptide precursor into the active protease subject to regulation by accessory gene products, the folding and activation of the precursor following its secretion are strongly influenced by environmental factors. The expression of the gene that encodes the protease is also subject to a multilayered regulatory program that includes growth phase, nutritional, and environmental cues. How all of these factors interact to control the expression of proteolytic activity during infection of host tissue is not well understood.
Previous studies have shown that activation of transcription of speB has an absolute dependence on the rgg family regulator ropB. Thus, understanding how ropB functions is essential for understanding the biogenesis of proteolytic activity, as well as for understanding how this widely distributed novel family of regulators has been adapted for control of transcription of other genes. Consistent with the complexity of the protease regulatory program, we have found in the present study that very few alterations can be made to the extensive ropB-speB intergenic region without affecting protease expression. The reason that this extensive regulatory region is required is not known.
When examining the repeats near the 5′ end of the ropB message, both inverted repeats in the RNA are followed by a run of U nucleotides (UAUUUUU, Fig. 1C) resembling the structure of a factor-independent transcription termination signal (5). Inverted repeats may play a key role in the regulation of genes by other rgg family members. Both rgg itself and gadR (35, 38) are located immediately upstream of their target genes. Unlike ropB, rgg and gadR are located in a direct orientation relative to their regulatory targets and have very short intergenic regions. Similar to ropB, the intergenic regions contain prominent rho-independent terminator-like structures that overlap the promoter elements of the target genes (35, 38). It has been proposed that Rgg influences the formation of this RNA secondary structure in order to promote the transcription and/or translation of the gftG message. The overlap of the ropB and speB promoters and the location of the terminator structures near both promoters strongly implicate these structures in the program for control of protease expression.
Similar to that of other promoters that are subject to activation, the −10 site of the speB promoter is quite close to consensus (TATgAT); however, the −35 site only matches at two of the six consensus nucleotides (aTGggt). In E. coli, many transcriptional activators bind to a specific site next to the −35 region of the promoter and interact with polymerase in order to compensate for the poor consensus −35 region and recruit polymerase to the promoter, thereby increasing the rate of transcription of the gene (4). The direct and inverted repeat sequences occurring directly upstream of the −35 site of the speB promoter fit into this category of regulatory sequences and resemble the palindromic sequences that are often bound by DNA-binding proteins to activate transcription. Consistent with this was the observation that RopB bound with the highest affinity to a DNA fragment that contained the repetitive region adjacent to P1.
The closest relative of ropB, lasX, regulates the expression of an operon for synthesis of the lantibiotic lactocin S in Lactobacillus sakei (36). Similar to the S. pyogenes cysteine protease, many lantibiotic operons are regulated in a growth phase-dependent pattern. In some of these operons, developmental control involves a bacterial cell density-dependent mechanism that involves direct monitoring of the concentration of the lantibiotic peptide itself by a two-component phosphorelay. However, for other operons, including lactocin S and mutacin of S. mutans, regulation is under the control of an rgg family regulator (32, 33, 36). Other similarities between lasX and ropB include the fact that both are located adjacent to their regulatory targets but are oriented in the opposite direction. Furthermore, both have intergenic regions containing several directly repeated sequences, both have promoters that overlap those of their regulatory targets, and lastly, regulation of both of their target genes is influenced by environmental cues such as pH (28). One difference is that, unlike that of speB and many other lantibiotics, expression of lactocin S is not subject to growth phase regulation (36). Nevertheless, the fact that ropB, lasX, and other members of the rgg family have many characteristics in common suggests that they use very similar mechanisms to interact with their regulatory programs to process information.
Regulation of speB is subject to both environmental and growth phase cues (7, 9, 12, 27). In the present report, we have shown that the program for expression of speB is not solely dependent on ropB, suggesting that other elements are involved. Prior studies have suggested that developmental control does not function through a mechanism that senses culture density (9) but may involve the two-component regulator csrRS/covRS since mutants with changes at this locus typically overexpress speB (22). Interestingly, ropB had a negative influence on transcription of a number of other virulence regulatory genes, including csrRS/covRS (10), suggesting a complex interplay between global regulators for control of S. pyogenes virulence. The details of these interactions remain to be elucidated and will be crucial for understanding the role that speB and other virulence factors of S. pyogenes contribute to interaction with the host.
Acknowledgments
This work was supported by Public Health Service grant A14643303 to M.C.
REFERENCES
- 1.Ashbaugh, C. D., H. B. Warren, V. J. Carey, and M. R. Wessels. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Investig. 102:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisno, A. L., and D. L. Stevens. 1996. Streptococcal infections of skin and soft tissues. N. Engl. J. Med. 334:240-245. [DOI] [PubMed] [Google Scholar]
- 3.Bjorck, L., P. Akesson, M. Bohus, J. Trojnar, M. Abrahamson, I. Olafsson, and A. Grubb. 1989. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature 337:385-386. [DOI] [PubMed] [Google Scholar]
- 4.Branden, C., and J. Tooze. 1991. Introduction to protein structure. Garland Publishing, Inc., New York, N.Y.
- 5.Brendel, V., G. H. Hamm, and E. N. Trifonov. 1986. Terminators of transcription with RNA polymerase from Escherichia coli: what they look like and how to find them. J. Biomol. Struct. Dyn. 3:705-723. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 7.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 68:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, J. O. 1969. Effect of culture medium composition and pH on the production of M protein and proteinase by group A streptococci. J. Bacteriol. 99:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara, T., T. Hoshino, T. Ooshima, S. Sobue, and S. Hamada. 2000. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 68:2475-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 18.Granok, A. B., A. Claiborne, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site of Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubba, S., D. E. Low, and J. M. Musser. 1998. Expression and characterization of group A streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect. Immun. 66:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanski, E., P. A. Horowitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser, A. R., and P. M. Schlievert. 1990. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J. Bacteriol. 172:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm, S. E., A. Norrby, A.-M. Bergholm, and M. Norgren. 1992. Aspects of pathogenesis of serious group A streptococcal infections in Sweden. J. Infect. Dis. 166:31-37. [DOI] [PubMed] [Google Scholar]
- 24.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inversed relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiliç, A. O., M. C. Herzberg, M. W. Meyer, X. Zhao, and L. Tao. 1999. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid 42:67-72. [DOI] [PubMed] [Google Scholar]
- 26.Lukomski, S., C. A. Montgomery, J. Rurangirwa, R. S. Geske, J. P. Barrish, G. J. Adams, and J. M. Musser. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion, and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortvedt-Abildgaard, C. I., J. Nissen-Meyer, B. Jelle, B. Grenov, M. Skaugen, and I. F. Nes. 1995. Production and pH-dependent bactericidal activity of lactocin S, a lantibiotic from Lactobacillus sake L45. Appl. Environ. Microbiol. 61:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neely, M. N., J. D. Pfeifer, and M. G. Caparon. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 70:3904-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Casal, J., J. A. Price, E. Maugin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 32.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlinson, E. L., I. F. Nes, and M. Skaugen. 2002. LasX, a transcriptional regulator of the lactocin S biosynthetic genes in Lactobacillus sakei L45, acts both as an activator and a repressor. Biochimie 84:559-567. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz, N., B. Wang, A. Pentland, and M. G. Caparon. 1998. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol. Microbiol. 27:337-346. [DOI] [PubMed] [Google Scholar]
- 35.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 36.Skaugen, M., E. L. Andersen, V. H. Christie, and I. F. Nes. 2002. Identification, characterization, and expression of a second, bicistronic, operon involved in the production of lactocin S in Lactobacillus sakei L45. Appl. Environ. Microbiol. 68:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. J. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olsen, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 38.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensson, M. D., D. A. Scaramuzzino, U. Sjobring, A. Olsen, C. Frank, and D. E. Bessen. 2000. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol. Microbiol. 38:242-253. [DOI] [PubMed] [Google Scholar]
- 40.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 41.Vickerman, M. M., P. E. Minick, and N. M. Mather. 2001. Characterization of the Streptococcus gordonii chromosomal region immediately downstream of the glucosyltransferase gene. Microbiology 147:3061-3070. [DOI] [PubMed] [Google Scholar]
- 42.Vickerman, M. M., M. C. Sulavik, and D. B. Clewell. 1995. Oral streptococci with genetic determinants similar to the glycosyltransferase regulatory gene, rgg. Infect. Immun. 63:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vickerman, M. M., M. Wang, and L. J. Baker. 2003. An amino acid change near the carboxyl terminus of the Streptococcus gordonii regulatory protein Rgg affects its abilities to bind DNA and influence expression of the glucosyltransferase gene gtfG. Microbiology 149:399-406. [DOI] [PubMed] [Google Scholar]