Abstract
The Vibrio cholerae SXT element is a conjugative self-transmissible chromosomally integrating element that encodes resistance to multiple antibiotics. SXT integrates in a site-specific fashion at prfC and excises from the chromosome to form a circular but nonreplicative extrachromosomal form. Both chromosomal integration and excision depend on an SXT-encoded recombinase, Int. Here we found that Int is necessary and sufficient for SXT integration and that int expression in recipient cells requires the SXT activators SetC and SetD. Although no xis-like gene was annotated in the SXT genome, Int was not sufficient to mediate efficient SXT chromosomal excision. We identified a novel SXT Xis that seems to function as a recombination directionality factor (RDF), facilitating SXT excision and inhibiting SXT integration. Although unrelated to any previously characterized RDF, Xis is similar to five hypothetical proteins that together may constitute a new family of RDFs. Using real-time quantitative PCR assays to study SXT excision from the chromosome, we determined that while SXT excision is required for SXT transfer, the percentage of cells containing an excised circular SXT does not appear to be a major factor limiting SXT transfer; i.e., we found that most cells harboring an excised circular SXT molecule do not act as SXT donors. In the absence of prfC, SXT integrated into several secondary attachment sites but preferentially into the 5′ end of pntB. SXT excision and transfer from a donor containing pntB::SXT were reduced, suggesting that the SXT integration site may also influence the element's transmissibility.
Integrative and conjugative elements (ICEs) are now recognized as a large and diverse class of mobile genetic elements in both gram-negative and gram-positive organisms (9). ICEs excise from the chromosomes of their hosts, transfer to a new host via conjugation, and then integrate into the chromosome again. These elements encode diverse excision, recombination, and conjugation systems, as well as many other properties, such as resistance to antibiotics (25, 30), nitrogen fixation (27), and degradation of aromatic compounds (24). ICE integration can be more or less site specific, and a large variety of sequences are used as targets for integration.
The SXT element is a Vibrio cholerae-derived ICE that has also been referred to as a conjugative transposon (29) and a constin (17). SXT was originally isolated in 1993 from MO10, a V. cholerae serogroup O139 clinical isolate. The MO10-derived SXT, SXTMO10, encodes resistance to sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin (29). After the extensive cholera outbreaks on the Indian subcontinent caused by V. cholerae O139 in late 1992 and 1993, V. cholerae O1 reemerged and was found to harbor an ICE, designated SXTET, that is very similar to SXTMO10 but contains different antibiotic resistance genes (15). Other SXT variants that lack antibiotic resistance genes have also been recognized in recent V. cholerae O139 isolates. SXT-related ICEs are currently found in virtually all V. cholerae clinical isolates from the Indian subcontinent. Also, SXT-related elements have been detected in recent V. cholerae clinical isolates from Mozambique and South Africa (11).
SXT-related elements are not unique to V. cholerae. We found SXT in Providencia alcalifaciens clinical isolates from patients in Bangladesh (15). R391, which mediates kanamycin and mercury resistance and was originally derived from a 1972 South African Providencia rettgeri isolate (10), and other IncJ elements are functionally and genetically related to SXT elements (14). In fact, our recent comparison of the complete DNA sequences of SXTMO10 and R391 revealed that both of these elements contain a highly conserved genetic backbone that mediates their regulation, excision-integration, and conjugative transfer (3, 4, 6). In the laboratory, SXT can be transferred by conjugation to a variety of gram-negative organisms (29). In addition, SXT can mediate the transfer of certain mobilizable plasmids and of chromosomal DNA in an Hfr-like fashion (16).
An autonomously replicating extrachromosomal form of SXTMO10 has not been isolated. Instead, this 99.5-kb ICE is found integrated into the 5′ end of the chromosomal gene prfC, which encodes the peptide chain release factor 3 (RF3) involved in translation regulation (17). SXT integration disrupts the 5′ end of prfC and provides a novel 5′ coding sequence for prfC and a promoter that leads to expression of a functional RF3. All the SXT-related ICEs that have been studied, including R391, are also integrated into pfrC (14). These elements encode a tyrosine recombinase, Int, which is related to the λ family of site-specific recombinases. In a previous study, it was shown that the excision and integration mechanisms of SXT resemble those of lambdoid phages (17). Like λ, the integrated SXT excises from the chromosome to form a circular extrachromosomal molecule. This episomal form of SXT appears to be an intermediate required for transfer. Recombination occurs between short sequences that are nearly identical on the circular form of the element (attP) and at the chromosomal site (attB) during integration and between attL and attR during excision. Int is necessary both in the donor cells for SXT excision and in the recipient cells for SXT integration. Both SXT excision and integration occur independent of recA.
The ∼99.5-kb DNA sequence of SXTMO10 was recently determined, and SXT genes involved in conjugative transfer and chromosomal excision were identified (4). SXT utilizes a conjugation system related to that encoded by the F plasmid; two SXT loci, setC and setD, encode transcriptional activators required for SXT excision and transfer. Although the annotation of the SXT genome did not identify any genes encoding a recombination directionality factor (RDF) (19), most of the integrating elements that have been studied, including phages and ICEs, encode an RDF. The λ RDF, Xis, is required during the Int-catalyzed recombination reaction between attL and attR to facilitate phage excision, and it inhibits recombination between attP and attB, preventing integration (2). λ Xis binds to and bends DNA in attP and attR to influence the architecture of the nucleoprotein complexes at these sites to either inhibit or favor recombination. In the present study, we investigated which SXT genes are required for the element's integration and excision by using real-time quantitative PCR to measure the frequency of SXT excision. While Int proved to be necessary and sufficient for integration, excision required a novel SXT-encoded Xis. The SXT Xis acts as an RDF, promoting SXT excision and inhibiting SXT integration. Determination of the percentage of cells containing an excised circular SXT revealed that the majority of such cells do not act as SXT donors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Bacterial strains were routinely grown in Luria-Bertani (LB) broth at 37°C on a roller drum incubator and were maintained at −80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 40 μg/ml; streptomycin, 200 μg/ml; sulfamethoxazole, 160 μg/ml; trimethoprim, 32 μg/ml; and tetracycline, 12 μg/ml.
TABLE 1.
E. coli K-12 derivative strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 | 26 |
| KB1 | MG1655 recA56 gutA52 gutR::Tn10 | K. Bettenbrock, unpublished data |
| HW220 | CAG18439 prfC::SXT | 17 |
| BI533 | MG1655 Nalr | 16 |
| BW25113 | lacIqrrnBt14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBAADLD78 | 12 |
| VI11 | CAG18439 prfC::SXT Δs002 | This study |
| VI38 | CAG18439 prfC::SXT Δs003 | This study |
| VI61 | CAG18439 ΔlacZ::attP-cat | This study |
| VI100 | CAG18439 prfC::SXT Δxis | This study |
| VI141 | CAG18439 ΔprfC pntB::SXT | This study |
| VI152 | KB1 ΔprfC ysgA::SXT | This study |
| SM10λpir | F−recA::RP4-2-Tc::Mu Km λpir | 20 |
| Plasmids | ||
| pGP704 | oriR6K mobRP4 Apr | 20 |
| pVI4 | pGP704 s003-int-attP | This study |
| pVI5A | pGP704 s003-s002-int-attP | This study |
| pVI6A | pGP704 s002-int-attP | This study |
| pVI6AM1 | pVI6A xis′ | This study |
| pVI6AM2 | pVI6A s002′ | This study |
| pVI8A | pCRII-TOPO attP | This study |
| pVI11 | pKD3 attP Cmr PCR template for one-step chromosomal gene activation | This study |
| pattP | pGP704 attP | This study |
| pInt33 | pBAD33 intSXT+ | B. Hochhut, unpublished data |
| pSetCD | pBAD-Topo setCD | 4 |
| pSetCD33 | pBAD33 setCD | J. Beaber, unpublished data |
| pXis | pBAD-Topo xis | This study |
| pKD3 | Cmr PCR template for one-step chromosomal gene activation | 12 |
| pKD4 | Kmr PCR template for one-step chromosomal gene activation | 12 |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Nalr, naladixic acid resistant.
Bacterial conjugation.
Conjugation assays were performed by mixing equal volumes of overnight cultures of donor and recipient strains. The cells were harvested by centrifugation and resuspended in 0.05 volume of LB broth. The cell suspensions were poured onto LB agar plates supplemented, when required, with 0.02% arabinose or 0.2% glucose. The conjugation mixtures were incubated at 37°C for 6 h. The cells were then resuspended from the plates in 1 ml of LB medium, and serial dilutions were plated on the appropriate selective media to determine the numbers of donors, recipients, and exconjugants.
Plasmid and strain construction.
Plasmids pVI4, pVI5A, and pVI6A (Table 1) were constructed by PCR amplification of the SXT attP region that resulted from excision of SXT with primers repKOF and orfX1 (Table 2) and template DNA from a Δs002 SXT (VI11), a wild-type SXT (HW220), and a Δs003 SXT (VI38), respectively. Then PCR products were purified from a 1% agarose gel by using a QIAquick gel extraction kit (Qiagen) and cloned into the TA cloning vector pCRII-TOPO (Invitrogen) according to the manufacturer's instructions. The subcloned fragments were removed from the pCRII-TOPO vector by digestion with KpnI/XbaI (pVI4) or EcoRI (pVI5A and pVI6A) and then ligated to KpnI/XbaI-digested or EcoRI-digested pGP704. pGP704 contains the RP4 mobilization locus, the R6K conditional origin of replication, and an ampicillin resistance gene (20). Frameshift mutations in s002 and xis were introduced by in vitro site-directed mutagenesis of pVI6A by using a QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The s002 frameshift mutation in pVI6AM2 was created with primers O2QCF and O2QCR, and the xis frameshift mutation in pVI6AM1 was created with primers MVI1F and MVI1R. pVI8A was constructed by PCR amplification of the SXT attP region with primers CAttPF and CAttPR. After purification, the PCR product was cloned into the TA cloning vector pCRII-TOPO. pVI11 was constructed by introducing the 600-bp BstBI attP-containing fragment of pVI8A into BstBI-digested pKD3. pKD3 contains a cat gene flanked by FLT recognition target (FRT) sites (12). pattP was constructed by introducing the SacI/XbaI fragment of pVI8A into SacI/XbaI-digested pGP704. pXis was constructed by cloning the xis gene amplified by PCR with primers XisTF and XisTR2 into the TA cloning expression vector pBAD-TOPO (Invitrogen) according to the manufacturer's instructions. The Δxis mutation was introduced into SXT by using the one-step chromosomal gene inactivation technique (12) with primers XisWF and XisWR and the pKD4 template as previously described (4). ΔprfC derivatives of Escherichia coli MG1655 were created by using the same technique with primers PrfCWF and PrfCWR and pKD3 as the template. Similarly, E. coli strain VI61, used as a calibrator for the real-time quantitative PCR, was constructed by using primers LacZWF and LacZWR and pVI11 as the template. The PCR product was introduced into the chromosome of E. coli CAG18439 as described by Datsenko and Wanner (12), generating a substitution of attP and cat in place of the lacZ gene.
TABLE 2.
DNA sequences of the oligonucleotides used in this study
| Primer | Nucleotide sequence (5′ to 3′) |
|---|---|
| repKOF | TTGCTCTGGTAAATCGGCTAGG |
| orfX1 | TACGCAGCCCCTTGCTAAAG |
| XisTF | TGATTTTCAGGAGAATGCAGA |
| XisTR2 | TCAGTCCTCTTTCTTCTCATAC |
| CAttPF | TTCGAAGGTTTAGCCACAGTTGTTTATGAGTG |
| CAttPR | TTCGAATATTCCGCTTTTGTAATGTCGAAA |
| LacZWF | TTGTGAGCGGATAACAATTTCACACAGGAAACAGCTGTGTAGGCTGGAGCTGCTTCG |
| LacZWR | GCGAAATACGGGCAGACATGGCCTGCCCGGTTATTACATATGAATATCCTCCTTA |
| XisWF | GATGATAGCTTTACATTGATTTTCAGGAGAATGCAGGTGTAGGCTGGAGCTGCTTCG |
| XisWR | CAACTGGCGTTTCGGACAATAATAAAAGAAAGAGGCCATATGAATATCCTCCTTA |
| PrfCWF | TAGCCGCAATTTTTCGTTTTCAACAAGCGCGGCGCGGTGTAGGCTGGAGCTGCTTCG |
| PrfCWR | CCGTAAGCGGCTAATAAGGAAGGGAAATTGACAGGGCATATGAATATCCTCCTTA |
| MVI1F | GATGTTAGAAAAGGGGTACCTGGTGCGCTACTG |
| MVI1R | CAGTAGCGCACCAGGTACCCCTTTTCTAACATC |
| O2QCF | GTTGTCGTCAACATTCCCCCTAGGGAGCCACCCTGAACTGC |
| O2QCR | GCAGTTCAGGGTGGCTCCCTAGGGGGAATGTTGACGACAAC |
| ARB1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
| ARB2 | GGCCACGCGTCGACTAGTAC |
| SXTR1 | GCCAATTACGATTAACACGACG |
| SXTR2 | CGGATTTGACAAGCGAAGAACTG |
| PrfCQF | GCTCAAAGGGCTGGTACAGC |
| PrfCQR | TTGTTGGAGATTGGACGGAAC |
| EattBF | GCCGCACTTTTGCCATTATT |
| EattBR | AGCAGCACCTTCTCGGTGAT |
| SXTJF | GCGAAGGACCTTTGCTATCATC |
| SXTJR | TGGTTTTAAGCGTTGAAAGGC |
Molecular biology methods.
Plasmid DNA was prepared with either a Qiaprep Spin miniprep kit or a Qiaprep miniprep kit (Qiagen), and chromosomal DNA was prepared with a G Nome DNA kit (Q-Biogene) as described in the manufacturer's instructions. Southern blotting was performed as described previously (28) with probes conjugated to horseradish peroxidase and detected with a chemiluminescent substrate (Amersham). The secondary integration sites of SXT were determined by performing arbitrary PCR as described by O'Toole and Kolter (22), with the following modifications. The first round of amplification was performed by using the arbitrary primer ARB1 (22) and the primer unique to the right end of SXT, SXTR1 (Table 2), in 50-μl PCR mixtures with a HotStarTaq Master Mix kit (Qiagen) and 30 ng of template DNA. The first-round PCR conditions were (i) 15 min at 95°C, (ii) 6 cycles of 20 s at 95°C, 30 s at 30°C, and 90 s at 72°C, (iii) 30 cycles of 20 s at 95°C, 30 s at 40°C, and 2 min at 72°C, and (iv) 5 min at 72°C. The second round of amplification was performed like the first round with 1 μl of the first-round reaction mixture as the template and primers ARB2 (22) and SXTR2, which was identical to the rightmost end of SXT (Table 2). The second-round amplification conditions were (i) 15 min at 95°C, (ii) 30 cycles of 20 s at 95°C, 30 s at 45°C, and 2 min at 72°C, and (iii) 5 min at 72°C. The purified PCR products were sequenced by using the SXTR2 primer at the Tufts Medical School DNA Sequencing Core Facility as described previously (29) and were compared with the GenBank DNA sequence database by using the BLASTN program (1).
Real-time quantitative PCR assays for relative quantification of attB and attP.
Real-time quantitative PCR assays were developed to measure the percentages of cells in a culture that contained unoccupied SXT attB sites and the SXT attP sequence (a measure of the amount of excised circularized SXT). The amounts of both attB and attP were normalized to the amount of chromosomal DNA in each sample; the latter quantity was determined by amplifying the 3′ end of prfC by using real-time quantitative PCR. An E. coli strain, VI61, containing single chromosomal copies of attB and attP, was constructed to calibrate the real-time quantitative PCR assays. Primer Express V1.5a (Applied Biosystems) was used to design primers EattBF and EattBR, primers SXTJ2F and SXTJ2R, and primers PrfCQF and PrfCQR, which were used for amplifying the 67-bp attB fragment, the 63-bp attP fragment, and the 66-bp prfC fragment, respectively (Table 2). The ABI PRISM 7700 sequence detection system (Applied Biosystems) was used to quantify the increase in fluorescence emission during PCR that resulted from binding of the dye SYBR Green I to double-stranded DNA. Sequence Detector software (version 1.7; Applied Biosystems) was used for data acquisition and analysis. Each 20-μl reaction mixture contained 10 μl of 2× SYBR Green PCR Master Mix (Applied Biosystems), each primer at a concentration of 500 nM, and 8 ng of the DNA template. The PCR conditions were (i) 2 min at 50°C, (ii) 10 min at 95°C, and (iii) 45 cycles of 15 s at 95°C and 1 min 60°C. Three reactions were performed for each sample. The mathematical model used for analysis of the data was developed by Pfaffl (23). Briefly, a standard curve was generated for each of the three primer sets by using VI61 DNA as the template by plotting the Ct values as a function of the concentration input of DNA (the Ct value was defined as the PCR cycle number at which the fluorescence rose above the background level). The PCR efficiencies (E) for particular primer sets were calculated by using the equation E = 10(−1/slope) (slopes were calculated from the standard curves). The relative ratio of the amplified att sequences was calculated based on E and the Ct deviation (ΔCt) of an experimental sample compared with the control sample (VI61 chromosomal DNA) and was expressed in comparison to the calibrator sequence (the 3′ end of prfC) by using the following equation:
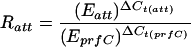 |
where Eatt is the PCR efficiency of the att site tested (attB or attP), EprfC is the PCR efficiency of the calibrator sequence (the 3′ end of prfC), ΔCt(att) is the difference between the Ct value of the reference strain and the Ct value of the strain tested [Ct(VI61) −Ct(sample)] for the att site tested, and ΔCt(prfC) is the difference between the Ct value of the reference strain and the Ct value of the strain tested [Ct(VI61) −Ct(sample)] for the calibrator sequence. For convenience, the ratio Ratt was expressed as a percentage (i.e., as the copy number of att sites per hundred chromosomes). The measured PCR efficiencies for the amplification of attB, attP, and prfC were 1.89, 2.00, and 1.97, respectively (ideally, the values are 2.00).
RESULTS
SetCD is required for int expression, and Int is sufficient to mediate SXT integration.
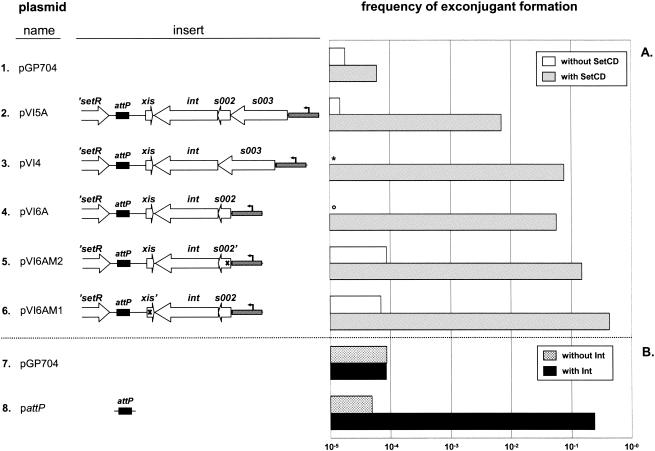
In previous studies, it was determined that int, which lies near one end of SXT, is required for SXT integration (17). int is part of a putative operon downstream of s002 and s003, two hypothetical genes that are apparently not required for SXT transfer (4, 17). We constructed several plasmids based on the mobilizable Apr suicide vector pGP704 (20) to investigate whether int is sufficient to mediate integration of the SXT attachment site (attP) into prfC. This vector requires the product of pir to replicate, so after its conjugative transfer from a pir+ host to a pir host, Apr exconjugants are isolated only following plasmid integration. The largest of the inserts in the plasmids (pVI5A) contained attP plus 3.6 kb of adjacent sequence, including int, s002, and s003 (Fig. 1A). The frequency of exconjugant formation for each of the plasmids tested did not differ from the background frequency of exconjugant formation observed with pGP704 (∼10−5) (Fig. 1A). This background frequency likely resulted from random integration of pGP704 into the recipient strain. Thus, the entire putative int-containing operon and attP were insufficient to promote site-specific integration of the plasmids into the chromosome. A control conjugation assay demonstrated that all the plasmids shown in Fig. 1 could be transferred at the same frequency to a pir+ host, indicating that their different inserts did not alter their capacities to be mobilized (data not shown).
FIG. 1.
Genes required for SXT integration. Integration of replication-deficient plasmids containing the SXT attP site was assessed by a conjugation assay. The inserts in these plasmids, all of which were derived from the mobilizable suicide vector pGP704, are indicated. In the diagrams of the inserts, open reading frames are indicated by arrows, the SXT attP site is indicated by a black box, the putative promoter region upstream of the int-containing operon is indicated by a gray box with an arrow, and the frameshift mutations are indicated by x's. The frequency of exconjugant formation was obtained by dividing the number of exconjugants (Tcr Apr CFU) by the number of recipients (Tcr CFU). In all the cases, the donor strain was E. coli SM10λpir and the recipient strains were E. coli CAG18439 derivatives. For the plasmids in panel A, the recipient cells harbored pSetCD33, which contained setCD under control of PBAD. For the plasmids in panel B, the recipient cells harbored pInt33, which contained int under control of PBAD. To induce expression of setCD or int, the conjugation assays were carried out by using media supplemented with 0.02% arabinose. The bars indicate the mean values obtained from two independent experiments. The asterisk and the circle in lines 3 and 4 indicate that the frequencies of exconjugant formation were less than 10−5 exconjugant/recipient; in line 3, the frequency was 9.7 × 10−6, and in line 4, the frequency was 7.0 × 10−6.
Since we previously reported that SetC and SetD stimulate the expression of s003 and presumably int as well (4), we hypothesized that expression of setCD in recipient cells might be required to promote integration of the plasmids described above. To test this idea, we introduced a SetCD expression vector into the recipient strain described above. When this new recipient was used, we observed an increase of nearly 3 orders of magnitude in the frequency of exconjugant formation with pVI5A, the plasmid containing the largest insert (Fig. 1A). Deletion of either or both of the hypothetical genes located 5′ of int, s002, and s003 did not reduce the SetCD-dependent formation of exconjugants (Fig. 1A, lines 3 to 5). In fact, pVI5A derivatives that lacked s002 and/or s003 appeared to integrate with greater efficiency than pVI5A, suggesting that s002 and/or s003 may decrease integration efficiency. Alternatively, expression of int from the presumed promoter upstream from s003 may have been altered in these plasmids. Overall, these results indicate that int-mediated integration of the SXT attP site requires SetCD in the recipient. This integration process is not dependent on homologous recombination in the recipient as similar results were obtained in a recA recipient (data not shown).
To assess whether SetC and SetD have a direct role in integration or function solely as regulators of int expression, we tested if the integrase itself, expressed in the recipient strain under control of an arabinose-inducible promoter, was sufficient to catalyze the integration of pattP, a pGP704-based plasmid containing a 600-bp fragment encompassing the SXT attP site. The frequency of exconjugant formation in a recipient expressing int was more than 3 orders of magnitude higher for pattP than for pGP704 (Fig. 1B). These results indicate that Int is the only SXT-encoded protein required to mediate SXT integration; the lack of integration initially observed in the experiments described above was most likely due to a lack of int expression. These observations also corroborate the idea that there is a SetCD-regulated promoter upstream of s003 that regulates int expression.
Int alone does not promote efficient excision.
Neither the annotated SXT (4) nor R391 (6) genome sequence was reported to encode a potential RDF (Xis-like protein). Since genes encoding Xis proteins are often located near int genes, we investigated whether pVI4, pVI5A, pVI6A, pVI6AM2, and pattP, once integrated into the chromosomes of the exconjugants described above, excised with equal efficiencies. To do this, we developed real-time quantitative PCR assays to measure the amounts of unoccupied attB sites and attP sequences (i.e., the amounts of excised circularized plasmid). In these assays, the amounts of attB and attP DNA were normalized to the copy number of chromosome DNA, which was determined by amplifying the 3′ end of prfC. The assays were also performed with a control SXT-free E. coli strain, strain VI61 (Table 1), which contains single chromosomal copies of attB and attP.
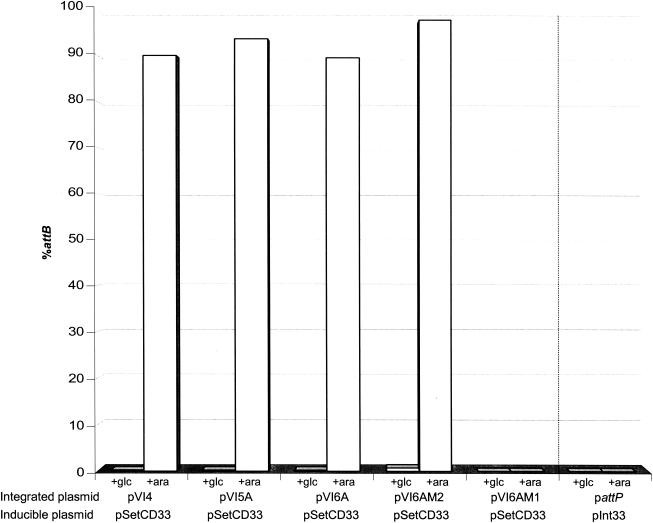
SetC and SetD were found to be required for excision of four of the five integrated plasmids, pVI4, pVI5A, pVI6A, and pVI6AM2 (Fig. 2). In the absence of these activators, virtually no attB was detected in cultures of cells containing the integrated forms of these plasmids. This was expected since int is required for SXT excision (17) and, as shown above, SetC and SetD are required for int expression. In the presence of SetCD, all four plasmids excised, and more than 90% of the cells contained an unoccupied attB (Fig. 2). Only a low percentage of attP was detected in these cells (data not shown), probably because the excised plasmids were lost by dilution due to an inability to replicate in a pir background. Thus, all four of these integrated plasmids contained the genes and sequences necessary and sufficient to mediate plasmid excision.
FIG. 2.
Xis augments excision of integrated SXT-derived plasmids. The amount of an unoccupied attB site was quantified by real-time quantitative PCR. The means of triplicate measurements performed with each sample are shown. E. coli CAG18439, which contained one copy of attB per chromosome, was used as a control strain. DNA templates were extracted from strains containing the integrated plasmids described in Fig. 1 and pSetCD33 or pInt33 expressing setCD or int under control of PBAD. Strains were grown overnight in LB medium supplemented either with 0.2% glucose (+glc) to repress expression or with 0.02% arabinose (+ara) to induce expression.
In marked contrast to the SetCD-activated excision of these four plasmids, we detected almost no Int-activated excision of integrated pattP. Int was necessary and sufficient for integration of this plasmid, which contained only the SXT attP site (Fig. 1B). The pronounced difference between the excision of pattP and the excision of the four plasmids discussed above suggests that Int alone is not sufficient to promote excision. Rather, a sequence absent from pattP and present in the other plasmids is necessary. Apparently, neither s002 nor s003 encodes this other function since pVI6AM2, which lacks both functional s002 and s003, excised at least as well as pVI5A (Fig. 2).
Identification of an SXT Xis.
The analyses described above suggested that a gene between the 3′ end of int and attL that facilitates SXT excision was overlooked in our annotation of the SXT genome. In fact, a small open reading frame of 195 nucleotides directed toward int was identified 14 bp downstream from the int stop codon by using the gene prediction program GeneMark 2.4 (7). This sequence is preceded by a suitable ribosome binding site (AGGAGA), which is located 6 bp upstream from the ATG start codon. The putative 64-amino-acid protein that we called Xis is predicted to have a basic pI (pI 8.10) and to contain a helix-turn-helix DNA-binding motif (Dodd-Egan score, 3.32 [13]).
We assessed the involvement of xis in SXT excision by introducing a frameshift mutation in xis in pVI6A, which yielded pVI6AM1. The frameshifted xis should have produced only the 11 N-terminal amino acids of the putative native Xis protein. In striking contrast to cells harboring integrated pVI6A, cells harboring integrated pVI6AM1 contained virtually no detectable unoccupied attB sites even in the presence of SetCD (Fig. 2). This observation demonstrated that xis plays a critical role in facilitating SXT excision. In addition, xis appeared to have a negative effect on integration, since pVI6AM1 formed exconjugants more efficiently than pVI6A formed exconjugants (Fig. 1A, lines 4 and 6). Together, these observations suggest that xis encodes an RDF.
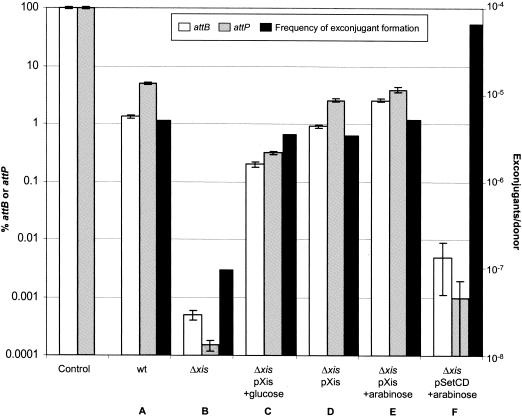
We further demonstrated that xis plays a critical role in excision of the entire SXT element. A Δxis derivative of SXT was constructed by using the one-step chromosomal gene inactivation technique (12). As shown in Fig. 3, the amount of attB and attP per chromosome was reduced by more than 3 orders of magnitude in the Δxis mutant of SXT compared to the amount in the wild-type SXT. Complementation analyses confirmed that xis promotes excision. As shown in Fig. 3, introduction of pXis, a plasmid containing xis under control of PBAD, into the Δxis SXT mutant markedly increased the percentage of cells containing the excised SXT element. Apparently, very few Xis molecules are required to enhance SXT excision, since the presence of pXis increased the percentage of cells containing attP and attB even in the absence of arabinose (Fig. 3). In contrast to the Xis-mediated increases in SXT excision observed in the Δxis SXT strain, overproduction of SetCD in this background led to only small increases in SXT excision. This suggests that activation of int expression is not sufficient to complement the excision deficiency in the Δxis background. Confirmation of this hypothesis was obtained when we found that overexpression of Int under control of PBAD did not enhance the frequency of excision of a wild-type SXT (data not shown).
FIG. 3.
Xis promotes excision and transfer of SXT. Real-time quantitative PCR was used to determine the percentage of unoccupied attB sites and attP sequences resulting from circularization of SXT. Triplicate measurements were obtained for each sample, and the mean and standard deviation are shown for each assay. The control strain E. coli VI61 (CAG18439 ΔlacZ::attP-cat) contained a single chromosomal copy of attB and attP. E. coli HW220 (CAG18439 prfC::SXT) was used as the wild-type (wt) strain. The Δxis strain was E. coli VI100 (CAG18439 prfC::SXT Δxis). DNA templates were prepared from overnight LB broth cultures containing 0.2% glucose or 0.02% arabinose where indicated. The frequencies of exconjugant formation were obtained by dividing the number of exconjugants (Nalr SXTr CFU) by the number of donor cells (Tcr CFU). In all the mating experiments, the recipient strain was E. coli BI533 (MG1655 Nalr). The solid bars indicate the mean values obtained from two independent mating experiments. The expression of xis by pXis and the expression of setCD by pSetCD in the donor strains were repressed with 0.2% glucose or were induced with 0.02% arabinose.
Deletion of xis from SXT or overexpression of xis in the recipient reduces SXT transfer.
The frequency of transfer of the Δxis SXT mutant was reduced by nearly 3 orders of magnitude compared to the wild-type SXT transfer frequency (Fig. 3), suggesting that circular excised SXT molecules are required for SXT transfer. However, unlike the frequency of transfer of a Δint mutant (17), the frequency of transfer was not zero for the Δxis mutant. This observation is consistent with the finding that there is a detectable, although greatly reduced, amount of attB in the Δxis SXT, suggesting that there is a very low level of Xis-independent SXT excision. The transfer frequency of the Δxis SXT could be restored to nearly wild-type levels by provision of Xis in trans from pXis (Fig. 3). However, there did not appear to be a direct correlation between the number of copies of excised circularized SXT per chromosome and the SXT transfer frequency. Overexpression of xis from pXis increased the percentage of cells containing detectable attB and attP to levels that were nearly 10-fold greater than those observed without xis expression, yet the SXT transfer frequency was increased only about twofold (Fig. 3C and E). In contrast, overexpression of setCD had a minimal effect on the percentage of cells containing attB and attP but led to a marked increase in SXT transfer (Fig. 3F). These results suggest that the frequency of SXT transfer is controlled both by regulation of SXT excision and by regulation, via SetCD, of the formation of the conjugal machinery.
The results of the plasmid transfer experiments discussed above suggested that Xis may lower the efficiency of exconjugant formation by reducing integration into the recipient chromosome (Fig. 1, lines 4 and 6). Further support for the idea that Xis negatively influences SXT integration was provided by experiments in which xis was overexpressed from pXis in recipient cells. We found that the presence of Xis in recipients reduced the frequency of exconjugant formation by about 700-fold (data not shown).
SXT integrates into alternate sites in the absence of prfC.
To examine whether SXT integration is limited to the prfC locus and whether integration at this site modulates SXT transfer, we constructed a ΔprfC mutant of E. coli MG1655. We first tested whether plasmid pattP (described in Fig. 1) integrates into this strain (Int was provided in trans in the recipient for these experiments). In the absence of prfC, pattP integration, which was measured by the frequency of exconjugant formation, was reduced by more than 2 orders of magnitude (Table 3). Despite this dramatic reduction in pattP integration into the ΔprfC background, this plasmid integrated more efficiently than pGP704 integrated (Table 3). The presence of recA in the recipient strain did not augment integration of pattP, suggesting that Int-mediated recombination of pattP with alternative attB sites accounts for integration of the plasmid in the ΔprfC background.
TABLE 3.
Effect of the ΔprfC mutation on the frequency of exconjugant formation
| Donor straina | Recipient strainb
|
Frequency of exconjugant formation (10−5)c | |
|---|---|---|---|
| recA | prfC | ||
| pGP704 | + | + | 8 |
| pattP | + | + | 25,000 |
| − | + | 43,000 | |
| − | − | 205 | |
| + | − | 78 | |
| SXT | + | + | 44 |
| + | − | 0.1 | |
The donor strain was E. coli SM10λpir containing either pGP704 or pattP or E. coli BW25113 prfC::SXT.
The recipient strains were the MG1655-derived strains CAG18439 (recA+) and KB1 (recA) and their ΔprfC derivatives. When pGP704 and pattP were used, the recipient strains expressed Int from pInt33, as shown in Fig. 1.
The frequency of exconjugant formation was calculated by determining the number of exconjugants per recipient and is the mean from two independent experiments. The mating experiments were carried out for 6 h at 37°C.
SXT integration into ΔprfC recipients was also reduced by more than 2 orders of magnitude (Table 3). Since SXT is unable to replicate autonomously, the exconjugants recovered in these experiments likely represented integration into alternative chromosomal attachment sites. The SXT integration sites in two of these exconjugants (VI141 and VI152) were determined by sequencing the junctions between the right end of SXT and the chromosome. The resulting sequences were then compared to the complete sequence of the genome of E. coli K-12 (5). In the case of VI152, SXT was integrated into the putative gene ysgA, which encodes a protein of unknown function. SXT integration totally disrupted the open reading frame of ysgA, suggesting that it was no longer functional in VI152. In the case of VI141, SXT was integrated into the 5′ end of pntB, the last gene of the pnt operon, which encodes a pyridine nucleotide transhydrogenase. As a result of the integration, 30 amino acids from the N terminus of the predicted protein should have been replaced by 21 amino acids provided by SXT. Interestingly, in the absence of prfC, the 5′ end of pntB appeared to be a preferred alternate SXT integration site, since Southern analysis revealed that SXT was integrated into this site in 5 of the 12 SXTr exconjugants tested; all the other alternate integration sites were unique (data not shown).
The frequency of SXT excision from the secondary integration site pntB in VI141 was about 3.5-fold lower than the frequency of SXT excision from prfC in HW220. Whereas HW220 contains about 2.19% circular SXT molecules, VI141 was found to contain only 0.62% circular SXT molecules. Thus, the frequency of SXT excision seems to be modulated by the sequence within which SXT is integrated. The reduction in SXT excision from VI141 may at least in part explain the 10-fold reduction in the frequency of SXT exconjugants observed when VI141 was used as a donor (3.3 × 10−5 exconjugant/donor) compared to the frequency when HW220 was used as a donor (3.0 × 10−4 exconjugant/donor). All eight SXT exconjugants derived from the VI141 donor were found to harbor SXT integrated into the 5′ end of prfC, its primary integration site (data not shown). Thus, integration of SXT into pntB in the donor did not alter the specificity for the 5′ end of prfC in the recipient strain.
DISCUSSION
SXT integration into and excision from the chromosome of host cells are critical steps in SXT maintenance and transmission. Int, the SXT integrase, is closely related to λ Int and like this phage recombinase was found to be necessary and sufficient for SXT integration. SetC and SetD, the SXT-encoded transcription activators, were also found to be required for int expression and thus for SXT integration. Int was also required for SXT excision (17), but it was not sufficient for this process. Instead, as is the case for numerous prophages and ICEs (19), another protein, designated Xis, was also needed for efficient SXT excision. Xis not only promoted SXT excision but also appeared to inhibit SXT integration, suggesting that this novel gene product functions as an RDF. Although SXT Xis is not related to any known RDF, like λ Xis it is predicted to be a small basic protein. It may therefore function like λ Xis to alter the architecture of the integrase-DNA complex. Xis-mediated SXT excision and SetCD-activated production of the SXT conjugal machinery appear to be independently controlled steps that regulate SXT transfer.
Int was necessary and sufficient to promote integration of mobilizable plasmids carrying the SXT attP site. s003 and s002, which are 5′ of int in SXT, were not required to promote integration of these plasmids, and their functions remain obscure. SetC and SetD were required in recipient cells to activate int expression from plasmids harboring attP, int, and the presumed SetCD-activated promoter upstream of s003; this result is consistent with the idea that s003, s002, and int constitute an operon. The requirement for SetCD in recipient cells to mediate integration of these plasmids suggests that SetCD-activated de novo expression of int from SXT transferred into recipient cells is required for SXT integration into the chromosome. Since it was previously shown that int is required in both donor and recipient cells for SXT transfer (17), such a requirement for int expression in recipient cells differs from the requirement for Tn916, where Int is thought to be transported along with Tn916 DNA through the mating pore into recipient cells (8). Our findings may suggest that setC and setD are transcribed very soon after SXT enters recipient cells. However, other experiments suggest that in the transfer of the entire SXT element, setC and setD are not required in the recipient cells (J. W. Beaber and M. K Waldor, unpublished results). This finding indicates that activation of int expression may result either from the transfer of SetCD from the donor to recipient cells or from the early transcription of another activator encoded by SXT and absent from the inserts in the plasmids used in this study.
Since the RDFs that have been characterized are a highly diverse group of small proteins (19), it is not surprising that we failed to annotate the SXT xis gene in our initial annotation of the SXT genome (4). The SXT Xis protein is unrelated to any previously described Xis protein and does not belong to any of the RDF subgroups outlined by Lewis and Hatfull (19). However, orf4 from R391, an ICE closely related to SXT (4, 6, 14), is virtually identical to xis and is located in the same relative position in the R391 genome as xis is in SXT. Our experimental analyses indicate that orf4 functions like SXT xis and that the SXT xis gene restored wild-type levels of excision and transfer to an R391 Δorf4 mutant (data not shown).
By using BlastP, five small hypothetical proteins from Mycobacterium leprae, Vibrio cholerae, Sinorhizobium meliloti, Mesorhizobium loti, and Brucella melitensis were found to be similar to Xis, exhibiting 34, 28, 45, 41, and 45% identity to Xis, respectively. Like Xis, all but one of these putative proteins was predicted to contain an N-terminal helix-turn-helix motif by the Dodd-Egan method (13). In addition, a ClustalW alignment of Xis with these proteins indicated that their predicted helix-turn-helix domains are related (Fig. 4). Since the genes encoding RDFs are often linked to genes encoding integrases in mobile elements, we looked for recombinase-encoding genes near the xis-like genes. Whereas no recombinase genes were close to ml2429 of M. leprae and vc1785 of V. cholerae, the three genes encoding the other three hypothetical Xis-like proteins were each closely linked to three different genes encoding related CP4-like tyrosine recombinases. This observation provides further evidence that these three xis-like genes encode RDFs. The xis-like gene of S. meliloti and the linked putative int gene are part of an unannotated 20,494-bp integrative element. This element is apparently integrated into the 3′ end of a tRNAThr gene, since 14 bp of the 16-bp direct repeat that flanks the element constitutes the 3′ end of this tRNA. The SXT Xis and the five Xis-like proteins that we identified may constitute a new subgroup of RDFs distinct from the 11 subgroups described by Lewis and Hatfull (19).
FIG. 4.
ClustalW alignment of SXT Xis with five hypothetical proteins from V.cholerae (VC1785), M.leprae (ML2429), S.meliloti (SMc02201), M.loti (msr5461), and B.melitensis (BMEI1700). Amino acid residues that are identical in at least two-thirds of the sequences are indicated by a black background, and residues that are similar in at least two-thirds of the sequences are indicated by a grey background. The bar indicates the predicted helix-turn-helix (HTH) DNA-binding motif in SXT Xis, VC1785, ML2429, SMc02201, msr5461, and BMEI1700. These peptides have Dodd-Egan scores of 3.32, 4.13, 4.24, 2.57, 4.70, and 2.26, respectively.
In most of the integrating mobile elements that have been studied, xis and int are part of the same operon and thus are cotranscribed. In SXT, xis and int are convergent genes that do not appear to be coregulated. While int expression requires SetCD, this does not seem to be the case for xis. The β-galactosidase activity of a lacZ fusion integrated at xis in SXT increased less than twofold following expression of setCD (data not shown).
As both Xis and Int are required for SXT excision, factors that influence the amounts and/or activities of these proteins are likely to control SXT excision, at least in part. As in SXT integration, s002 and s003 do not appear to have a significant effect on SXT excision. However, our data suggest that there may be an SXT-encoded factor that reduces Xis production or activity. We observed a much larger SetCD-mediated increase in the percentage of cells containing an unoccupied attB in the experiments with integrated plasmids (Fig. 2) than in the experiments with the entire integrated SXT (Fig. 3 and data not shown). A potential explanation for this difference is that SXT encodes a factor that is absent from the integrated plasmids that negatively regulates the Xis function. In λ, Xis levels are controlled by the Lon and FtsH proteases (18). Interestingly, SXT encodes a Lon orthologue (s039), but we do not know yet whether this presumed protease affects Xis levels. Besides the amounts and activities of Xis and Int, the site of SXT integration also appears to influence SXT excision. We observed a small reduction in SXT excision (3.5-fold) when SXT was integrated at pntB in a ΔprfC host.
While SXT excision is required for SXT transfer, the number of excised circular SXT molecules does not ordinarily appear to be a major factor limiting SXT transfer. We found that about 5% of cells grown in rich medium contained the SXT attP, yet the frequency of SXT transfer from these cells was less than 10−5 (Fig. 3A). The discrepancy between these frequencies indicates that most cells that harbor an excised circular SXT molecule do not act as SXT donors. Our data suggest that the factors that strongly influence SXT transfer are SetCD regulated since overexpression of these transcription activators greatly enhanced SXT transfer, even in the Δxis SXT background (Fig. 3F), in which the percentage of cells harboring the SXT attP was greatly reduced. Presumably, the SetCD-regulated factor is the conjugative machinery, as all of the SXT tra genes that have been characterized are regulated by SetCD (4). The apparently distinct control of the formation of circular SXT molecules and the SXT conjugative machinery contrasts with the control of Tn916 excision and conjugation. In this well-characterized ICE, transcription of xis, int, and tra genes is usually coupled, since tra gene expression requires Tn916 excision (21).
It was hypothesized previously that SXT integration into prfC may influence SXT function since prfC encodes RF3, a protein involved in regulation of translation (17). Integration of the E. coli pathogenicity island Pai I into selC (encoding a selenocysteine-specific tRNA) has been shown to have functional implications for the regulation of Pai I-encoded genes (25a). We found that SXT has a relaxed specificity of integration when prfC, the primary integration site, is absent. At least nine different secondary SXT integration sites were detected, and one of them, the 5′ end of the pntB gene, is a preferred target. SXT integrated at pntB in a ΔprfC background was still transmissible, indicating that RF3 is not required for either SXT excision or SXT transfer. However, RF3 may have subtle effects on SXT transfer. The strain harboring SXT integrated into pntB exhibited about a 10-fold reduction in the ability to transfer SXT compared to the ability of a strain containing SXT integrated into prfC. It is possible that the absence of RF3 from the pntB::SXT strain may account for this reduction. The site of integration of other ICEs, such as Tn916, has also been shown to influence the transmissibility of these elements (21).
The existence of alternative SXT attachment sites has potential implications for the transmissibility of SXT, as well as chromosomal sequences. Alternative attachment sites may broaden the SXT host range to include organisms that do not have conserved prfC orthologues. Finally, since SXT is able to mobilize chromosomal DNA in an Hfr-like manner, the existence of several alternative SXT integration sites may allow mobilization of genes located at virtually all positions in many prokaryotic genomes.
Acknowledgments
We are grateful to B. Davis, S. McLeod, A. Kane, and J. Beaber for critically reading the manuscript.
This work was supported by funds from NIH grant AI42347, the Howard Hughes Medical Institute, and the NEMC GRASP Center (grant P30DK-34928).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Azaro, M. A., and A. Landy. 2002. The integration/excision cycle of lambda and other bacteriophages, p. 117-148. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambovitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 3.Beaber, J. W., V. Burrus, B. Hochhut, and M. K. Waldor. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene prediction for both DNA strands. Comput. Chem. 17:123-133. [Google Scholar]
- 8.Bringel, F., G. L. Van Alstine, and J. R. Scott. 1992. Conjugative transposition of Tn916: the transposon int gene is required only in the donor. J. Bacteriol. 174:4036-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 10.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 11.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48:827-838. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 18.Leffers, G. G., Jr., and S. Gottesman. 1998. Lambda Xis degradation in vivo by Lon and FtsH. J. Bacteriol. 180:1573-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullany, P., A. P. Roberts, and H. Wang. 2002. Mechanism of integration and excision in conjugative transposons. Cell. Mol. Life Sci. 59:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed]
- 24.Ravatn, R., S. Studer, A. J. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Ritter, A., G. Blum, L. Emody, M. Kerenyi, A. Bock, B. Neuhieri, W. Rabsch, F. Scheutz, and J. Hacker. 1995. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol. Microbiol. 17:109-121. [DOI] [PubMed] [Google Scholar]
- 26.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldor, M. K., H. Tschape, and J. J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]