Abstract
Clostridium thermocellum produces an extracellular multienzyme complex, termed the cellulosome, that allows efficient solubilization of crystalline cellulose. The complex is organized around a large noncatalytic protein subunit, termed CipA or scaffoldin, and is found either free in the supernatant or cell bound. The binding of the complex to the cell is mediated by three cell surface anchoring proteins, OlpB, Orf2p, and SdbA, that interact with the CipA scaffoldin. The transcriptional level of the olpB, orf2, sdbA, and cipA genes was determined quantitatively by RNase protection assays in batch and continuous cultures, under carbon and nitrogen limitation. The mRNA level of olpB, orf2, and cipA varied with growth rate, reaching 40 to 60 transcripts per cell under carbon limitation at a low growth rate of 0.04 h−1 and 2 to 10 transcripts per cell at a growth rate of 0.35 h−1 in batch culture. The mRNA level of sdbA was about three transcripts per cell and was not influenced by growth rate. Primer extension analysis revealed two major transcriptional start sites, at −81 and −50 bp, upstream of the translational start site of the cipA gene. The potential promoters exhibited homology to the known sigma factors σA and σL (σ54) of Bacillus subtilis. Transcription from the σL-like promoter was found under all growth conditions, whereas transcription from the σA-like promoter was significant only under carbon limitation. The overall expression level obtained in the primer extension analysis was in good agreement with the results of the RNase-protection assays.
Clostridium thermocellum, a thermophilic, gram-positive, anaerobic bacterium, produces a highly active cellulase complex, termed the cellulosome, that mediates the efficient hydrolysis of crystalline cellulosic substrates (5, 6, 24, 44, 45). The enzymatic subunits of the cellulosome are incorporated into a scaffolding protein or scaffoldin, named CipA, which also promotes binding of the complex to cellulose (15, 16, 35, 42, 48). The CipA protein contains nine highly similar cohesin domains that recognize and bind tightly to complementary dockerin domains harbored by each of the catalytic subunits (13, 16, 47).
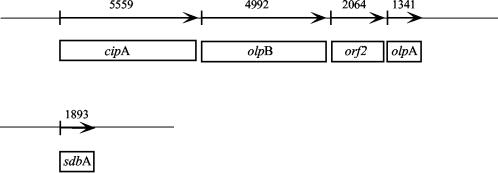
The cellulosome is present both in an exocellular form, distributed in protuberance-like structures located on the outer layer of the bacterial cell surface, and in an extracellular form, released into the culture medium in the latter stages of growth (3, 4, 9, 20, 32). Attachment of the cellulosome to the cell surface is thought to be mediated by at least three anchoring proteins, OlpB, Orf2p, and SdbA (14, 29, 40). These polypeptides contain a triplicate motif at their C-terminal region, which is similar to the S-layer homology domain shown previously to be responsible for the attachment of various proteins to the cell surface of gram positive bacteria (30, 31). In the case of OlpB, the involvement of the S-layer homology domain in the attachment to the cell surface was verified experimentally (29). On this basis, it was also deduced that Orf2p and SdbA would be similarly located on the cell surface (27, 28). SdbA, Orf2p, and OlpB carry one, two and four cohesin domains, respectively, at their N-termini (14, 27). This type of cohesin binds to the dockerin present on CipA (27, 28), but fails to bind to the enzyme-borne dockerin domains (41). The interaction of the CipA dockerin with the cohesins of the anchoring proteins was referred to as type II, in order to distinguish it from the type I interaction that serves to incorporate the enzyme subunits into the scaffoldin subunit (27). The cipA, olpB, and orf2 genes are located in tandem on the C. thermocellum chromosome (14), as opposed to sdbA, which is located elsewhere on the genome (Fig. 1).
FIG. 1.
Organization of C. thermocellum scaffoldin-related genes. cipA and the three genes olpB, orf2, and olpA are located in tandem within a gene cluster. sdbA is located on a distinct region of the chromosome. The proteins OlpB, Orf2p, and SdbA serve as anchoring proteins that connect the CipA scaffoldin to the cell surface. In contrast, OlpA selectively binds individual cellulases to the cell surface. The numbers above the gene in the scheme refer to its size (in base pairs).
Previous studies have demonstrated that cellulosomal enzymes are regulated by the growth conditions that influence the activity of the entire complex (4, 11, 12, 17, 22, 25, 34, 39). Moreover, differences in the disposition of cellulosomes on the cell surface of cellulosome-producing bacteria have been observed as a function of the medium or growth conditions (2, 8, 32). It has also been noted that the cellulosomal enzymes of C. thermocellum were invariably present in the complexed form (4). It thus follows that the scaffoldin and anchoring proteins may be regulated by the growth conditions, and that these structural cellulosomal components may subsequently influence the overall ability of the bacteria to degrade crystalline cellulosic substrates.
In this work, we studied the regulation of expression of the scaffolding gene, cipA, and the three anchoring proteins olpB, orf2, and sdbA at the transcriptional level, under different growth conditions. It was found that in addition to the scaffoldin gene, two of the genes for the anchoring proteins (olpB and orf2) are regulated by growth rate.
MATERIALS AND METHODS
Organism, substrates, and culture conditions.
C. thermocellum YS was originally isolated from soil samples obtained at the hot springs of Yellowstone National Park in North Dakota (1, 25, 26). Cells were grown in batch culture at 60°C in Duran anaerobic bottles (Schott, Mainz, Germany), in medium containing the following (per liter): 0.65 g of K2HPO3.3H2O, 0.5 g of KH2PO4, 1.3 g of (NH4)SO4, 42 g of morpholinepropanesulfonic acid (MOPS), 5 g of yeast extract, 1 g of cysteine, 0.5 g of MgCl2, and 2 mg of resazurin. The medium included 1.0% of the desired carbon source—either cellobiose from Acros Organics (Geel, Belgium) or microcrystalline cellulose (Avicel), obtained from Teva-Pharmaceutical Industries (Kfar Sava, Israel). Before measuring turbidity, cellulose-grown cultures were vortexed vigorously and centrifuged at 100 × g for 1 min to remove the insoluble substrate. Continuous cultures were performed in a BIOFLO 3000 fermentor (New Brunswick Scientific, Edison, N.J.) in 1.5-liter working volume, at 60°C. pH was maintained at 7.2 by automatic addition of 5 N NaOH. Agitation was kept constant at 100 rpm. To maintain anaerobic conditions, the headspace of the bioreactor contained 99.99% CO2 for initial growth and then changed to 99.99% N2 while starting the continuous process. Continuous cultures were operated under conditions of either cellobiose (2 g/liter) or nitrogen [(NH4)SO4, 0.04 g/liter] limitation, whereby C. thermocellum was adapted to different growth rates. The residual cellobiose concentration at steady state was assessed by the reducing sugar assay using the dinitrosalicylic reagent (33). The cellobiose concentration correlated, as expected, to the dilution rate. Typical values were 0.22, 0.17 and 0.14 g/liter at dilution rates of 0.23, 0.19, 0.15 h−1, respectively. Note that C. thermocellum cannot utilize nitrogen or carbon from the yeast extract. The establishment of steady-state conditions was assumed when the culture had been growing for a period of at least three generations, whereby the cell density (monitored spectrophotometrically) remained unchanged for at least one generation.
RNA extraction.
Precipitated cells, derived from 10 ml of culture medium, were washed twice with 10 mM Tris-HCl buffer, pH 7.5. The pellets were snap-frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted from cells, using the RNeasy kit (Qiagen GmbH, Hilden, Germany) with minor modifications as described earlier (11). The RNA concentration was estimated by measuring A260, and the resultant RNA preparations were stored in aliquots at −80°C.
RPA.
The RNase protection assay (RPA) was performed using the RPAII kit (Ambion Inc., Austin, Tex.). Different amounts of RNA were hybridized with 32P-antisense probes, and the protected RNAs were placed directly in a scintillation counter for quantification. For visualization, the protected RNA was then separated on 5% polyacrylamide gels, containing 7 M urea, and visualized using a phosphorimager system. The probes were labeled using the Maxiscript in vitro transcription kit (Ambion), with T7 RNA polymerase. The appropriate fragments were cloned either into pGEM (Promega, Madison, Wis.) or pBluescript II KS(+) (Stratagene, La Jolla, Calif.) vectors. The different probes included the following DNA fragments (position relative to the initial ATG): The orf2 probe (from −160 to + 234 bp), the sdbA probe (from −115 to + 245 bp), the olpB probe (from −129 to + 399 bp), and the cipA probe (from + 1226 to 1495 bp).
Determination of the amount of RNA per cell.
The amount of RNA was established for each of the cultures wherein the transcript level of the genes was determined. The pellet derived from a 30-ml culture was washed with 10 mM Tris-HCl, pH 7.6, and resuspended in 10 ml of 10% cold trichloroacetic acid (TCA). The suspension was kept on ice for 30 min. The supernatant fluids were discarded after 10 min of centrifugation at 10,000 × g. Cells were then resuspended in 5% TCA, centrifuged, and the pellet was dissolved gently in 1.5 ml 0.1 N NaOH using a sealed Pasteur pipette. The samples were incubated overnight at 37°C to allow complete hydrolysis of RNA. The solution was neutralized by 1 ml of 10% TCA, incubated 15 min on ice, and centrifuged. To determine the RNA concentration in the sample, 1 volume of appropriate diluted supernatant fluid was mixed with 1 volume of orcinol reagent (1% orcinol dissolved in 0.1% FeCl3 in concentrated HCl), and the solution was boiled for 15 min. After the tubes were cooled under running tap water, 2 volumes of distilled water was added. The absorbance was recorded at two wavelengths—600 nm (background) and 660 nm (background plus green complex). To determine the RNA or nucleotide concentration, a deoxyadenosine solution of 50 μg/ml was used as a standard. Since the results are obtained as purine riboside equivalents of RNA, the values were doubled to obtain nucleotide equivalents (purines plus pyrimidines) and multiplied by the average nucleotide molecular weight. To determine the amount of total RNA per cell, cells were counted in a microscope using a Petroff-Hausser Counter Chamber. The number of transcripts per cell of each gene was then calculated from the resultant values of counts per minute per μg of total RNA, the specific activity of the probe, the molecular weight of the probe, and the average amount of total RNA determined per cell.
Northern blotting.
Northern hybridization of the desired-mRNA was preformed according to Sambrook et al. (43). The RNA was denatured in the presence of formamide and formaldehyde; 15 μg of total RNA was loaded and subjected to electrophoresis on a 1% agarose gel containing 6% formaldehyde. The separated RNA was blotted onto a nitrocellulose membrane (Schleicher & Schuell, Inc., Keene, N.H.). The radioactive probes were prepared by random-primer labeling, using the construct described above for RPA.
Primer extension.
Primer extension with reverse transcriptase (avian myeloblastosis virus reverse transcriptase in the presence of RNasin RNase inhibitor [Promega]) was performed according to the manufacturer's instructions. A synthetic oligonucleotide probe (5′-CATCTACCATTCCTCCC-3′) was end labeled by T4 polynucleotide kinase (Fermentas, Hanover, Md.) and hybridized to a complementary coding region downstream of the ATG translation start site of the cipA gene. A 50-μg sample of mRNA, obtained from the desired cell culture, was heated for 1 min at 90°C and hybridized for 2 min at 60°C with approximately 500,000 cpm of the labeled probe. Extension with reverse transcriptase was performed essentially as described by Moran (36). Products of primer extension were analyzed by 6% acrylamide-8 M urea sequencing gel, together with sequencing reactions derived from the same primer.
RESULTS
The effect of carbon source and growth phase on the transcript level of the genes for scaffoldin and anchoring proteins.
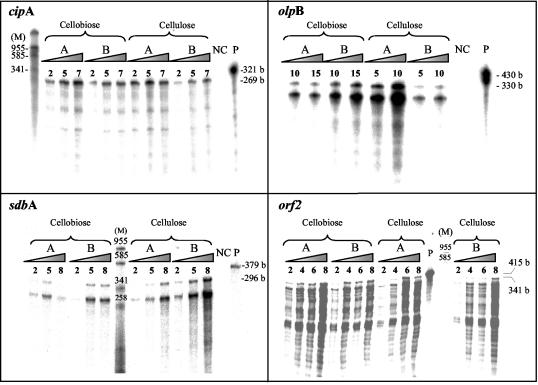
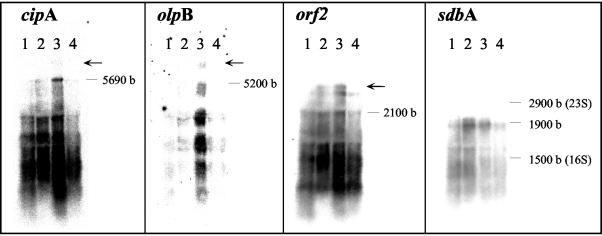
The expression of the scaffolding gene, cipA, and the genes for the three anchoring proteins, olpB, orf2, and sdbA, was determined at the transcriptional level in cultures grown either on cellobiose or cellulose. Cells were harvested during the exponential and the late-exponential phases, when culture turbidities (A660) reached 0.85 and 1.8, respectively, for cellobiose-grown cells, and 0.65 and 2.0, respectively, for cellulose-grown cells. RPA was used to assess the mRNA level of each gene. In order to quantify the results and to ensure that the amount of the probe in the hybridization reaction was not limiting, different amounts of the isolated RNA were hybridized with each type of 32P-labeled antisense probes. Following digestion with RNases, the RPA product was precipitated, the supernatant fluids were removed, and the radioactivity was assessed in a scintillation counter. The protected product was separated on a polyacrylamide gel for visualization by a phosphorimager system (Fig. 2). A linear correlation was obtained between the amount of total RNA used in the assay and the radioactivity of the protected product (data not shown). The number of transcripts per cell of each gene was calculated, and the results are presented in Table 1. To confirm that the antisense RNA probes used in the RPA interacted with the expected transcribed genes, Northern blot analyses were performed with DNA-labeled probes. Appropriate mRNA transcripts of 5,559 nt, 5,200 nt, 2,100 nt, and 1,900 bases were labeled with the probes for cipA, olpB, orf2, and sdbA, respectively (Fig. 3). In addition, larger-than-anticipated transcripts were labeled with both the olpB probe and especially the orf2 probe. In the case of olpB, these results may reflect read-through via the cipA transcriptional terminator. Indeed, a similarly sized, faint transcript can be discerned with the cipA probe. In the case of orf2, the larger transcript observed supports earlier evidence for cotranscription of orf2 together with the olpA gene located immediately downstream (14).
FIG. 2.
Transcript levels of the cipA gene and the genes coding for the cipA-specific anchoring proteins, olpB, orf2, and sdbA. RPAs were performed with the relevant 32P-labeled antisense probe as described earlier (11), using RNA from exponential (A) or stationary (B) phase cultures, grown on either cellobiose or crystalline cellulose. Different amounts (in micrograms) of RNA hybridized with the probes are indicated at the top of each lane. Autoradiographs of the respective RPA products were visualized using a phosphorimager system. Negative control lanes (NC) contained RNA from yeast. Lane M corresponds to markers (in base pairs). The full-length probe is shown in lane P. Values at right indicate the sizes of the undigested full-length antisense probes and the estimated sizes of the protected products in bases (b).
TABLE 1.
Transcript levels of the scaffoldin gene, cipA, and the genes encoding for anchoring proteins olpB, orf2p, and sdbA in C. thermocellum cultures.
| Gene | No. of transcripts/cella
|
|||
|---|---|---|---|---|
| Cellobiose
|
Crystalline cellulose
|
|||
| Exponential | Late exponential | Exponential | Late exponential | |
| cipA | 9 | 8 | 23 | 7 |
| olpB | 2 | 4 | 11 | 3 |
| orf2p | 13 | 14 | 19 | 11 |
| sdbA | 3 | 4 | 4 | 6 |
Values represent the average ± 15% standard error of at least three determinations of the mRNA level.
FIG. 3.
mRNA expression of cipA and scaffoldin-related genes as determined by Northern blot analysis. Northern blot analysis was performed using total RNA isolated from cultures grown on either cellobiose (lanes 1 and 2) or crystalline cellulose (lanes 3 and 4), during exponential (lanes 1 and 3) and stationary (lanes 2 and 4) phases of growth. The isolated RNA (15 μg) was loaded and separated on agarose gels and then transferred to nitrocellulose membranes. The blot was hybridized subsequently with cipA, olpB, sdbA, and orf2 probes. The sizes of the16S and 23S rRNA and the estimated sizes of the transcripts are indicated in bases (b). Arrows denote larger-than-expected sized transcripts labeled by the designated probe (see text for details).
During the exponential phase of growth, comparison of cellulose- and cellobiose-grown cells revealed that the expression of cipA and olpB was more than two- and fivefold higher, respectively, in the case of the former substrate. For cipA, the transcript level was 23 transcripts per cell in cellulose-grown cells versus 9 in cellobiose-grown cells, whereas the corresponding levels of olpB were 11 and 2. The difference in the transcript level of orf2, however, was less pronounced on cellulose- than on cellobiose-grown cells (i.e., 19 transcripts/cell versus 13 transcripts/cell). In contrast, no significant difference was observed in the transcript level of sdbA under the latter growth conditions. Apparently the expression of the cipA, olpB, and orf2 genes is influenced by the availability of the carbon source or the growth rate. Maximal growth rates achieved on cellobiose and cellulose were 0.35 h−1 and 0.23 h−1 (i.e., doubling times of 2 and 3 h), respectively. Accordingly, it would be anticipated that the decrease in cellobiose concentration (i.e., growth rate) at the late exponential phase would be accompanied with a further increase in the transcript level of these genes. However, the level of transcripts failed to increase when cells entered the stationary growth phase in both cellobiose- and cellulose-grown cells. The results contradict the likelihood that transcription of cipA and orf2 or even olpB is regulated solely by cellobiose concentration (i.e., growth rate), and other factors may contribute to transition-state regulation. Since in batch cultures, the growth rate and the composition of the medium change constantly when cells enter the stationary phase, this approach would limit our capacity to identify the factor(s) involved in gene regulation. Consequently, we resorted to the use of continuous culture that allows us to obtain defined growth conditions at steady state.
Transcript level in chemostat limited on cellobiose.
To further investigate the transcriptional regulation of genes involved in the cell-surface anchoring of the cellulosome in C. thermocellum, chemostat studies were conducted. Using this system, the expression of cipA and the genes encoding for the various anchoring proteins was determined under defined steady-state conditions. The continuous culture was operated under limiting concentrations of cellobiose at dilution rates between 0.04 to 0.21 h−1 at cell turbidities (A660) in the range of 0.5 to 0.7.
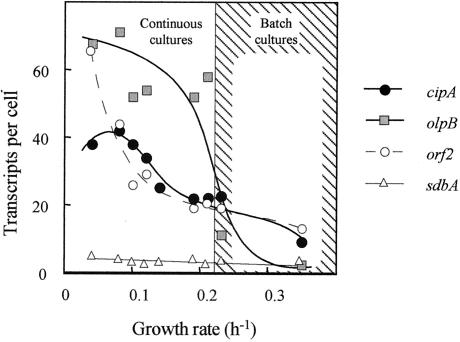
As shown in Fig. 4, the level of transcripts obtained for olpB was about fivefold higher in continuous culture operating at 0.04 to 0.21 h−1 than that of cells grown on cellulose during the exponential phase at a growth rate of 0.23 h−1. In the continuous culture, only slight differences in the cellular transcript level were observed at the different dilution rates. It appears that at a critical rate of growth—in the region of 0.21 h−1—the transcription of the olpB gene approaches its maximum level (about 60 transcripts per cell). In contrast to olpB, the transcript level of cipA and orf2 increased gradually with dilution rate. Thus, from a dilution rate of 0.21 h−1 to 0.04 h−1, the expression level of cipA increased from 22 to 38 transcripts per cell, and that of orf2 increased from 19 to 66 transcripts per cell (Fig. 4). In addition, no difference in the expression level of cipA and orf2 was observed between cellulose-grown cells and cells grown in continuous culture on cellobiose at the highest dilution rate (0.21 h−1). These results indicate that each of the genes can respond in a different manner to the environmental conditions of the growth culture. The transcript level of sdbA remained constant, independent of cellobiose concentration or growth rate. As in the batch cultures, the expression level was low (in the range of three to five transcripts per cell at the different dilution rates) compared to those observed for the other genes studied in this work.
FIG. 4.
Transcript levels of cipA and scaffoldin-related genes as a function of growth rate. The amounts of the respective mRNA were determined by RPA and converted to the number of transcripts per cell based on the average measured amount of total RNA in a single cell (0.18 pg/cell). Values represent the average of several (at least 3) measurements at an accuracy of ± 25%. Continuous cultures were operated under carbon (cellobiose) limitation at dilution rates between 0.04 ≤ μ ≤ 0.21; batch cultures were grown to exponential phase on either cellulose or cellobiose at rates of μ = 0.23 and 0.35 h−1, respectively.
Transcript level in continuous cultures under nitrogen limitation.
The results of the previous section indicate that the expression of cipA, olpB, and orf2 increases with a decrease in dilution rate. Since the change in dilution rate necessarily dictates corresponding changes in the cellobiose concentration in the fermentor, it cannot be ruled out that these genes are actually regulated by cellobiose concentration (i.e., catabolite repression). In order to exclude the possible influence of cellobiose concentration on the expression of these genes and to emphasize the influence of growth rate per se, continuous cultures were operated under conditions of nitrogen limitation. In this case, the carbon source (cellobiose) is present in excess independent of dilution rate. Indeed, the expression of cipA, olpB, and orf2 increases with the reduction of dilution rate (Table 2) —similar to that observed above for cultures under cellobiose limitation. The level of cipA and olpB increased more than threefold (8 versus 28 transcripts/cell and 9 versus 31 transcripts/cell, respectively) as a function of decreased growth rate from 0.14 h−1 to 0.07 h−1. As observed above, the expression of sdbA under nitrogen limitation was again not substantially influenced by the dilution rate.
TABLE 2.
Transcript levels of designated genes during continuous culture under nitrogen limitation
| Gene | No. of transcripts per cella at dilution rate (h−1)
|
||
|---|---|---|---|
| 0.14 | 0.11 | 0.07 | |
| cipA | 8 | 14 | 28 |
| olpB | 9 | 17 | 31 |
| orf2p | 10 | 14 | 25 |
| sdbA | 3.9 | 3.5 | 5 |
Values represent the average ± 15% standard error of at least three determinations of the mRNA level.
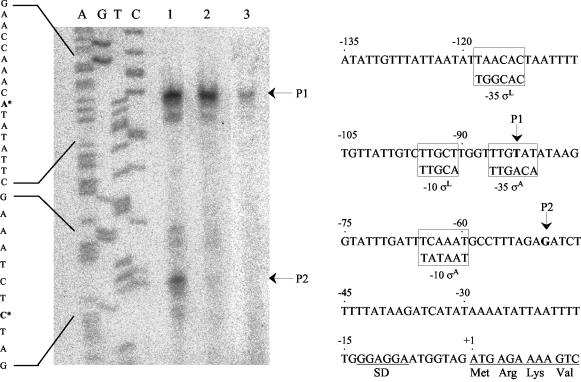
Transcriptional start site of cipA.
Primer extension analysis was used to locate the start site of transcription. As shown in Fig. 5 a major cipA transcript was detected in cells grown on either cellobiose (lane 3) or cellulose (lane 2), during the exponential phase of growth. In continuous cultures under conditions of cellobiose limitation, an additional transcript was observed (lane 1). The major transcription start site is nucleotide T located 81 bp upstream of the start codon. Potential binding sequences of σL are present upstream of the cipA transcriptional start site. The level of this transcript was higher in cellulose-grown cells than in cellobiose-grown cells, and similar to levels obtained in cells grown in continuous culture. These results are compatible with the RPA data. The additional transcript detected in the continuous culture starts with nucleotide G, located 50 bp upstream of the start codon. Homologous consensus sequences to the binding site of the Bacillus subtilis vegetative transcription factor (σA) were found upstream of this transcription start site.
FIG. 5.
Mapping of the 5′ terminus of cipA by primer extension analysis. (A) 32P-labeled oligonucleotide was hybridized to mRNA obtained from C. thermocellum grown under the following conditions: continuous culture limited on cellobiose, diluted to a rate of 0.1 h−1 (lane 1); exponential-phase culture grown on Avicel (lane 2); and exponential-phase culture grown on cellobiose (lane 3). Dideoxynucleotide sequence reactions were performed using the same primers employed in the reverse transcriptase reactions. The positions of the transcriptional start points are indicated by asterisks on the inferred non-template-strand sequences. The products of primer extension are indicated by P1 and P2. (B) Sequence data for the regulatory region of cipA. The respective transcriptional start points (P1 and P2) are indicated. The consensus B. subtilis σA and σL promoter sequences are framed at the homologous sites of the C. thermocellum sequence. The proposed Shine-Dalgarno (SD) site and the initiating ATG codon are indicated.
DISCUSSION
Previous studies have demonstrated that the expression of selected cellulosomal enzymatic subunits is influenced by growth conditions (4, 11, 34, 37), such that growth on cellobiose versus crystalline cellulose results in a different profile of cellulosomal components. In turn, the altered complement of enzymes appeared to affect the overall activity of the entire complex, since the cellulase activity of cellulose-grown cells was shown to be higher than that of cellobiose-grown cells (12, 17, 22, 38, 39). Our recent report (11) demonstrated that the transcription of the major enzymatic component of the complex, the exoglucanase CelS, is affected by the growth rate rather than cellobiose concentration per se. The results of the present study indicate further that the primary scaffoldin gene, cipA, and two of the three genes that encode for anchoring proteins (i.e., the genes for OlpB and Orf2p, responsible for attaching CipA to the bacterial cell-surface) are also regulated by growth rate at the transcriptional level.
The results of the cell growth experiments in chemostats under cellobiose limitation and in batch cultures would appear to support the notion that expression of the designated genes depends on growth rate and/or cellobiose concentration. Under conditions of exponential growth on cellobiose, the transcript levels of cipA, olpB, and orf2 were 9, 2, and 13 transcripts per cell, respectively, reaching corresponding maximal levels of 38, 60, and 66 transcripts per cell under conditions of cellobiose limitation. In other words, catabolite repression by cellobiose could conceivably be considered an additional mechanism by which transcription of those genes is governed.
Catabolite repression in gram-positive bacteria is mediated via binding of the CcpA repressor to the CRE consensus sequence TGWAAR(C/G)YTWNCW located in the promoter regions of the genes that are regulated by this system. A homologous CRE sequence could only be detected in the putative promoter region of orf2. The sequence TGTAAT(C/G)ATTTAT is located 420 bp upstream of the orf2 start codon. In the promoter region of olpB, only partial homology TGTACT(C/G)ACAACTT was found in the complement DNA strand (3′→5′), 18 bp upstream of the start codon. In this regard, we failed to amplify a ccpA gene from C. thermocellum genomic DNA using degenerated primers via PCR. BLAST analysis against the emerging genomic sequence of C. thermocellum (http://genome.ornl.gov/microbial/cthe/) again failed to detect a highly homologous ccpA gene, although several LacI/GalR homologous proteins were found. Taken together, it is likely that the known mechanisms of catabolite repression in gram-positive cells do not play a role in regulation of the currently studied cellulosome-related genes. However, it should be mentioned that the RegA protein in Clostridium acetobutylicum, although deviating from the CcpA consensus sequence (23), is capable of complementing a ccpA mutant in B. subtilis (10). Further support for regulation by growth rate comes from continuous culture under nitrogen limitation, whereby an increase in transcript level was still obtained with decrease in dilution rate. In fact, the final transcript levels were relatively high (28, 31, and 25 transcripts per cell).
Taken together with our previous report on the regulation of the celS gene (11), it appears that under conditions that support low growth rate, the bacterium employs a combined strategy to control cellulosomal components and their attachment to the cell surface, in order to improve its ability to utilize the preferred substrate. The first is accomplished by increasing the level of critical catalytic subunit(s) and the second by promoting the formation of new cell surface complexes by increasing the levels of CipA, OlpB, and Orf2p.
It should be noted that the three prospective anchoring proteins contain unequal numbers of cohesins that could interact with the CipA dockerin. OlpB harbors four cohesins, compared with the two of Orf2p and the single cohesin of SdbA. Under conditions of full occupancy, the latter anchoring proteins would presumably have the capacity to bind to the corresponding numbers of CipA molecules and its nine associated enzymes. However, perusal of our data indicates a striking disparity in the number of cipA transcripts versus the number of cohesins presumably available for the anchoring of CipA to the cell surface. Based on the relative number of transcripts and the number of cohesins carried by each anchoring protein (assuming a direct relationship between mRNA and protein levels), there is a 10-fold excess of cohesins under conditions of very low growth rate.
When cells were cultivated under conditions of high growth rates (i.e., exponential growth on cellobiose) the Orf2p anchoring component appeared to be the most prominent among the known anchoring proteins. Its transcript level was found to be about four- to sixfold higher than those of the other two anchoring genes. The expression of olpB and orf2 increases with a decrease in growth rate, ultimately reaching a similar maximum level. In contrast, the transcript level of the other known anchoring protein, SdbA, remains at a relatively low basal level independent of growth rate. Interestingly, C. thermocellum strain AD2, an adhesion-defective mutant, fails to adhere to cellulose when it grows on cellobiose (1). Nevertheless, the property of cellulose adhesion is regained, after a defined 2- to 4-h lag period, when the mutant is grown on cellulose. On the basis of the results presented here, the most plausible explanation would be that mutation in orf2 could confer the above-described phenotype, since its deficiency would severely impede anchoring of the cellulosome to the cell surface and would consequently impair cell adhesion to cellulose. Upon growth of the cells on cellulose, the deficiency of orf2 may be compensated by an increase in the transcript level of olpB. This hypothesis is supported by the previous observation that the cell surface of cellobiose-grown mutant AD2 is essentially devoid of cellulosome whereas extracellular cellulosome could still be detected (4).
In the early work (15) that first described the sequencing of the three tandem genes (olpB, orf2, and olpA) downstream of cipA, the relationship among them was unclear. It was suggested that cipA and olpB would be transcribed from the same promoter and that transcription of olpB would result from a read-through past the transcriptional terminator located immediately downstream of cipA. The results of Northern blot analysis in the present work indicate that the predominant transcript obtained upon hybridization with the olpB probe is of a size consistent with that expected for the olpB gene alone. In addition, under certain conditions, the transcript level of olpB is nearly twofold higher than that of cipA, indicating that olpB is monocistronic and possesses its own promoter. On the other hand, Northern blot analysis of orf2 supports the previous premise (15) that orf2 and olpA can also be cotranscribed.
Inspection of the sequence regions upstream of the two transcription start sites of cipA (Fig. 5) revealed in one of them similarity to the known σA (σ70) promoters of B. subtilis and Escherichia coli. The presence of a σA promoter sequence associated with cipA and other cellulosome-related genes lends further credence to the notion that σA functions as the primary sigma factor in clostridia (7, 18, 34), as observed in B. subtilis and other bacteria (19, 21). However, only minor levels of transcription were obtained from this apparent promoter, and the majority of the cipA mRNA would be obtained from the second transcription start site. The sequence region upstream of the latter showed partial resemblance to the promoter of the σL transcription factor, which also resembles sequences upstream of the olpB and orf2 genes (Table 3). σL is reportedly implicated in expressing genes whose products show a wide range of different functions (46). It is unlikely, however, that the above-described C. thermocellum genes are regulated by a σL-like transcription factor, since the degree of sequence conservation between σL-RNA polymerase-dependent promoters is high, even among distantly related bacteria (46). Therefore, it is tempting to speculate that the similarity in sequences upstream of the C. thermocellum genes of interest represents a common regulatory element involved in their transcription.
TABLE 3.
Proposed promoter sequences in the regions upstream of selected cellulosome-related genes
Acknowledgments
This research was supported by the Israel Science Foundation (grants 771/01, 446/01, and 250/99); the United States-Israel Binational Agricultural Research and Development Fund (BARD Research Grant 3106-99C); and a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. Additional support was provided by the Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation (Munich, Germany), and funds from the Technion-Niedersachsen Cooperation (Hannover, Germany).
REFERENCES
- 1.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, E. A., and R. Lamed. 1986. Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J. Bacteriol. 167:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, E. A., E. Morag, Y. Shoham, J. Tormo, and R. Lamed. 1996. The cellulosome: a cell-surface organelle for the adhesion to and degradation of cellulose, p. 155-182. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley-Liss, Inc., New York, N.Y.
- 4.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, E. A., L. J. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes: structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. The cellulosome—an exocellular organelle for degrading plant cell wall polysaccharides, p. 387-439. In R. J. Doyle (ed.), Glycomicrobiology. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 7.Béguin, P., M. Rocancourt, M.-C. Chebrou, and J.-P. Aubert. 1986. Mapping of mRNA encoding endoglucanase A from Clostridium thermocellum. Mol. Gen. Genet. 202:251-254. [DOI] [PubMed] [Google Scholar]
- 8.Blair, B. G., and K. L. Anderson. 1999. Regulation of cellulose-inducible structures of Clostridium cellulovorans. Can J. Microbiol. 45:242-249. [DOI] [PubMed] [Google Scholar]
- 9.Coughlan, M. P., K. Hon-Nami, H. Hon-Nami, L. G. Ljungdahl, J. J. Paulin, and W. E. Rigsby. 1985. The cellulolytic enzyme complex of Clostridium thermocellum is very large. Biochem. Biophys. Res. Commun. 130:904-909. [DOI] [PubMed] [Google Scholar]
- 10.Davison, S. P., J. D. Santangelo, S. J. Reid, and D. R. Woods. 1995. A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology 141:989-996. [DOI] [PubMed] [Google Scholar]
- 11.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujino, T., P. Béguin, and J.-P. Aubert. 1992. Cloning of a Clostridium thermocellum DNA fragment encoding polypeptides that bind the catalytic components of the cellulosome. FEMS Microbiol. Lett. 94:165-170. [DOI] [PubMed] [Google Scholar]
- 14.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujino, T., S. Karita, and K. Ohmiya. 1993. Nucleotide sequences of the celB gene encoding endo-1,4-β-glucansase-2, ORF1 and ORF2 forming a putative cellulase gene cluster of Clostridium josui. J. Ferment. Bioeng. 76:243-250. [Google Scholar]
- 16.Gerngross, U. T., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 17.Halliwell, G., T. M. Philips, and N. Halliwell. 1995. Microcrystalline forms of cellulose as substrates for strains of Clostridium thermocellum and cellulase formation. Proc. Biochem. 30:243-250. [Google Scholar]
- 18.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley, C. B., and R. P. Renolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hon-nami, K., M. P. Coughlan, H. Hon-nami, and L. G. Ljungdahl. 1986. Separation and characterization of the complexes constituting the cellulolytic enzyme system of Clostridium thermocellum. Arch. Biochem. Biophys. 145:13-19. [Google Scholar]
- 21.Jarmer, H., T. S. Larsen, A. Krogh, H. H. Saxild, S. Brunak, and S. Knudsen. 2001. Sigma A recognition sites in the Bacillus subtilis genome. Microbiology 147:2417-2424. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, E. A., F. Bouchot, and A. L. Demain. 1985. Regulation of cellulase formation in Clostridium thermocellum. J. Gen. Microbiol. 131:223-232. [Google Scholar]
- 23.Kraus, A., E. Kuster, A. Wagner, K. Hoffman, and W. Hillen. 1998. Identification of a co-repressor binding site in catabolite control protein CcpA. Mol. Microbiol. 30:955-963. [DOI] [PubMed] [Google Scholar]
- 24.Lamed, R., and E. A. Bayer. 1988. The cellulosome concept: exocellular/extracellular enzyme reactor centers for efficient binding and cellulolysis, p. 101-116. In J.-P. Aubert, P. Beguin, and J. Millet (ed.), Biochemistry and genetics of cellulose degradation. Academic Press, London, United Kingdom.
- 25.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Technol. 7:37-41. [Google Scholar]
- 26.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibovitz, E., H. Ohayon, P. Gounon, and P. Béguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Béguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matuschek, M., G. Burchhardt, K. Sahm, and H. Bahl. 1994. Pullulanse of Thermoanaerobacter thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene composite structure of the enzyme, and the common model for its attachment to the cell surface. J. Bacteriol. 176:3295-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer, F., M. P. Coughlan, Y. Mori, and L. G. Ljungdahl. 1987. Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53:2785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, G. L. R., W. E. Blum, and A. L. Burton. 1960. Measurements of carboxymethylcellulase activity. Anal. Biochem. 2:127-132. [Google Scholar]
- 34.Mishra, S., P. Béguin, and J. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morag, E., E. A. Bayer, and R. Lamed. 1992. Unorthodox intra-subunit interactions in the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 33:205-217. [Google Scholar]
- 36.Moran, C. P. 1990. Measuring gene expression in Bacillus, p. 267-294. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley & Sons, Chichester, United Kingdom.
- 37.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 68:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, T. K., T. K. Weimer, and J. G. Zeikus. 1977. Cellulolytic and physiological properties of Clostridium thermocellum. Arch. Microbiol. 114:1-7. [DOI] [PubMed] [Google Scholar]
- 39.Nochur, S. V., A. L. Demain, and M. F. Roberts. 1990. True cellulase production by Clostridium thermocellum grown on different carbon sources. FEMS Microbiol. Lett. 71:199-204. [Google Scholar]
- 40.Salamitou, S., M. Lemaire, T. Fujino, H. Ohayon, P. Gounon, P. Béguin, and J.-P. Aubert. 1994. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salamitou, S., O. Raynaud, M. Lemaire, M. Coughlan, P. Béguin, and J.-P. Aubert. 1994. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-integrating protein CipA. J. Bacteriol. 176:2822-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salamitou, S., K. Tokatlidis, P. Béguin, and J.-P. Aubert. 1992. Involvement of separate domains of the cellulosomal protein S1 of Clostridium thermocellum in binding to cellulose and in anchoring of catalytic subunits to the cellulosome. FEBS Lett. 304:89-92. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratories, Cold Spring Harbor, N.Y.
- 44.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 45.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 46.Studholme, D. j., and M. Buck. 2000. The biology of enhancer-dependent transcriptional regulation in bacteria: insight from genome sequences. FEMS Microbiol. Lett. 186:1-9. [DOI] [PubMed] [Google Scholar]
- 47.Tokatlidis, K., P. Dhurjati, J. Millet, P. Béguin, and J. P. Aubert. 1991. High activity of inclusion bodies formed in Escherichia coli overproducing Clostridium thermocellum endoglucanase D. FEBS Lett. 282:205-208. [DOI] [PubMed] [Google Scholar]
- 48.Tokatlidis, K., S. Salamitou, P. Béguin, P. Dhurjati, and J.-P. Aubert. 1991. Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 291:185-188. [DOI] [PubMed] [Google Scholar]