Abstract
Exposure of cellular DNA to reactive oxygen species generates several classes of base lesions, many of which are removed by the base excision-repair pathway. However, the lesions include purine cyclodeoxynucleoside formation by intramolecular crosslinking between the C-8 position of adenine or guanine and the 5′ position of 2-deoxyribose. This distorting form of DNA damage, in which the purine is attached by two covalent bonds to the sugar-phosphate backbone, occurs as distinct diastereoisomers. It was observed here that both diastereoisomers block primer extension by mammalian and microbial replicative DNA polymerases, using DNA with a site-specific purine cyclodeoxynucleoside residue as template, and consequently appear to be cytotoxic lesions. Plasmid DNA containing either the 5′R or 5′S form of 5′,8-cyclo-2-deoxyadenosine was a substrate for the human nucleotide excision-repair enzyme complex. The R diastereoisomer was more efficiently repaired than the S isomer. No correction of the lesion by direct damage reversal or base excision repair was detected. Dual incision around the lesion depended on the core nucleotide excision-repair protein XPA. In contrast to several other types of oxidative DNA damage, purine cyclodeoxynucleosides are chemically stable and would be expected to accumulate at a slow rate over many years in the DNA of nonregenerating cells from xeroderma pigmentosum patients. High levels of this form of DNA damage might explain the progressive neurodegeneration seen in XPA individuals.
Keywords: endogenous DNA lesions, xeroderma pigmentosum, neurodegeneration
The nucleotide excision-repair (NER) system appears to be present in all organisms and serves to remove major helix-distorting lesions from DNA (1–3). The most important source of such cellular damage is exposure to the UV component of sunlight, which generates dipyrimidine adducts in DNA. Environmental mutagens and carcinogens, such as benzpyrene and aflatoxin, also may cause the formation of bulky DNA lesions after their metabolic activation, and those lesions are corrected by NER. On the other hand, endogenously formed DNA damage produced by reactive oxygen species, hydrolysis, and methylating agents is removed primarily by the base excision-repair (BER) pathway (2). In consequence, NER-defective xeroderma pigmentosum (XP) patients exhibit strongly increased sensitivity to UV light exposure but close to normal resistance to ionizing radiation or simple alkylating agents.
Seven genetic complementation groups, XP-A to XP-G, correspond to different proteins involved in NER (1–3). XP patients of the relatively common genetic complementation group A are totally defective in NER because of disruptions in the function of XPA protein, which binds to damaged DNA and to other proteins of the NER complex. Such XP-A individuals show no general alterations in gene expression or embryological development. Nevertheless, they progressively become dysfunctional and suffer massive loss of neurons with age, possibly because of slow accumulation of unrepaired DNA damage, leading to neurodegeneration (3, 4). We previously provided some experimental support for this model by showing that a minor subclass of base lesions in DNA exposed to γ-irradiation or treatment with H2O2/Cu2+ could not be removed by the known DNA glycosylases that initiate the BER pathway by excising oxidized purines and pyrimidines from DNA. Instead, NER enzymes in human cell extracts were required for repair, which consequently did not occur in extracts of XP cells (5). In a related study (6), it was shown that the purified Escherichia coli UvrABC nuclease was able to incise DNA at the same subclass of oxidative lesions. However, the chemical structure of the lesion(s) was not defined.
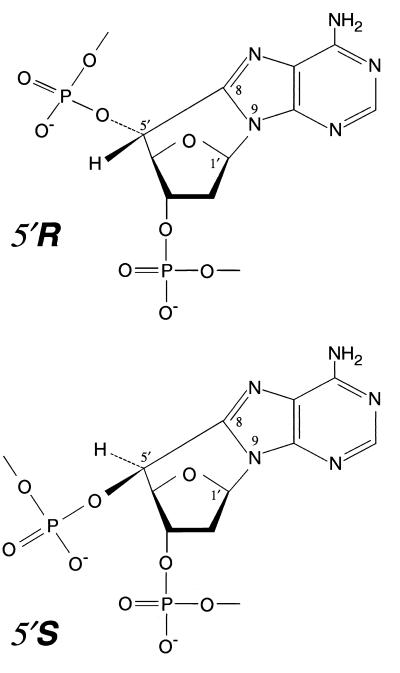
One important candidate for such a bulky DNA lesion generated by oxygen free radicals was a purine cyclodeoxynucleoside (cyPu), but in the absence of defined DNA substrates, this model could not be readily tested (5). When DNA is exposed to hydroxyl radicals, abstraction of the hydrogen from the C-5′ position of 2-deoxyribose occurs as a minor damaging event, and in the absence of oxygen the C-5′ sugar radical can add to C-8 of adenine or guanine to generate a cyclopurine deoxynucleoside (7–10). Formation of this covalent bond between a purine C-8 moiety and the deoxyribose-phosphate backbone causes local distortion of the DNA structure. Action of a DNA glycosylase would not be expected to release such adducts, because the purine would remain attached by the 5′,8 carbon–carbon bond even after cleavage of the 1′,9 glycosyl bond (Fig. 1).
Figure 1.
The 5′R and 5′S diastereoisomers of 5′,8-cyclo-2′-deoxyadenosine in DNA damaged by hydroxyl radicals. In these unusual lesions, there are two covalent linkages of the purine base to the sugar-phosphate backbone.
Cyclopurine deoxynucleosides are formed in two configurations (Fig. 1) with generation of the 5′R diastereoisomer being predominant to that of the 5′S analogue in single-stranded DNA, whereas both diastereoisomers are produced in similar amounts in double-stranded DNA. Cyclodeoxyadenosines are the major lesions, being more abundant than cyclodeoxyguanosines (8–11). Until recently, the difficult synthesis of oligonucleotides containing cyPu residues had not been achieved. However, oligodeoxyribonucleotides containing a single 5′S diastereoisomer of either cyclodeoxyadenosine or cyclodeoxyguanosine have now been synthesized by using modified phosphoramidite chemistry (12), and development of new methodology for epimerization of the C-5′ carbon also allowed for the synthesis of oligodeoxyribonucleotides with site-specific introduction of the 5′R diastereoisomer (13). In the present work, these oligonucleotides have been incorporated into covalently closed circular DNA plasmids and evaluated as potential substrates for the human NER enzyme complex.
Materials and Methods
Construction of Covalently Closed Circular DNA Substrates Containing a Site-Specific cyPu Residue.
Oligodeoxyribonucleotides containing a 5′R,8-cyclo-2′-deoxyadenosine, 5′S,8-cyclo-2′-deoxyadenosine (Fig. 1), or 5′S,8-cyclo-2′-deoxyguanosine residue were prepared by using solid-phase synthesis, as described (12, 13). The lesions were incorporated into the sequence 5′-CACTTCGGXTCGTGACTGATCT-3′, where X is 5′R-cyclo-dA or 5′S-cyclo-dA, or into the sequence 5′-TCTTCTTCTGTXCACTCTTCTTCT-3′, where X is 5′S-cyclo-dG. The latter sequence was also used to incorporate a 1,3-intrastrand d(GpTpG)-cisplatin crosslink (Pt-GTG) as a positive control (14). The R form of cyclo-dG was not available. Oligonucleotides were subsequently analyzed by reverse-phase HPLC and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry for structural confirmation, as described (12, 13).
To generate a substrate suitable for DNA repair by the human NER enzyme complex, oligonucleotides containing a cyPu residue were 5′ phosphorylated by using nonradioactive ATP or [γ-32P]ATP and T4 polynucleotide kinase and incorporated into covalently closed circular DNA, as described (14). A control circular DNA substrate was synthesized by using an oligonucleotide without a cyPu lesion. The plasmid M13mp18 cyclo-dA (see below, and see Fig. 4a) was constructed by replacing the 44-bp PstI–EcoRI fragment of M13mp18GTGx (14) with a synthetic DNA duplex formed by annealing appropriate oligonucleotides. Purities of DNA substrates were assessed by agarose gel electrophoresis. To confirm proper insertion of cyPu-containing oligonucleotides into the M13mp18 substrate, DNA (50 ng) was digested with XhoI and labeled with T7 DNA polymerase in the presence of [α-32P]dCTP, as described (14). Subsequently, DNA was digested with AlwI, and the fragments were separated on a denaturing 14% polyacrylamide gel. Inhibition of AlwI digestion indicated the presence of a cyPu lesion in the substrate (data not shown).
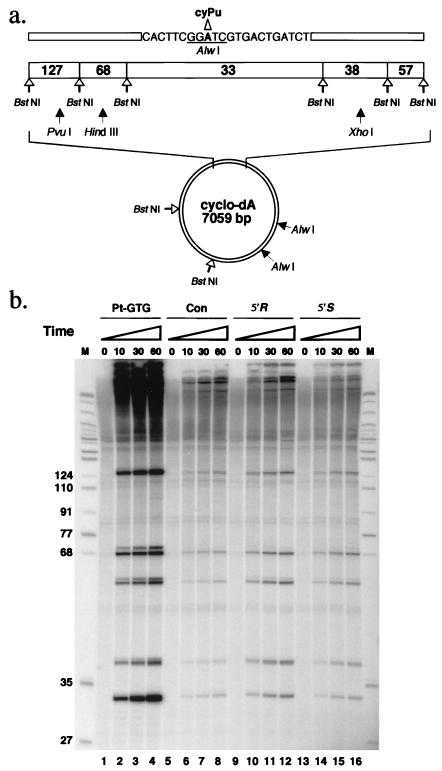
Figure 4.
DNA repair synthesis in response to a cyPu lesion. (a) The plasmid M13mp18 cyclo-dA is shown diagrammatically. Eight BstNI restriction sites and the three AlwI restriction sites are indicated. One AlwI site (underlined) overlaps the region containing the cyPu residue. Unique PvuI, HindIII, and XhoI restriction enzyme sites are also indicated. (b) Autoradiograph after denaturing 14% PAGE, demonstrating DNA repair synthesis in the region containing the cyPu lesion. Plasmid DNA was incubated with HeLa cell extracts and radioactively labeled deoxynucleoside triphosphates (see Materials and Methods) and subsequently digested with BstNI before electrophoresis. Lanes 1–4, Pt-GTG substrate as positive control; lanes 5–8, control DNA without any lesion; lanes 9–12, 5′R-cyclo-dA substrate; lanes 13–16, 5′S-cyclo-dA substrate. M, size markers as in Fig. 2.
Primer Extension Assays.
For primer extension, plasmid DNA (50 ng) was digested with XhoI and PvuI, and subsequently heat-denatured in the presence of a 32P end-labeled oligonucleotide primer with the sequence 5′-CAGGAAACAGCTATGAC-3′. Calf thymus DNA polymerase δ (Pol δ; a gift from U. Hübscher, University of Zürich) was purified as described (15), and human proliferating cell nuclear antigen (PCNA) was produced in Escherichia coli (16, 17). After annealing to the template, the primer was extended with T7 DNA polymerase (Sequenase version 2.0, United States Biochemical) or Pol δ with PCNA, as described (18). Reactions were terminated with sequencing stop buffer (98% deionized formamide/25 mM Tris-borate-EDTA/0.025% bromophenol blue/0.025% xylene cyanol) and loaded onto a 14% denaturing polyacrylamide gel for electrophoresis.
DNA–Repair Synthesis.
For NER synthesis assays (19), reaction mixtures (10 μl) contained 100 ng of plasmid containing either 5′R-cyclo-dA, 5′S-cyclo-dA or 5′S-cyclo-dG or, as positive control, 50 ng of plasmid with the efficiently repaired Pt-GTG lesion (14, 20) and 60 μg of HeLa cell extract protein in reaction mixtures containing 32P-labeled dCTP and 32P-labeled dTTP, as described (17, 20). DNA was recovered (20) and digested with BstNI at 60°C for 1 h, then analyzed by denaturing 14% PAGE.
DNA Dual-Incision Assay.
Reaction mixtures (50 μl) contained 750 ng of plasmid with an incorporated 32P-labeled oligonucleotide containing 5′R- or 5′S-cyclo-dA and 300 μg of HeLa cell extract protein in repair buffer containing 45 mM Hepes-KOH (pH 7.8), 50 mM KCl, 5 mM MgCl2, 0.9 mM DTT, 0.4 mM EDTA, 2 mM ATP, 20 μM each of dATP, dCTP, dGTP, and dTTP, 3.4% glycerol, and 18 μg of BSA. Cell extract supplemented with antiserum against XPA protein (21) and purified recombinant XPA protein (22) as indicated were preincubated in repair buffer at 30°C for 10 min. DNA substrate was then added, and incubation continued at 30°C for 30 min. The neutralizing XPA rabbit antiserum against full-length recombinant human XPA protein was kindly provided by K. Tanaka (Osaka University, Osaka). Purified DNA was separated by electrophoresis on a denaturing 16% polyacrylamide gel, and excised fragments were quantified with a phosphorimager.
DNA Glycosylase and Lesion Reversal Assays.
Reaction mixtures (50 μl) contained 22-bp oligonucleotides having either a 5′R or 5′S diastereoisomer of 5′,8-cyclodeoxyadenosine in the 5′-32P end-labeled strand, 60 μg HeLa cell extract protein, 20 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, and 3–50 mM KCl in different experiments. The mixtures were incubated at 37°C for 30 min. Oligonucleotides were recovered by ethanol precipitation, resuspended in 70 μl 1 M piperidine, and incubated at 90°C for 30 min. Piperidine was removed by evaporation. In a control reaction, uracil-DNA glycosylase activity in the cell extract was measured with the uracil-containing oligonucleotide 5′-CACTTCGGUTCGTGACTGATCT-3′, annealed with 5′-AGATCAGTCACGAGCCGAAGTG-3′.
For the lesion reversal assay, after incubation with HeLa cell extract as above and ethanol precipitation, cyclodeoxyadenosine containing 32P-labeled oligonucleotides were digested with 4 units of Sau3AI (New England Biolabs) at 37°C for 5 h, under conditions recommended by the manufacturer. The presence of a cyPu residue in the Sau3AI recognition site prevented restriction enzyme cleavage. In all assays, oligonucleotides were analyzed by electrophoresis on a denaturing 14% polyacrylamide gel, followed by autoradiography.
Results
cyPu Block DNA Synthesis by Mammalian DNA Pol δ and T7 DNA Polymerase.
Primer extension assays using the mammalian replication enzyme Pol δ in the presence of PCNA, or T7 DNA polymerase, were performed to determine whether cyPu lesions block synthesis by DNA polymerases. Fig. 2 shows strong termination of the major primer extension product at the cyPu site in templates containing either 5′R- or 5′S-cyclodeoxyadenosine, although the primer was efficiently extended to the end of the fragment with the undamaged control template. T7 DNA polymerase was able to incorporate a residue opposite 5′S- and 5′R-cyclodeoxyadenosine, and also showed slight but significant read-through activity with synthesis of the complete fragment, especially with the R rather than the S diastereoisomer in the template. In contrast, Pol δ ceased DNA synthesis at the template residue immediately before the cyPu lesion, and no read-through activity was detected. These results demonstrate that cyPu moieties effectively block DNA replication by polymerases in vitro, and, in consequence, these lesions would be expected to be highly cytotoxic in the absence of DNA repair or efficient bypass polymerases.
Figure 2.
Inhibition of primer extension by T7 DNA polymerase and mammalian DNA Pol δ by cyPu. Autoradiograph after denaturing 14% PAGE showing the inhibition of primer extension by 5′R- and 5′S-cyclodeoxyadenosine. A 32P end-labeled primer was annealed to cyPu-containing plasmid DNA predigested with PvuI and XhoI, then extension reactions were performed with either T7 DNA polymerase or purified recombinant human DNA Pol δ with PCNA. Lanes 1–3, T7 DNA polymerase; Lane 1, control plasmid without cyclo-dA; lane 2, 5′R-cyclo-dA; lane 3, 5′S-cyclo-dA. Lanes 4–6, Pol δ/PCNA. Lane 4, control without cyclo-dA; lane 5, 5′R-cyclo-dA; lane 6, 5′S-cyclo-dA. M, size markers (3′-32P-labeled MspI digest of pBR322).
Search for DNA Glycosylases or Damage Reversal Activities.
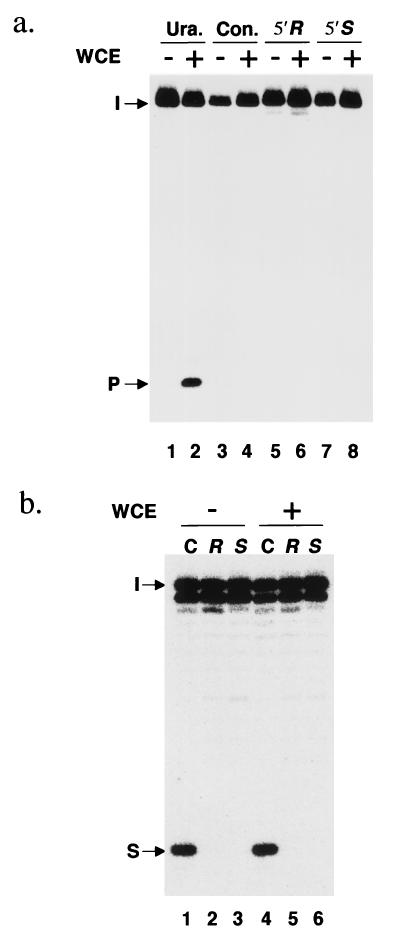
We investigated whether a detectable human DNA glycosylase might catalyze the cleavage of the regular 1′,9 glycosyl bond of a cyPu residue (Fig. 1). After incubation of duplex oligonucleotides containing a single cyPu residue in the 5′-32P-labeled strand with HeLa cell extracts, the substrate was treated with piperidine at 90°C to generate a chain break at any site where a base-sugar bond had been cleaved, followed by denaturing gel electrophoresis. No indications of any DNA glycosylase active on cyPu residues were obtained under a variety of experimental conditions, whereas DNA strand cleavage of a control oligonucleotide containing a uracil residue was readily detected (Fig. 3a). These data are in agreement with previous results by the same experimental approach, which showed that the E. coli DNA glycosylases Fpg and Nth, which are active on other oxidative DNA lesions, failed to cleave base-sugar bonds in cyPu residues (13). Although this hypothetical cleavage reaction would not have released a free base from DNA (Fig. 1), it could have made DNA susceptible to further repair in vivo by incision and excision nucleases active on abasic sites.
Figure 3.
Search for a DNA glycosylase or lesion reversal enzyme acting on cyclopurine deoxynucleosides. (a) DNA glycosylase assay. Lanes 1 and 2, duplex oligonucleotide containing a uracil instead of cyPu at the same position; lanes 3 and 4, control oligonucleotide without any lesion; lanes 5 and 6, 5′R-cyclodeoxyadenosine-containing oligonucleotide; lanes 7 and 8, 5′S-cyclodeoxyadenosine-containing oligonucleotide. These oligonucleotides were incubated with (lanes 2, 4, 6, and 8) or without (lanes 1, 3, 5, and 7) HeLa whole cell extract (WCE) and then treated with 1 M piperidine at 90°C. The arrows mark the positions of intact oligonucleotide (I), and product (P) after DNA glycosylase action followed by piperidine treatment. (b) Lesion reversal assay. Lanes 1 and 4, control oligonucleotide without any lesion; lanes 2 and 5, 5′R-cyclodeoxyadenosine-containing oligonucleotide; lanes 3 and 6, 5′S-cyclodeoxyadenosine-containing oligonucleotide. The duplex oligonucleotides were incubated with (lanes 4–6) or without (lanes 1–3) HeLa cell extract and then digested with Sau3AI. The arrows mark the positions of intact oligonucleotide (I), and product of Sau3AI cleavage (S). Only partial restriction enzyme cleavage of the short control oligonucleotide was observed, even at high Sau3AI concentrations. (a and b) The relevant, lesion-containing strand had been 5′-32P-labeled, and 14% polyacrylamide gels are shown.
Enzymatic cleavage of the 5′,8 bond of a cyPu residue would immediately regenerate the intact purine deoxynucleoside residue without any need for excision of the residue (Fig. 1). However, we were unable to detect any such reversion activity in HeLa cell extracts that would convert the cyPu-containing Sau3AI recognition sequence to a form sensitive to restriction enzyme cleavage (Fig. 3b). Supplementation of reaction mixtures with Mg2+ and ATP did not help to reveal any such lesion reversal activity. Thus, the cyPu 5′,8 carbon-carbon bond, which is chemically very stable, also seems refractory to cleavage by human enzyme activities.
NER Synthesis in the Region of cyPu Damage.
To evaluate whether NER of cyPu lesions occurs, a 7,059-bp plasmid, M13mp18 cyclo-dA, containing a 5′-R or 5′-S site-specific cyPu, was utilized as substrate (Fig. 4a). On attempted digestion with AlwI, the 5′R-cyclo-dA and 5′S-cyclo-dA substrates were resistant to cleavage, confirming the presence of the lesion. Closed circular substrates were incubated with HeLa cell extracts in the presence of radioactively labeled deoxyribonucleoside triphosphates to analyze NER synthesis (20) in DNA substrates containing a cyPu lesion. The cisplatin adduct Pt-GTG already known to be efficiently repaired by NER (14) was used as a positive control lesion. After incubation in reaction mixtures, the DNA was purified, digested with BstNI, and analyzed by denaturing PAGE. DNA synthesis in a 33-bp BstNI fragment containing the Pt-GTG lesion was readily detected (Fig. 4b). Both 5′-R and 5′-S cyPu-containing substrates also showed specific DNA repair synthesis, by comparison with the undamaged control substrate (Fig. 4b), but the signals were considerably weaker than those observed with the Pt-GTG DNA substrate. Repair synthesis was more efficient in the plasmid substrate containing the 5′R-cyclo-dA lesion than in the substrate with the 5′S-cyclo-dA lesion (Fig. 4b). Similar results were obtained with the 5′S diastereoisomer of cyclo-dG by using partly fractionated HeLa cell extracts. This weak but significant DNA repair synthesis with 5′S-cyclo-dG containing substrate was confirmed to depend on the single-stranded DNA binding protein RPA (1, 2), a known NER factor (data not shown). These results indicate that cyPu lesions are removed by localized excision and replacement by resynthesis in human cell extracts.
Oligonucleotides Formed by Dual Incision of DNA Containing a cyPu and Dependence on XPA.
A dual-incision assay was performed to determine whether cyPu lesions in DNA are repaired by NER enzymes (14, 20). Closed circular DNA containing a cyPu residue was prepared with a 32P-labeled nucleotide eight residues upstream of the cyPu residue (Fig. 4a). Fig. 5a shows that radioactively labeled oligonucleotides 20–28 nt in length were generated by dual incision on incubation of both 5′-R and 5′-S cyclo-dA substrates with HeLa cell extracts. The predominant incision products were 23–26 nt long. Dual-incision products from 5′R-cyclo-dA were three to four times more abundant than those from 5′S-cyclo-dA, in agreement with the data in Fig. 4b showing that the R form was more efficiently repaired than the S form (Fig. 5 a and b). These observations indicate that the stereospecific differences between 5′R- and 5′S diastereoisomers of cyclo-dA affect the dual-incision activity.
Figure 5.
Initiation of nucleotide excision-repair by dual incision at cyPu lesions in DNA by human cell extracts. (a) HeLa whole cell extract (WCE) was incubated with plasmid DNA containing a 5′,8-cyclo-2′-deoxyadenosine residue in either the R or S stereoisomeric form, as indicated (lanes 1 and 4). Excision products are indicated by the bracket. Addition of 2 μl neutralizing XPA antiserum to the 50 μl reaction mixture was used in lanes 2, 3, 5, and 6. In addition, lanes 3 and 6 show reaction mixtures supplemented with 450 ng purified XPA protein to reverse the inhibitory effect of the antiserum. The M1 and M2 markers are oligonucleotides of 22 residues containing a single cyPu residue in the 5′R or 5′S form, respectively. An autoradiograph of a 16% polyacrylamide gel is shown after denaturing gel electrophoresis. (b) Quantification of the excision products in the 20–28 nt range by phosphorimager analysis.
To confirm that NER enzymes generated the observed incision product ladder (Fig. 5), the same extracts were incubated with anti-XPA antibody. This neutralizing antibody obliterates the capacity of HeLa cell extracts to carry out NER (21). Here, addition of the antibody strongly inhibited formation of dual incision products at cyPu residues (Fig. 5). Subsequent reconstitution of antibody-containing extracts with an excess of purified XPA protein (22) effectively restored the dual-incision activity (Fig. 5). These complementation results show that generation of incision products by the extracts was XPA-dependent, which is characteristic of NER. The dual-incision efficiencies around the 5′R- and 5′S-cyclo-dA residues (Fig. 5b) were low, about 40 and 150 times less, respectively, than that of a highly susceptible control plasmid with a radioactively labeled Pt-GTG substrate, which was cleaved with a 15% dual-incision efficiency (data not shown).
Discussion
cyPu formation in DNA as a consequence of hydroxyl radical damage in vivo and in vitro has been demonstrated by gas chromatography/mass spectrometry analysis and by HPLC of hydrolysates (8–11). The cyPu lesions appear to be relatively abundant forms of DNA damage after exposure to reactive oxygen species, introduced at 20–30% of the levels of the major lesions 8-hydroxyguanine and thymine glycol, although the relative rates of formation vary with experimental conditions (11, 23). Until recently, it has not been feasible to synthesize oligodeoxyribonucleotide substrates containing this unique type of cyclic lesion. The availability of such substrates (12, 13) has now allowed for the demonstration that they are efficient blocking lesions both to mammalian and microbial replicative DNA polymerases (Fig. 2). This is the expected consequence of intramolecular covalent crosslink formation between the purine base and 2-deoxyribose moieties, and suggests that cyPu residues would be cytotoxic DNA lesions in vivo.
The presence of distinct diastereoisomers of both 5′,8 cyclo-2′-deoxyadenosine and 5′,8-cyclo-2′-deoxyguanosine made it necessary to assess separately their recognition by repair enzymes. By comparison, the Sp diastereoisomer of a DNA methyl phosphotriester is efficiently recognized and repaired by the E. coli Ada protein, thereby generating the signal for induction of the adaptive response to alkylating agents, whereas the Rp isomer is not repaired at all by the same protein (24, 25). The 5′R diastereoisomer of 5′,8-cyclo-2′-adenosine 5′-diphosphate acts as a substrate for pyruvate kinase, whereas the 5′S isomer is inactive (26). Experiments here with an oligonucleotide containing a cyPu as substrate indicated that neither the S nor the R diastereoisomer is recognized by human DNA glycosylases active in the base excision-repair pathway or by hypothetical factors for direct enzymatic reversion of the lesion (Fig. 3). Moreover, the lesions remained as cyPu residues after incubation with high concentrations of the bacterial reagent DNA glycosylases Fpg and Nth that act on oxidative DNA damage, without cleavage of the 1′,9 glycosyl bond (13). Consequently, the putative role of NER in the repair of cyPu lesions was investigated. It was demonstrated that such repair occurs (Figs. 4 and 5) and that the 5′R diastereoisomer was corrected more efficiently than the 5′S analogue by the human NER system. However, both diastereoisomers were relatively poor NER substrates compared with the 1,3-intrastrand d(GpTpG)-cisplatin crosslink, which is in a well-repaired class of lesions that include (6–4) pyrimidine photoproducts and acetylaminofluorene-G lesions (27, 28). In this regard, the cyPu resemble cyclobutane pyrimidine dimers, which are also only sluggishly repaired by the NER system (27, 28). Nevertheless, both diastereoisomers of cyPu residues can clearly be removed from DNA by NER. While this work was being completed, Brooks et al. (29) reported in an abstract that the 5′S diastereoisomer of cyclodeoxyadenosine can be repaired by an NER process in Chinese hamster ovary cell extracts.
Several abundant DNA lesions, such as 7-methyldeoxyguanosine, methyl phosphotriesters, and deoxyguanosine adducts generated by malondialdehyde produced from lipid peroxidation (30), are chemically relatively labile and could not accumulate in DNA for more than a few days in vivo before a steady-state of simultaneous generation and decay would occur. In contrast, cyclic residues formed by generation of a stable 5′,8 carbon–carbon bond could persist and accumulate in DNA for very long time periods unless removed by NER. These damaged residues also induce local destabilization of duplex DNA structure (12) and act as blocking lesions (Fig. 2). Moreover, cyPu lesions have been detected in DNA exposed to moderate doses of ionizing radiation (8–10). For this reason, cyPu residues may be considered leading contenders to account for long-term intracellular DNA damage caused by endogenously formed reactive oxygen species in repair-deficient cells. Other forms of bulky DNA damage have been detected after exposure to high doses of ionizing radiation, such as tandem base lesions including intrastrand d(GpT) dimers with a covalent link between the C-8 carbon of guanine and the C-5 carbon of an adjacent thymine (31). Such lesions, as well as cyPu residues, may have contributed in previous experiments demonstrating NER of DNA exposed to oxidative damage (5, 6). In contrast, a suggestion (32) that cytotoxic thymine glycol lesions could build up in DNA because of endogenous oxidative damage seems unlikely, because those lesions are excised by the hNTH1 DNA glycosylase and are corrected by the BER pathway both in normal human cells and XP cells, preventing their accumulation. A specific defect in repair of oxidized pyrimidine residues by XP group G cells remains possible, as the XPG protein may contribute to BER of such lesions by stimulating hNTH1 (33).
The brain has a high level of oxygen metabolism, and neurons differ from most other cells in their extremely limited capacity to regenerate. Thus, the central nervous system is a likely target for slow accumulation of nonrepairable DNA damage generated as an unfortunate side effect of oxygen metabolism. There are no clear indications that such formation of nonrepairable DNA damage is a physiological problem in normal individuals. In contrast, XP patients unable to perform NER could suffer progressive formation of critical, cytotoxic DNA lesions in this way, and this scenario provides a likely explanation for the increasingly serious neurodegeneration seen in XP-A individuals (3, 4). After slow accumulation of cyPu in neural DNA over decades, gene expression in the nonreplicating neurons might gradually deteriorate, ultimately leading to cytotoxicity. A direct search by analytical chromatography/mass spectrometry methods for cyPu residues in DNA of nonproliferative tissues of XP patients currently is hampered by the lack of accessible human tissue banks with suitable biopsy or autopsy material.
Acknowledgments
I.K. was a recipient of a postdoctoral fellowship from Japan Society for the Promotion of Science. C.B. was a recipient of a postdoctoral fellowship from the American Cancer Society (PF-006-01-CNE). This work was supported by the Imperial Cancer Research Fund and by grants (to J.C.) from the French Atomic Energy Commission and the Comité de Radioprotection d'Electricité de France.
Abbreviations
- NER
nucleotide excision-repair
- BER
base excision-repair
- XP
xeroderma pigmentosum
- cyPu
purine cyclodeoxynucleoside
- Pol δ
DNA polymerase δ
- PCNA
proliferating cell nuclear antigen
- Pt-GTG
1,3-intrastrand d(GpTpG)-cisplatin crosslink
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070471597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070471597
References
- 1.de Laat W L, Jaspers N G J, Hoeijmakers J H J. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver J E, Kraemer K H. In: The Metabolic Basis of Inherited Disease. 7th Ed. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 1995. pp. 4393–4419. [Google Scholar]
- 4.Robbins J H. J Am Med Assoc. 1988;260:384–388. [Google Scholar]
- 5.Satoh M S, Jones C J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roldán-Arjona T, Sedgwick B. Mol Carcinog. 1996;16:188–196. doi: 10.1002/(SICI)1098-2744(199608)16:4<188::AID-MC2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Keck K. Z Naturforsch B. 1968;23:1034–1043. [PubMed] [Google Scholar]
- 8.Mariaggi N, Cadet J, Téoule R. Tetrahedron. 1976;32:2385–2387. [Google Scholar]
- 9.Fuciarelli A F, Shum F Y, Raleigh J A. Biochem Biophys Res Commun. 1986;134:883–887. doi: 10.1016/s0006-291x(86)80502-2. [DOI] [PubMed] [Google Scholar]
- 10.Dizdaroglu M. Biochem J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirksen M L, Blakely W F, Holwitt E, Dizdaroglu M. Int J Radiat Biol. 1988;54:195–204. doi: 10.1080/09553008814551631. [DOI] [PubMed] [Google Scholar]
- 12.Romieu A, Gasparutto D, Molko D, Cadet J. J Org Chem. 1998;63:5245–5249. [Google Scholar]
- 13.Romieu A, Gasparutto D, Cadet J. Chem Res Toxicol. 1999;12:412–421. doi: 10.1021/tx9802668. [DOI] [PubMed] [Google Scholar]
- 14.Moggs J G, Yarema K J, Essigmann J M, Wood R D. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 15.Weiser T, Gassmann M, Thömmes P, Ferrari E, Hafkemeyer P, Hübscher U. J Biol Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 16.Fien K, Stillman B. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biggerstaff M, Wood R D. In: DNA Repair Protocols: Eukaryotic Systems. Henderson D S, editor. Vol. 113. Totowa, NJ: Humana; 1999. pp. 357–372. [Google Scholar]
- 18.Shivji M K K, Ferrari E, Ball K, Hübscher U, Wood R D. Oncogene. 1998;17:2827–2838. doi: 10.1038/sj.onc.1202352. [DOI] [PubMed] [Google Scholar]
- 19.Wood R D, Robins P, Lindahl T. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 20.Shivji M K K, Moggs J G, Kuraoka I, Wood R D. In: DNA Repair Protocols: Eukaryotic Systems. Henderson D S, editor. Vol. 113. Totowa, NJ: Humana; 1999. pp. 373–392. [Google Scholar]
- 21.Miura N, Miyamoto I, Asahina H, Satokata I, Tanaka K, Okada Y. J Biol Chem. 1991;266:19786–19789. [PubMed] [Google Scholar]
- 22.Jones C J, Wood R D. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 23.Fuciarelli A F, Wegher B J, Blakely W F, Dizdaroglu M. Int J Radiat Biol. 1990;58:397–415. doi: 10.1080/09553009014551761. [DOI] [PubMed] [Google Scholar]
- 24.Weinfeld M, Drake A F, Saunders J K, Paterson M C. Nucleic Acids Res. 1985;13:7067–7078. doi: 10.1093/nar/13.19.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo I, Sedgwick B, Kilpatrick M W, McCarthy T V, Lindahl T. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 26.Haromy T P, Raleigh J, Sundaralingam M. Biochemistry. 1980;19:1718–1722. doi: 10.1021/bi00549a031. [DOI] [PubMed] [Google Scholar]
- 27.Szymkowski D E, Lawrence C W, Wood R D. Proc Natl Acad Sci USA. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 29.Brooks P J, Wise D S, Berry D A, Somers R L, Mackie H, Robbins J H. J Invest Med. 1999;47:A130. [Google Scholar]
- 30.Mao H, Schnetz-Boutaud N C, Weisenseel J P, Marnett L J, Stone M P. Proc Natl Acad Sci USA. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Box H C, Budzinski E E, Dawidzik J B, Gobey J S, Freund H G. Free Radical Biol Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 32.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klungland A, Höss M, Gunz D, Constantinou A, Clarkson S G, Doetsch P W, Bolton P H, Wood R D, Lindahl T. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]