Abstract
Folate deficiency leads to neural tube defects and other human diseases, and is a global health problem. Because plants are major folate sources for humans, we have sought to enhance plant folate levels (biofortification). Folates are synthesized from pteridine, p-aminobenzoate (PABA), and glutamate precursors. Previously, we increased pteridine production in tomato fruit up to 140-fold by overexpressing GTP cyclohydrolase I, the first enzyme of pteridine synthesis. This strategy increased folate levels 2-fold, but engineered fruit were PABA-depleted. We report here the engineering of fruit-specific overexpression of aminodeoxychorismate synthase, which catalyzes the first step of PABA synthesis. The resulting fruit contained an average of 19-fold more PABA than controls. When transgenic PABA- and pteridine-overproduction traits were combined by crossing, vine-ripened fruit accumulated up to 25-fold more folate than controls. Folate accumulation was almost as high (up to 15-fold) in fruit harvested green and ripened by ethylene-gassing, as occurs in commerce. The accumulated folates showed normal proportions of one-carbon forms, with 5-methyltetrahydrofolate the most abundant, but were less extensively polyglutamylated than controls. Folate concentrations in developing fruit did not change in controls, but increased continuously throughout ripening in transgenic fruit. Pteridine and PABA levels in transgenic fruit were >20-fold higher than in controls, but the pathway intermediates dihydropteroate and dihydrofolate did not accumulate, pointing to a flux constraint at the dihydropteroate synthesis step. The folate levels we achieved provide the complete adult daily requirement in less than one standard serving.
Keywords: metabolic engineering, vitamin
Tetrahydrofolate (THF) and its derivatives (folates) are essential cofactors in one carbon transfer reactions in almost all organisms, being required for synthesis of glycine, serine, methionine, purines, and thymidylate (1, 2). Plants and microorganisms are able to synthesize folates, but humans lack this capacity and require a dietary supply. Folates in human diets come mainly from plant foods (2). However, many plants, including tubers, cereals, and most fruits contain far less folate than green leafy vegetables, the best source (2, 3).
Folate deficiency is a worldwide health problem associated with spina bifida and other birth defects, megaloblastic anemia, cardiovascular diseases, and some cancers (4–7). Currently, the United States and other western countries mandate fortification of grain products with synthetic folic acid to help their populations reach the recommended dietary allowance, which is 400 μg/day for adults and 600 μg/day for pregnant women (5, 8). However, food fortification can be difficult to implement in developing countries due to recurrent costs, distribution inequities, and lack of an industrial food system (9). In these areas, folate deficiency causes at least 200,000 severe birth defects every year (10). In addition, epidemiological data suggest that folate malnutrition is the main cause of anemia in at least 10 million pregnant women in developing countries (11, 12). Moreover, concerns have arisen about the effects of chronic exposure of western populations to synthetic folic acid (7, 13, 14). Such concerns are based primarily on the ability of folic acid, at high levels of intake, to bypass the usual physiological control of folate distribution and metabolism, causing entry of unreduced folic acid into the systemic circulation. Consequently, dietary tetrahydrofolates are predicted to have a higher margin of safety. Folate enhancement in plant foods (biofortification) through metabolic engineering therefore represents an attractive alternative strategy to increase the intake of natural folates in rich and poor countries alike (2, 15, 16).
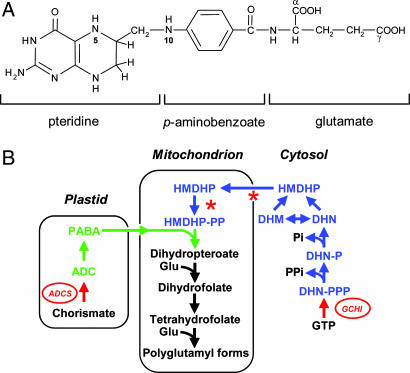
Folates are tripartite molecules consisting of pteridine, p-aminobenzoate (PABA), and glutamate moieties usually with a short, γ-linked chain of additional glutamates attached to the first glutamate (Fig. 1A). Folate biosynthesis in plants is highly compartmentalized, with pteridines synthesized in the cytosol and PABA produced in plastids. These moieties are condensed in mitochondria forming dihydropteroate, which is glutamylated to form folates (Fig. 1B). Previous engineering work in tomato fruit and Arabidopsis involved the overexpression of GTP cyclohydrolase I (GCHI) (17, 18), which catalyzes the first committed step of pteridine biosynthesis (Fig. 1B) (19). Although GCHI overexpression greatly increased the flux to pteridines, folate levels were raised only 2- to 4-fold (17, 18). This modest increase in folate was associated with severely depleted PABA pools in the engineered plants, suggesting that the PABA supply had become limiting for folate synthesis (17). This problem was successfully addressed in the present study by engineering PABA production in ripening tomato fruit, and combining this trait with pteridine overproduction. This work demonstrates the feasibility of developing crops with enough folate to supply the adult recommended dietary allowance in a single standard serving (100 g/serving). It also sheds light on the control of the folate biosynthesis pathway in plants.
Fig. 1.
Structure and biosynthesis of folates. (A) Chemical structure of THF, monoglutamyl form. Plant folates have γ-linked polyglutamyl tails of up to approximately six residues attached to the first glutamate. One-carbon units at various levels of oxidation are attached to N-5 and/or N-10. The pteridine ring of folates and free pteridines can exist in tetrahydro-, dihydro-, and fully oxidized forms. (B) The plant folate biosynthesis pathway. Pteridines are in blue. PABA and its precursor aminodeoxychorismate (ADC) are in green. Red arrows are the engineered GTP cyclohydrolase I (GCHI) and ADC synthase (ADCS) reactions. Asterisks show two possible constraints in the pathway in engineered fruit (pteridine transport and phosphorylation). DHN, dihydroneopterin; -P, monophosphate; -PP, pyrophosphate -PPP, triphosphate; DHM, dihydromonapterin; HMDHP, hydroxymethyldihydropterin.
Results
Aminodeoxychorismate Synthase (ADCS) Overexpression Elevates PABA Production.
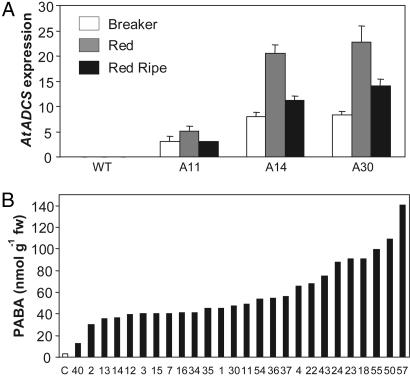
Tomato was transformed with the Arabidopsis aminodeoxychorismate synthase (AtADCS) coding sequence (including its native chloroplast targeting peptide) under the control of the fruit ripening-specific E8 promoter (20). Genomic DNA was extracted from tissue culture-generated plantlets and PCR-screened for the AtADCS gene. Twenty-five independent transformants were obtained. Fruits from three transformants were sampled for expression analysis. Relative AtADCS expression was estimated by real-time PCR using the 2(−ΔΔCT) method (21) with an actin gene as benchmark (22). In all AtADCS+ transformants, RNA levels were highest in fruit at the red stage; no signal was present in wild-type fruit (Fig. 2A). Total PABA levels were determined in red ripe fruit from all 25 independent lines generated, and in five empty vector controls. On average, transgenic fruit accumulated 56 nmol PABA/g fresh weight, which is 19-fold more than empty vector controls at red ripe stage (Fig. 2B). Almost all of the overproduced PABA (>85%) was present as glucose ester as previously reported for plant tissues (23). There was no difference in folate levels between PABA overproducers and controls (data not shown).
Fig. 2.
AtADCS overexpression increases PABA content in tomato fruit. (A) Levels of AtADCS mRNA relative to actin in ripening tomato fruit, determined by real-time PCR (red and red ripe stages are 3 and 7 days after breaker stage, respectively). Data are means and SE of 2(−ΔΔCT) values calculated from three PCR replicates for the AtADCS and actin genes. Transgenic fruits were sampled from T1 AtADCS+ plants. (B) Total PABA accumulation in red ripe fruit from 25 independent AtADCS+ primary transformants (T0) analyzed by HPLC with fluorescence detection. Control value is the mean of five independent vector-alone transformants. Values for each AtADCS+ line are from single fruits. WT, wild type; fw, fresh weight.
Coexpression of AtADCS and GCHI Leads to PABA, Pteridine, and Folate Hyperaccumulation.
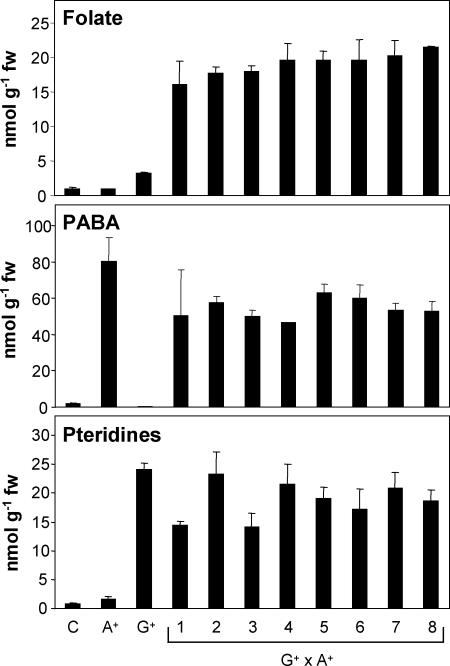
Hemizygous T0 AtADCS+ plants were crossed with one-insertion, hemizygous T1 GCHI+ plants (17) (Table 1). Genomic DNA from the progenies was screened by PCR for double transgenics, i.e., plants carrying both transgenes. Neither plant nor fruit development in double transgenics differed from controls (nontransgenic segregants from the same progenies). Fruits from transgenics and controls were sampled at late red ripe stage for folate, pteridine, and PABA analysis. GCHI+/AtADCS+ transgenic fruit accumulated up to 25-fold more folate (25 nmol/g fresh weight) than controls (Fig. 3). In the same transgenic fruit, total pteridine and PABA levels were up to 30-fold higher than in controls (Fig. 3). However, neither dihydropteroate nor pteroate (its oxidized form) was detected. In addition, no accumulation of hydroxymethyldihydropterin pyrophosphate was observed (data not shown). Taken together, these data point to an additional constraint in the biosynthesis pathway (discussed below).
Table 1.
Independent transgenic lines crossed to generate G+ × A+ (GCHI+/AtADCS+) double transgenic tomato plants
| G+ × A+ plants | Transgenic lines crossed* |
|---|---|
| 1 | G067 × A14 |
| 2 | G067 × A23 |
| 3 | G067 × A30 |
| 4 | G102 × A14 |
| 5 | G102 × A24 |
| 6 | G102 × A30 |
| 7 | G075 × A55 |
| 8 | G102 × A11 |
*The following generations were used as parental lines: T1 G+ as the maternal line and T0 A+ as the paternal line.
Fig. 3.
Accumulation of folate, PABA, and pteridines in G+ × A+ (GCHI+/AtADCS+) late red ripe tomatoes. Values for controls (nontransgenic segregants) and for single G+ and A+ transgenics are shown for comparison. Controls are mean values of seven fruits from nontransgenic segregants for folate data and three fruits for PABA and pteridine data. G+, A+, and G+ × A+ are mean values from three fruits per independent plant (except for lines 5–7 in which data come from two fruits). Total folate contents were analyzed by HPLC with electrochemical detection. Total PABA (free PABA plus PABA-glucose) contents were analyzed by HPLC with fluorescence detection. Total pteridine contents are expressed in hydroxymethylpterin equivalents and were analyzed by HPLC with fluorescence detection.
One-Carbon Substituents and Polyglutamylation of Accumulated Folates.
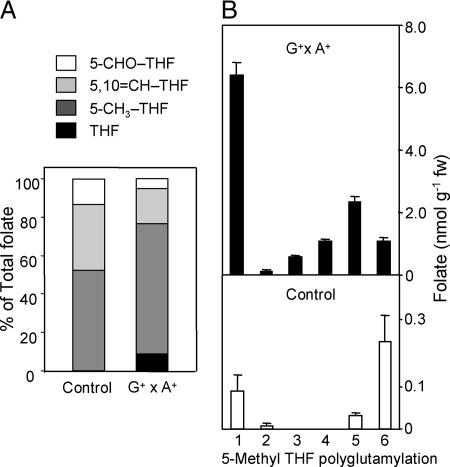
The main folate derivative in GCHI+/AtADCS+ fruit and controls was 5-methyl-THF (68% of the total folate pool), which is consistent with the pattern found in wild-type and GCHI+ pteridine overproducers (17, 19). 5,10-Methenyl-THF, 5-formyl-THF, and THF were also present, but neither dihydrofolate (DHF) nor its oxidized form, folic acid, were detected (Fig. 4A). 5-Methyl-THF monoglutamate was predominant in GCHI+/AtADCS+ fruit, whereas the most abundant polyglutamyl folate in controls and fruit overexpressing GCHI+ alone was the 5-methyl-THF hexaglutamate (Fig. 4B) (17).
Fig. 4.
Analysis of folate derivatives in G+ × A+ and control fruits by HPLC with electrochemical detection. (A) Percentages of one-carbon substituents of folates. Data are averages of all of the fruit analyzed. (B) Polyglutamyl tail lengths of the most abundant folate, 5-methyl-THF. Data are averages of three fruits and SE. 5-CHO-THF, 5-formyltetrahydrofolate; 5,10=CH-THF, 5,10-methenyltetrahydrofolate; 5-CH3-THF, 5-methyltetrahydrofolate; THF, tetrahydrofolate.
Folate Levels in Transgenic Fruit Increase Continuously Throughout Ripening.
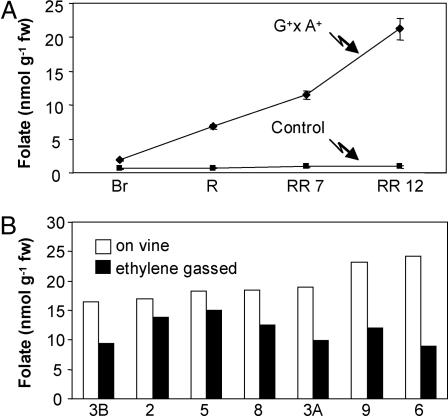
Folates were quantified in control and transgenic fruit harvested at various ripening stages. Although total folate levels remained constant during ripening in nontransgenic controls (Fig. 5A), those in GCHI+/AtADCS+ fruit increased continuously throughout ripening, consistent with the reported action of the E8 promoter that was used to drive the expression of both transgenes (24, 25). All folate classes increased during ripening in folate hyperaccumulators; polyglutamylation reached a plateau at red stage, subsequent folate accumulation being of monoglutamyl forms [supporting information (SI) Fig. 6].
Fig. 5.
Changes in total folate content in G+ × A+ and control fruits during ripening. (A) Changes during ripening on the vine. Ripening stages are: Br, breaker; R, red (Br + 3 days); RR 7, red ripe (Br + 7 days); RR 12, late red ripe (Br + 12 days). Data are mean and SE values for three fruits. (B) Differences in folate content between fruit ripened on the vine versus fruit detached at the mature green stage and ethylene-gassed until breaker stage. Numbers in the x axis correspond to the crosses specified in Table 1. Detached fruit was allowed to ripen at 20°C for 12 days after breaker stage. Data are values from single matching fruits from the same plant.
Folates Still Hyperaccumulate in Ethylene-Ripened Fruit.
In the vegetable industry, tomatoes are normally harvested when mature green, stored, and then ethylene-gassed to induce ripening (26). We mimicked this process with our transgenic fruit to test whether folate hyperaccumulation still occurs. Transgenic fruits were harvested at mature green stage, ethylene-gassed until they reached breaker stage, then allowed to ripen for 12 days. Folate contents were then compared with fruit from the same plant ripened on the vine (Fig. 5B). Ethylene-treated fruit accumulated folates up to 15 nmol/g fresh weight, 79% of the average folate levels detected at late red ripe stage in vine-ripened fruit (Fig. 5B). In ethylene-treated fruit, the folate classes and polyglutamylation profiles were very similar to those in vine-ripened fruit (Fig. 4 and SI Fig. 7). These data show that little of the engineering benefit is lost by detaching the fruit.
Discussion
This work establishes that the engineering of both pteridine and PABA branches of the synthesis pathway is necessary to substantially enhance folate production in plants. On average, double transgenic tomato fruit expressing GCHI and AtADCS genes contained 19- and 6-fold more folate than controls and single transgenic GCHI+ fruit, respectively. The GCHI+/AtADCS+ transgenic fruit accumulated enough folates (840 μg per 100 g) to provide the whole recommended dietary allowance for a pregnant woman in less that one standard serving. This folate overproduction was not associated with a change in fruit size or appearance. To our knowledge, the folate levels we achieved are the highest reported for plants; our tomatoes accumulate up to seven times more folates than green leafy vegetables, which are considered rich folate sources.
The engineering results offer insight into in vivo regulation of the folate synthesis pathway. The marked PABA accumulation obtained in AtADCS+ fruit (Fig. 2A) confirms the high availability of chorismate in plastids, as indicated by the engineering of another chorismate-using enzyme (UbiC) in tobacco plastids (27). The proportions of folate classes in GCHI+/AtADCS+ fruit were typical of plant tissues; however, their extent of polyglutamylation was less than in controls. Glutamate availability is unlikely to limit folate polyglutamylation, because free glutamate is abundant in red stage fruit (2.8 μmol/g fresh weight) (28). Therefore, we infer that the high folate production in transgenic fruit exceeded the capacity of folylpolyglutamate synthase. Similar results were obtained when folate biosynthesis was engineered in the dairy bacterium Lactococcus lactis, where the folates overproduced were mainly monoglutamates unless folylpolyglutamate synthase was overexpressed (29, 30).
Despite their use for folate synthesis, high levels of pteridines and PABA were still present in the GCHI+/AtADCS+ fruit (Fig. 3), implying an additional constraint on folate synthesis flux. The lack of dihydropteroate or DHF accumulation shows that the limitation cannot be at the DHF synthase or DHF reductase steps, but must lie upstream of them, at the dihydropteroate synthesis level. Dihydropteroate is synthesized in plant mitochondria by the bifunctional enzyme hydroxymethyldihydropterin pyrophosphokinase–dihydropteroate synthase (31). Both enzyme activities are inhibited by the products of the reactions and some folate forms (32). Because we found no accumulation of hydroxymethyldihydropterin pyrophosphate, the constraint presumably lies at the pteridine phosphorylation step; two possibilities are lack of pyrophosphokinase activity or lack of sufficient pteridine transport activity into the mitochondria (Fig. 1B, asterisks). A third possibility is the oxidation state of hydroxymethylpterin, which cannot be fully established by fluorometric HPLC. The oxidized form of hydroxymethyldihydropterin, hydroxymethylpterin, cannot be phosphorylated by hydroxymethyldihydropterin pyrophosphokinase (HPPK) (33), and high proportions of the pteridines in ripe GCHI+ fruit are oxidized (17). It is also possible that plants, like bacteria (34), can reduce dihydropteridines to the tetrahydro state, and hydroxymethyltetrahydropterin may not be a substrate for HPPK.
Our results establish that all enzymes of the folate biosynthesis pathway are active in the engineered fruit during ripening. However, GCHI protein and ADC synthase and lyase mRNAs are undetectable in wild type fruit at late ripening stages (19, 35, 36). Although very low levels of enzymes might have been sufficient to sustain the observed fluxes because pterin and PABA levels were so high, it is also possible that hyperaccumulation of pteridines and PABA induced the expression of downstream enzymes in the biosynthesis pathway. Gene expression profiling in folate overproducers will be required to investigate this possibility.
PABA is a food additive classified as GRAS (generally recognized as safe) by the U.S. Food and Drug Administration when <30 mg/day is ingested. In addition, PABA has been evaluated in human clinical trials and found to have low toxicities at intakes of up to 12 g per day (37, 38). Because GCHI+/AtADCS+ fruit would provide <1 mg of PABA per 100-g portion, consumption of biofortified food plants with the kind of levels accumulated in our engineered fruit should not be a major concern. On the other hand, nothing is known about pteridine contents of plant foods or pteridine intake by humans. However, mammals synthesize the pteridine tetrahydrobiopterin, and also catabolize pteridines (39). Engineered fruit accumulated free pteridines in the order of 0.36 mg per portion, which would represent <10% of the pteridines that humans excrete in the urine per day (40, 41). In addition, it is reported that the medicinal legume, velvet bean, contains ≈470 nmol/g fresh weight of total pteridines in the pericarp (42), 25-fold more than our engineered fruit. These considerations suggest that the pteridine dose in a serving of transgenic tomato fruit might not be outside the normal range of human consumption and metabolism, but more research is clearly required to assess the safety of pteridines accumulated in transgenic fruit and in other plants.
Finally, it should be noted that the success of our two-gene biofortification strategy is likely to be repeatable in other plants. Specifically, overexpression of GCHI alone in Arabidopsis leaves increased folate levels up to three-fold, similar to the situation in GCHI+ tomatoes (17, 18). When these engineered Arabidopsis were supplied with PABA, folate accumulation was enhanced a further 7-fold (43); the same could presumably be achieved by engineering an increased PABA supply. We conclude that the technology for plant folate enhancement is in place, and may well be transferable to other food plants such as tubers and cereal grains.
Materials and Methods
Expression Vector Construction.
A modified pMON10086 vector (17) containing the long (2-kb) form of the tomato E8 promoter (20) was used. The AtADCS coding sequence, At2g28880, was obtained by PCR by using as template the cDNA clone RAFL09-32-D4. The plant consensus leader sequence TAAACA (44) was added before the start codon. The modified sequence was cut with NotI and AscI and ligated into the vector. The construct was amplified and sequence verified before introduction into Agrobacterium tumefaciens strain ABI by electroporation.
Transgenic Plants.
Tomato (Lycopersicon esculentum Mill., cv. MicroTom) was transformed by A. tumefaciens harboring the AtADCS construct, and plantlets were generated and selected as described (17, 45). Kanamycin-resistant transformants were screened for the AtADCS construct by PCR with a forward primer located in the E8 promoter (5′-CTTAATCAGACGTATTGG-3′) and a reverse primer from the AtADCS cDNA (5′-CATTAACATGGGACGAAG-3′). PCR-positive plants were transplanted to soil and grown to maturity in a growth chamber as described (17).
Transgenic Plant Crosses.
T1 progenies from GCHI+ plants were tested for segregation pattern by PCR. The lines chosen segregated in a 3:1 (transgene/wild type) ratio, indicative of one transgene insertion event. Five independent hemizygous T0 AtADCS+ plants were crossed with three T1 GCHI+ lines (17) as indicated in Table 1. Pollen was collected from the AtADCS+ plants, and pollination of GCHI+ emasculated flowers was conducted as described (46). Fruit from the pollinated flowers was harvested at red ripe stage, and their seeds were planted. Seedlings were PCR-screened for the presence of both transgenes by using the AtADCS primers given above and the GCHI primer set specified previously (17). Segregation patterns suggested that the T0 AtADCS+ plants used for crossing had one insertion event. Plantlets containing both transgenes and nontransgenic segregants (controls) were allowed to set fruit. Late red ripe fruits (12 days after breaker) were harvested, frozen in liquid N2, and stored at −80°C until analysis.
RNA Isolation and Expression Analysis.
Representative segments from three frozen tomato fruits were pooled and ground in liquid N2 for each reported sample. Total RNA was isolated by using a RNeasy Plant Mini kit and DNase-treated (RNase-Free DNase Set; Qiagen, Valencia, CA). RNA samples (1 μg) were reverse-transcribed to cDNA by using the RevertAid first-strand cDNA synthesis kit (Fermentas, Hanover, MD) with random hexamers. The resulting reaction mixture was diluted with 4 volumes of water. PCR primers for AtADCS were 5′-GGGACTTTATCTCCACCTGAGAGA-3′ and 5′-CGAAGAGCATAAAGACAGATTTGC-3′. For the benchmark actin gene (GenBank accession no. U60480), the primers used were as described (22). Amplicon lengths were 91 bp for both genes. PCR was conducted by using the GeneAmp 5700 System (Applied Biosystems, Foster City, CA). SYBR GREEN PCR Master Mix (Applied Biosystems) was used in 25-μl mixtures containing 5 μl of the cDNA dilution and a 300 nM concentration of each primer. PCR conditions were: denaturation at 95°C for 10 min, followed by 40 cycles of two-step PCR at 95°C for 15 s and 72°C for 60 s. A dissociation step that increased the temperature gradually from 60°C to 95°C was inserted after PCR cycle. Data were analyzed by using SDS 2.2.1 software (Applied Biosystems). Expression values were normalized to that of actin and calculated by the difference between the crossing thresholds (CT) of AtADCS and actin for each sample (21). To confirm the specificity of the amplification, control plasmids containing AtADCS and actin genes were used as templates in the same run; the dissociation curves obtained from all amplicons matched those from the corresponding controls.
Folate Analysis.
Folates were extracted from ≈1 g of fruit as described (17) with the following modifications: (i) the volume loaded on the folate affinity columns was reduced to half for the GCHI+/AtADCS+ extracts; (ii) the potentials set for the four-channel electrochemical detector were 100, 300, 500, and 750 mV. The detector was calibrated with THF, DHF, 5-methyl-THF, 5,10-methenyl-THF, 5-formyl-THF, folic acid, and pteroate standards (Schircks). For analysis of polyglutamates, 5-methyl-THF tri- and pentaglutamate were used as standards.
Dihydropteroate and pteroate were recovered from the folate affinity columns. Recovery studies showed that dihydropteroate underwent oxidation and gave a final recovery of 10%, whereas pteroate recovery was 100%. Detection limits were 60 pmol/g fresh weight for pteroate and 600 pmol/g fresh weight for dihydropteroate.
The affinity-purified fraction from GCHI+ and double transgenic fruits showed an unknown peak in the HPLC/electrochemical chromatograms. Its molecular ion [M + H]+ was m/z = 298, ruling out a folate. This mass and the fragmentation pattern (which included neutral elimination of an m/z = 78 fragment) were consistent with that of a 2-mercaptoethanol adduct of an oxidation product of tetrahydroneopterin (47). The adduct was shown to form when authentic tetra- or dihydroneopterin was boiled in 10 mM 2-mercaptoethanol.
Pteridine and PABA Analysis.
Representative fruit segments (1 g) were polytron-homogenized in 7 ml of methanol. A 600-μl aliquot was sampled for pteridine analysis and processed as described (17). Pteridine contents were expressed as hydroxymethylpterin (HMPT) equivalents after calibration with authentic HMPT (Schirks) (fluorescence responses of the various pteridines differ by <40%). HMPT-pyrophosphate was estimated from the difference in HMPT values before and after treatment of the pteridine extract with alkaline phosphatase. For PABA analysis, 5 ml of the remaining extract was processed and analyzed by HPLC with fluorescence detection as described (17, 23). For total PABA, the sample was hydrolyzed to release the PABA from PABA-glucose. PABA-glucose was estimated from the increase in free PABA after hydrolysis.
Ethylene Treatment.
GCHI+/AtADCS+ fruits were harvested at mature green stage and placed in a 3-liter closed container with a constant ethylene/air flow. The flow rate was 0.5 liter/min, and the ethylene/air ratio was adjusted to maintain an ethylene concentration of 100 μl/liter inside the container. Fruit reached the breaker stage after 72 h of ethylene treatment. Breaker stage fruits were placed in a tray, partially covered to minimize water loss, and stored in a chamber at 20°C for 12 days. Samples were then frozen in liquid N2 and stored at −80°C until analysis.
Supplementary Material
Acknowledgments
We thank Dr. G. Henderson for help with LC-MS analysis, Dr. S. Sargent and A. Berry for help with ethylene treatments, N. Contreras and T. Theodoris for technical assistance, and Monsanto for pMON10086. This work was supported in part by National Science Foundation Grant MCB0443709, by an endowment from the C. V. Griffin Sr. Foundation, and by a fellowship from the Mexican National Council for Science and Technology (CONACyT) (to R.I.D.d.l.G.).
Abbreviations
- PABA
p-aminobenzoate
- ADCS
aminodeoxychorismate synthase.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3675.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700409104/DC1.
References
- 1.Hanson AD, Roje S. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:119–137. doi: 10.1146/annurev.arplant.52.1.119. [DOI] [PubMed] [Google Scholar]
- 2.Scott J, Rebeille F, Fletcher J. J Sci Food Agric. 2000;80:795–824. [Google Scholar]
- 3.Holland B, Unwin ID, Buss DH. The Composition of Foods. Cambridge, UK: R Soc Chem and Ministry of Agriculture, Fisheries and Food; 1991. [Google Scholar]
- 4.Ames BN. Ann NY Acad Sci. 1999;889:87–106. doi: 10.1111/j.1749-6632.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 5.Bailey LB, Gregory JF., III J Nutr. 1999;129:779–782. doi: 10.1093/jn/129.4.779. [DOI] [PubMed] [Google Scholar]
- 6.Lucock M. Mol Genet Metab. 2000;71:121–138. doi: 10.1006/mgme.2000.3027. [DOI] [PubMed] [Google Scholar]
- 7.Stover PJ. Nutr Rev. 2004;62:S3–S12. doi: 10.1111/j.1753-4887.2004.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 8.Bailey LB. Nutr Rev. 2004;62:S14–S20. doi: 10.1111/j.1753-4887.2004.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 9.Bouis HE. J Nutr. 2002;132:491S–494S. doi: 10.1093/jn/132.3.491S. [DOI] [PubMed] [Google Scholar]
- 10.Adamson P UNICEF. Vitamin & Mineral Deficiency. Oxfordshire, UK: P&LA; 2004. The Micronutrient Initiative; pp. 20–40. [Google Scholar]
- 11.Rush D. Am J Clin Nutr. 2000;72:212S–240S. doi: 10.1093/ajcn/72.1.212S. [DOI] [PubMed] [Google Scholar]
- 12.Sifakis S, Pharmakides G. Ann NY Acad Sci. 2000;900:125–136. doi: 10.1111/j.1749-6632.2000.tb06223.x. [DOI] [PubMed] [Google Scholar]
- 13.Lucock M, Yates Z. Nat Rev Genet. 2005;6:235–240. doi: 10.1038/nrg1558. [DOI] [PubMed] [Google Scholar]
- 14.Rothenberg SP. Semin Hematol. 1999;36:65–74. [PubMed] [Google Scholar]
- 15.Storozhenko S, Ravanel S, Zhang GF, Rebeille F, Lambert W, Van Der Straeten D. Trends Food Sci Technol. 2005;16:271–281. [Google Scholar]
- 16.Nestel P, Bouis HE, Meenakshi JV, Pfeiffer W. J Nutr. 2006;136:1064–1067. doi: 10.1093/jn/136.4.1064. [DOI] [PubMed] [Google Scholar]
- 17.Diaz de la Garza R, Quinlivan EP, Klaus SM, Basset GJ, Gregory JF, III, Hanson AD. Proc Natl Acad Sci USA. 2004;101:13720–13725. doi: 10.1073/pnas.0404208101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hossain T, Rosenberg I, Selhub J, Kishore G, Beachy R, Schubert K. Proc Natl Acad Sci USA. 2004;101:5158–5163. doi: 10.1073/pnas.0401342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basset G, Quinlivan EP, Ziemak MJ, Diaz de la Garza R, Fischer M, Schiffmann S, Bacher A, Gregory JF, III, Hanson AD. Proc Natl Acad Sci USA. 2002;99:12489–12494. doi: 10.1073/pnas.192278499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deikman J, Kline R, Fischer RL. Plant Physiol. 1992;100:2013–2017. doi: 10.1104/pp.100.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Coker JS, Vian A, Davies E. Physiol Plant. 2005;124:311–322. [Google Scholar]
- 23.Quinlivan EP, Roje S, Basset G, Shachar-Hill Y, Gregory JF, III, Hanson AD. J Biol Chem. 2003;278:20731–20737. doi: 10.1074/jbc.M302894200. [DOI] [PubMed] [Google Scholar]
- 24.Deikman J, Xu R, Kneissl ML, Ciardi JA, Kim KN, Pelah D. Plant Mol Biol. 1998;37:1001–1011. doi: 10.1023/a:1006091928367. [DOI] [PubMed] [Google Scholar]
- 25.Kneissl ML, Deikman J. Plant Physiol. 1996;112:537–547. doi: 10.1104/pp.112.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasmire RF, Kader AA. Outlook. 1978;5:5–12. [Google Scholar]
- 27.Viitanen PV, Devine AL, Khan MS, Deuel DL, Van Dyk DE, Daniell H. Plant Physiol. 2004;136:4048–4060. doi: 10.1104/pp.104.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boggio SB, Palatnik JF, Heldt HW, Valle EM. Plant Sci. 2000;159:125–133. doi: 10.1016/s0168-9452(00)00342-3. [DOI] [PubMed] [Google Scholar]
- 29.Sybesma W, Starrenburg M, Kleerebezem M, Mierau I, De Vos WM, Hugenholtz J. Appl Environ Microbiol. 2003;69:3069–3076. doi: 10.1128/AEM.69.6.3069-3076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sybesma W, Van Den Born E, Starrenburg M, Mierau I, Kleerebezem M, De Vos WM, Hugenholtz J. Appl Environ Microbiol. 2003;69:7101–7107. doi: 10.1128/AEM.69.12.7101-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebeille F, Macherel D, Mouillon JM, Garin J, Douce R. EMBO J. 1997;16:947–957. doi: 10.1093/emboj/16.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouillon JM, Ravanel S, Douce R, Rebeille F. Biochem J. 2002;363:313–319. doi: 10.1042/0264-6021:3630313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsuda H, Suzuki Y. Biochem Biophys Res Commun. 1969;36:1–6. doi: 10.1016/0006-291x(69)90640-8. [DOI] [PubMed] [Google Scholar]
- 34.Ikemoto K, Sugimoto T, Murata S, Tazawa M, Nomura T, Ichinose H, Nagatsu T. Biol Chem. 2002;383:325–30. doi: 10.1515/BC.2002.035. [DOI] [PubMed] [Google Scholar]
- 35.Basset GJ, Quinlivan EP, Ravanel S, Rebeille F, Nichols BP, Shinozaki K, Seki M, Adams-Phillips LC, Giovannoni JJ, Gregory JF, III, Hanson AD. Proc Natl Acad Sci USA. 2004;101:1496–14501. doi: 10.1073/pnas.0308331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basset GJ, Ravanel S, Quinlivan EP, White R, Giovannoni JJ, Rebeille F, Nichols BP, Shinozaki K, Seki M, Gregory JF, III, Hanson AD. Plant J. 2004;40:453–461. doi: 10.1111/j.1365-313X.2004.02231.x. [DOI] [PubMed] [Google Scholar]
- 37.Xavier S, Macdonald S, Roth J, Caunt M, Akalu A, Morais D, Buckley MT, Liebes L, Formenti SC, Brooks PC. Int J Radiat Oncol Biol Phys. 2006;65:517–527. doi: 10.1016/j.ijrobp.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Hoagland RJ. Am J Med. 1950;9:272–274. doi: 10.1016/0002-9343(50)90054-4. [DOI] [PubMed] [Google Scholar]
- 39.Witter K, Werner T, Blusch JH, Schneider EM, Riess O, Ziegler I, Rodl W, Bacher A, Gutlich M. Gene. 1996;171:285–290. doi: 10.1016/0378-1119(95)00886-1. [DOI] [PubMed] [Google Scholar]
- 40.Messahel S, Pheasant AE, Pall H, Ahmed-Choudhury J, Sungum-Paliwal RS, Vostanis P. Neurosci Lett. 1998;241:17–20. doi: 10.1016/s0304-3940(97)00976-2. [DOI] [PubMed] [Google Scholar]
- 41.Messahel S, Pheasant AE, Pall H, Kerr AM. Eur J Paediatr Neurol. 2000;4:211–217. doi: 10.1053/ejpn.2000.0308. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida T, Akino M. Experientia. 1980;36:639–640. [Google Scholar]
- 43.Schubert KR, Hossain T, Quinlivan E, Rosenberg J, Selhub J, Hanson AD, Gregory JF., III Pteridines. 2005;16:79. [Google Scholar]
- 44.Koziel MG, Carozzi NB, Desai N. Plant Mol Biol. 1996;32:393–405. doi: 10.1007/BF00039392. [DOI] [PubMed] [Google Scholar]
- 45.Tieman DM, Ciardi JA, Taylor MG, Klee HJ. Plant J. 2001;26:47–58. doi: 10.1046/j.1365-313x.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- 46.Rick CM. In: Hybridization of Crop Plants. Fehr W, Hadley H, editors. Madison, WI: Am Soc Agronomy/Crop Sci Soc Am; 1980. pp. 669–680. [Google Scholar]
- 47.Rembold H, Gyure WL. Angew Chem Int Ed Engl. 1972;11:1061–1072. doi: 10.1002/anie.197210611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.