Abstract
The external layer of the Gram-negative bacterial outer membrane is primarily composed of a protective, selectively permeable LPS. The biosynthesis of LPS relies on UDP-3-O-acyl-glucosamine N-acyltransferase (LpxD), which transfers 3-hydroxy-arachidic acid from acyl carrier protein to the 2′ amine of UDP-3-O-myristoyl glucosamine in Chlamydia trachomatis. Our crystallographic study reveals that LpxD is a homotrimer, each subunit of which is constructed from a novel combination of an N-terminal uridine-binding domain, a core lipid-binding domain, and a C-terminal helical extension. Highly conserved residues dominate nucleotide binding. Phe-43 and Tyr-49 form π-stacking interactions with uracil, and Asn-46 and His-284 form hydrogen bonds with the phosphate groups. These interactions place the glucosamine moiety at the catalytic center formed by two adjacent subunits. Here His-247 and His-284 contribute to a mechanism involving nucleophilic attack by the amine of one substrate on the carbonyl carbon of an acyl carrier protein thioester conjugate. Serendipitously, our study reveals a fatty acid (FA) binding groove near the catalytic center. MS elucidated the presence of a FA mixture binding to LpxD, with palmitic acid the most prevalent. The placement of UDP-N-acetylglucosamine and the FA provides details of N-acyltransferase ligand interactions and allows for a description of structure and reactivity at an early stage of LPS assembly.
Keywords: Chlamydia trachomatis, enzyme structure, fatty acid binding, enzyme mechanism
Lipopolysaccharide (LPS) forms the amphipathic interface between Gram-negative bacteria and their environment and contributes protection against antibiotics and the complement system. The alternative name, endotoxin, is indicative of the capacity to cause septic shock by hyperstimulation of the immune system (1, 2). LPS consists of three components: core polysaccharide, O-antigen, and lipid A. The core polysaccharide is a branched structure of 9–12 sugar units and contains the unusual 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) whereas the O-antigen is a linear polysaccharide consisting of 50–100 repeating saccharide units with four to seven sugars per unit. Lipid A, the potent macrophage-activating component primarily responsible for endotoxin activity, is a phosphorylated glucosamine disaccharide carrying long-chain saturated fatty acid (FA) substituents that anchor LPS in the outer membrane. Lipid A is similar for all Gram-negative Enterobacteriaceae, and synthetic lipid A produces effects identical to that isolated from Escherichia coli in both in vitro and in vivo endotoxin tests (3). In addition, only lipid A and two Kdo moieties from the core are essential to support E. coli growth (4).
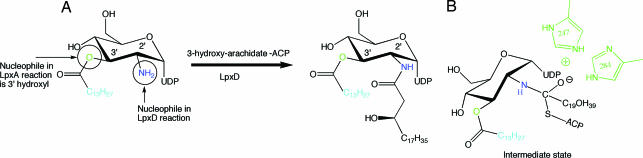
The biosynthesis of Kdo2-lipid A is best characterized in E. coli (4). The first three enzymes in the pathway are essential for generation of the outer membrane and for bacterial viability and so offer potential as therapeutic targets. Indeed, the treatment of infection using an inhibitor of UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC), the second enzyme in the pathway, has been successful in mice (5). LpxC and the first enzyme in lipid A biosynthesis, UDP-N-acetylglucosamine (UDP-GlcNAc) O-acyltransferase (LpxA), have been structurally characterized (6–10), but no such data are available for the third enzyme, UDP-3-O-acyl-glucosamine N-acyltransferase (LpxD). In E. coli, LpxD catalyzes the transfer of 3-hydroxy-myristic acid from acyl carrier protein (ACP) to UDP-3-O-[3-hydroxymyristoyl] glucosamine (11) whereas 3-hydroxy-arachidic acid is transferred onto UDP-3-O-myristoyl glucosamine in Chlamydia trachomatis (Fig. 1) (12).
Fig. 1.
Reaction catalyzed by C. trachomatis LpxD. The reaction is based on orthology to E. coli LpxD and the relative abundance of FAs attached to the 2′-N and 3′-OH positions of UDP-GlcNAc from C. trachomatis lipid A. Sites relevant to LpxA and LpxD reactivity are colored green and blue, respectively.
LpxD belongs to the left-handed β-helix family of proteins, of which LpxA is the founding member (8, 13). Each coil of the β-helix is constructed from three hexapeptide repeats of the consensus sequence [(Ile,Val,Leu)GlyXXXX]. Both LpxA and LpxD acylate a UDP-GlcNAc derivative (11, 14), and the amino acid sequences of the pair of enzymes in E. coli and C. trachomatis are 28% and 25% identical, respectively. Residues of LpxA likely to interact with UDP-GlcNAc and FA have only been inferred (10, 15–17) because the appropriate enzyme–ligand complexes have not been obtained.
We present crystallographic analyses of recombinant C. trachomatis LpxD in complex with UDP-GlcNAc, which represents a fragment of substrate, and FA serendipitously extracted from the bacterial expression system. These studies permit the correlation of structure with reactivity at an early stage in lipid A biosynthesis.
Results and Discussion
General Comments.
The structure of LpxD has been determined with experimental phases derived from a single-wavelength anomalous dispersion experiment applied to a selenomethionine derivative (Se-LpxD) (Table 1). Structures in the presence of 25 and 100 mM UDP-GlcNAc were also determined and are named complex I and complex II, respectively. A third structure, native LpxD, was solved in the absence of UDP-GlcNAc. Complex I provides the basis for analysis and figures, unless otherwise stated, because it is The most highly resolved structure. These structures are very similar, and the largest difference involves residues 43–52 of each subunit: The rmsd for overlay of main-chain atoms of complex I with II is 0.44 Å, and the rmsd for overlay of native LpxD with complex I and complex II 0.48 Å and 0.61 Å, respectively. Gel filtration chromatography (data not shown) indicates that LpxD forms a trimer of ≈121.5 kDa. The asymmetric unit consists of such a trimer with subunits A, B, and C of the model comprising residues 1–346, 1–345, and 3–348, respectively.
Table 1.
Crystallographic statistics
| Structure | Complex I | Complex II | LpxD | Se-LpxD |
|---|---|---|---|---|
| Unit cell, a, b, and c, Å | 98.8, 98.8, 283.1 | 98.9, 98.9, 283.0 | 98.7, 98.7, 284.5 | 98.5, 98.5, 283.4 |
| Data collection | ESRF ID14-EH4 | Rigaku RU200 | Rigaku Micromax | ESRF ID14-EH4 |
| Detector/λ, Å | ADSC Q4/0.97563 | R-Axis IV/1.5418 | R-Axis IV++/1.5418 | ADSC Q4/0.97930 |
| Cryoprotectant | 1.6 M (NH4)2SO4, 0.1 M Hepes (pH 7.5), 0.1 M NaCl, 20% ethylene glycol | 1.3 M (NH4)2SO4, 0.1 M MES (pH 6.5), 2% dioxane, 87.5 mM UDP-GlcNAc, 20% ethylene glycol | 3 M Li2SO4 | 1.3 M (NH4)2SO4, 0.1 M MES (pH 6.5), 2% dioxane, 20% ethylene glycol |
| Oscillation range/Δ, ° | 48.4/0.2 | 69/0.5 | 90/0.5 | 90/1.0 |
| Resolution range, Å | 50.0–2.2 | 28.5–3.1 | 40.0–2.7 | 50.0–2.5 |
| Reflections/multiplicity | 65,635/4.3 | 26,254/5.1 | 38,280/4.7 | 49,280/7.2 |
| Completeness, % | 91.1 (95.1) | 98.9 (99.9) | 96.5 (90.4) | 100 (100) |
| 〈I/σ (I)〉 | 11.9 (1.8) | 11.1 (2.1) | 11.3 (2.0) | 6.0 (1.0) |
| Rsym, % | 7.7 (61.4) | 15.3 (79.0) | 11.1 (50.8) | 10.6 (67.1) |
| Wilson B, Å2/DPI, Å | 47.8/0.21 | 92.2/0.49 | 53.7/0.35 | 50.8/- |
| Protein residues | 1,037 | 1,037 | 1,037 | |
| Waters/sulfates/FA | 496/6/3 | 95/6/3 | 155/8/3 | |
| UDP-GlcNAc | 1 | 2 | — | |
| Rwork/Rfree, % | 20.6/25.6 | 22.1/28.1 | 20.9/27.3 | |
| Average B values (subunit A/B/C), Å2 | 61.9/61.1/61.9 | 76.2/77.0/76.7 | 39.4/39.5/45.6 | |
| Overall | ||||
| Main/side chain | 61.4/61.9 | 76.5/76.8 | 40.7/42.4 | |
| Waters/sulfates | 64.5/69.9 | 58.3/75.7 | 37.2/39.5 | |
| UDP-GlcNAc/FA | 67.4/64.7 | 77.4/47.7 | -/59.7 | |
| B43–52 loop | 64.0 | 79.0 | 70.7 | |
| rms bond lengths, Å | 0.011 | 0.007 | 0.011 | |
| rms bond angles, ° | 1.344 | 1.039 | 1.387 | |
| PDB ID code | 2IU8 | 2IU9 | 2IUA | |
Values in parentheses pertain to the highest-resolution shell (width = 0.1 Å). B is the isotropic thermal parameter. ADSC, Area Detector Systems Corporation; ESRF, European Synchrotron Radiation Facility; DPI, diffraction-component precision index (43).
Overall Structure.
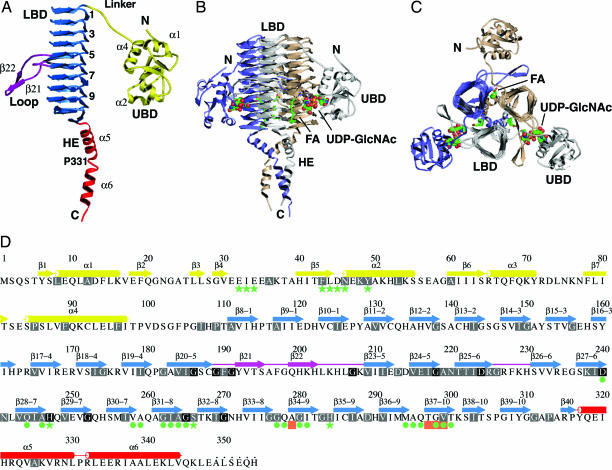
Each LpxD subunit (Fig. 2), with dimensions of ≈100 Å × 45 Å × 45 Å, is composed of two domains and a C-terminal helical extension (HE). The globular N-terminal uridine-binding domain (UBD) comprises residues 1–97, which form a five-stranded β-sheet (β2 and β4-β7) surrounded by four helices (α1–α4) and a short, two-stranded β-sheet (β1 and β3). Residues 98–110 form a linker section, ≈27 Å in length, leading to the lipid-binding domain (LBD), which is a left-handed β-helix structure constructed from 10 coils, each composed of three hexapeptide repeats (18). Four residues in this repeating sequence form a β-strand, and the remaining two form a 120° turn into the next strand. The LBD has the appearance of an elongated prism of length 53 Å, with an equilateral triangle of edge length 18 Å as the base. The continuity of the β-helix is disrupted in coils five and six by extruding loops. The first loop (β21 and β22) extends ≈25 Å from the prism, and the second extends 14 Å from the prism. From the LBD beginning at residue 317, a HE nearly 45 Å in length is observed. Pro-331 creates a kink of ≈30° and splits this section into two distinct helices (α5 and α6). Architectural comparisons with known structures (19) reveal that LpxD constitutes a unique combination of domains.
Fig. 2.
Structure of LpxD. (A) Ribbon diagram of a subunit. The UBD is yellow, the LBD is blue, loops are magenta, and the HE is red. Selected elements of secondary structure are labeled, and the coils are numbered. (B) The trimer. The view is parallel to the noncrystallographic symmetry threefold axis. Subunits are colored gray, wheat, and slate, and the domains of the gray subunit are labeled. UDP-GlcNAc (complex II) is represented by spheres, and palmitic acid is represented by sticks. The atoms of the ligands are colored as follows: C, green; N, blue; O, red; P, yellow. (C) Orthogonal view of the trimer. (D) Primary and secondary structure. β-strands are depicted by arrows, and α-helices are depicted by cylinders. Colors are as described in A. Disordered residues at the C terminus are marked by dots. Highly conserved residues (>60% identity in 85 sequences) are highlighted in gray, and strictly conserved residues (100% identity) are in black. Green stars and circles indicate residues that interact with UDP-GlcNAc and palmitic acid, respectively. Salmon boxes represent sites of conditionally lethal point mutations in E. coli and S. typhimurium LpxD.
Although the domain combination of LpxD is distinctive, the LBD is very similar to that of LpxA [supporting information (SI) Fig. 5]. Quantification of the structural relationship between the LpxD LBD and E. coli LpxA subunit yields an rmsd of 1.1 Å and a Z-score of 26.9 for the overlay of 191 Cα atoms. (Z-score measures the statistical significance of the best alignment. Typically, dissimilar structures present a Z-score of <2.0.) Both of these LBDs comprise 10 coils and are almost identical in length: In the superposition of the two domains, the final coils of the C terminus are in the same location whereas the N-terminal coil of LpxD has an extra 1.5 hexapeptide repeats. In addition, the loops that disrupt the left-handed β-helix extend from overlapping coils of the two proteins. Both LpxA and LpxD form trimers, and, in their quaternary structures, these loops interact with adjacent subunits to form an active site cleft.
The LpxD trimer is an equilateral-triangular prism with protrusions on each edge and a helical bundle at one end (Fig. 2B). The subunit-subunit interface, comprising 65% nonpolar residues, extends for almost the full length of the trimer with nearly one third of the accessible surface area (5,600 Å2) of each subunit occluded by oligomerization. The three HE sections form an amphipathic helical bundle. However, this feature does not contribute significantly to oligomer stability because truncated constructs lacking residues 319–354 or 331–354 still retain a trimeric structure as indicated by gel filtration chromatography (data not shown).
Three active sites are created near the inter-subunit regions. Here LpxD must bind the acceptor UDP-3-O-(3-hydroxymyristoyl)-glucosamine and an acyl-ACP thioester as the substrate donor (Fig. 1). Our structures identify FA and UDP-GlcNAc binding sites; the ACP binding site is inferred.
LpxD–UDP-GlcNAc Interactions.
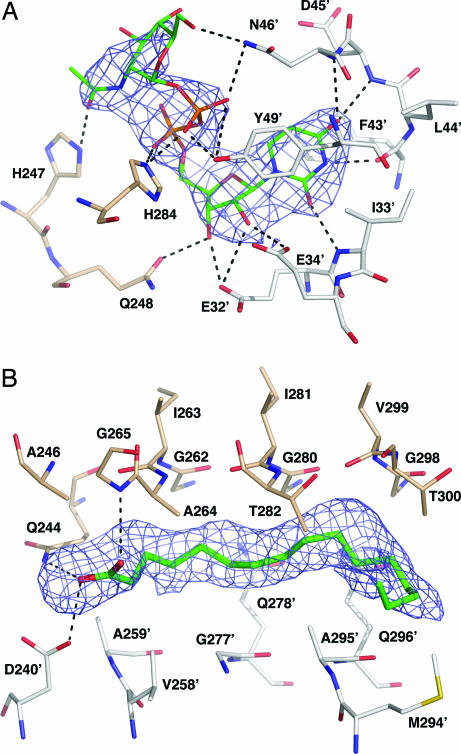
UDP-GlcNAc binds with the nucleotide interacting with a UBD of one subunit and the glucosamine moiety with the LBD of an adjacent subunit; the diphosphate bridges the two domains. The ligands are assigned to two subunits, the first being the one to which UDP binds and the second the one to which GlcNAc binds. In the A-C binding pocket, the residues 46–52 of the UBD contact crystallographic symmetry related molecules, which appear to preclude ligand binding (data not shown). The B-A binding pocket was 50% occupied by UDP-GlcNAc in complex I; in complex II, the B-A and C-B binding pockets were 75% and 50% occupied, respectively (Table 1). There is a gradient of order within this ligand as evidenced by the thermal parameter distribution and the electron density. Uracil is the most ordered component whereas the GlcNAc moiety displays higher thermal parameters and more diffuse density (Fig. 3A and SI Fig. 6).
Fig. 3.
LpxD–ligand interactions. (A) UDP-GlcNAc binding site from B-A binding pocket of complex II and omit difference electron density map associated with the ligand. The map, shown as blue mesh, has been calculated with coefficients |Fo − Fc| and αc and contoured at 3σ. Fo represents the observed structure factors, Fc represents the calculated structure factors, and αc represents the calculated phases for which ligand contributions were omitted. Protein residues within a 3.85-Å radius of the FA are labeled, and putative hydrogen bonds are shown as dashed lines. A prime symbol in the residue label signifies a residue from a partner subunit. The Cα atoms of chains B and A are shown in gray and wheat, respectively, and other atoms are colored as described in Fig. 2B. (B) FA omit noncrystallographic symmetry-averaged difference electron density map contoured at 3σ of the B-A binding pocket of complex I. Protein residues within 3.85 Å of the FA are labeled.
Identical binding interactions between LpxD and uracil are observed in both complexes but there is some variation with respect to the rest of the ligand. We confine discussion to the most reliable electron density, that observed in the B-A binding pocket of complex II (Fig. 3A and SI Fig. 6). Electron density for the polypeptide of residues 43–49 of the UBD is poor in the absence of any ligand and refined thermal parameters greatly exceed the average observed for the protein structure (Table 1). When UDP-GlcNAc is bound, well defined electron density for these residues is observed (data not shown) and the thermal parameters become comparable to the average for the protein. The pyrimidine is stacked between Phe-43 and Tyr-49, forming π-bond interactions. An aromatic residue at one of these positions is strictly conserved in LpxD sequences, suggesting a critical contribution to substrate binding. Hydrogen bonds between uracil and main chain groups of Ile-33, Phe-43, Leu-44, and Asp-45 further stabilize the complex (Fig. 3A and SI Fig. 6). The ribose hydroxyl groups form hydrogen bonds with the carboxylate groups of Glu-32 and Glu-34 from one subunit and the side chain of Gln-248 from a partner subunit. Although not strictly conserved, Glu-34 is a glutamate, aspartate, or glutamine in 65% of LpxD sequences, and Gln-248 is either glutamine or asparagine in 90% of LpxD sequences (data not shown). Conservation of polar or charged side chains at these sites suggests that two of the hydrogen bonds with the ribose could be preserved. The highly conserved Asn-46 and His-284 donate hydrogen bonds to the ligand phosphates, thereby orienting the GlcNAc moiety into a pocket created by the extending loops of the LBD from one subunit and the UBD of a partner subunit (Fig. 2B). The side chain of Asn-46 forms a hydrogen bond with 6′-OH of GlcNAc whereas His-247, located at the base of the pocket, participates in a hydrogen bond with the acetyl oxygen.
There are structural and functional similarities between LpxD and LpxA. The similarity in mechanism between LpxA and LpxD is that in each case a nucleophilic addition occurs: the distinction is that substitution occurs at different chemical groups and positions on the glucosamine (Fig. 1). The nucleophile is the C3′ hydroxyl in LpxA and the 2′ primary amine in LpxD. Chemical modification and site-directed mutagenesis of E. coli LpxA suggest that acylation of UDP-GlcNAc proceeds through a general base-catalyzed mechanism with His-125, a strictly conserved and essential residue, assigned as the base (16). A structural overlay superimposes E. coli LpxA His-125, the assigned general base, and C. trachomatis LpxD His-247 (SI Fig. 5), suggesting an important contribution to this enzyme's function. Furthermore, His-247 is strictly conserved among sequences of LpxD. Structural homology and sequence identity indicate that His-247 of LpxD can contribute to a general base mechanism by acquiring an amine proton before the nucleophilic attack.
An additional role for His-247 is also suggested. His-247 is one of a pair of histidines that flank the acetyl group of UDP-GlcNAc (Fig. 3A and SI Fig. 6). Once protonated, His-247 can, in conjunction with His-284, stabilize the negatively charged transition state (Fig. 1B). Conservation of this histidine pair emphasizes its potential importance in catalysis. His-284 is conserved in 84 of the 85 LpxD sequences compared (Fig. 2C). The exception, in Anabaena sp. PCC7120, is an alteration to a glutamine that could function in the same capacity as a histidine to stabilize a carboxyanion intermediate. The second stage of catalysis, elimination, follows: breakage of the reactive C-S bond occurs, and the carbonyl π-bond is reformed. ACP is released, and the nucleotide derivative is now left carrying two lipid tails.
ACP delivers acyl groups and FA derivatives in numerous metabolic pathways, including lipid A biosynthesis. ACP is highly acidic and requires an electropositive surface on its cognate partners for binding. Chemical modification of arginine or lysine residues on LpxA compromises the acyl transferase activity (16), and, in FA biosynthesis, interactions of ACP with FabG, FabH, and AcpS are dependent on specific arginine residues (20–22). Like other ACP-targeted enzymes, LpxD presents an electropositive surface near the active site (SI Fig. 7). Several basic residues are proximal to the active site His-247. In particular, the highly conserved Lys-48 is a possible binding partner.
FA Identification.
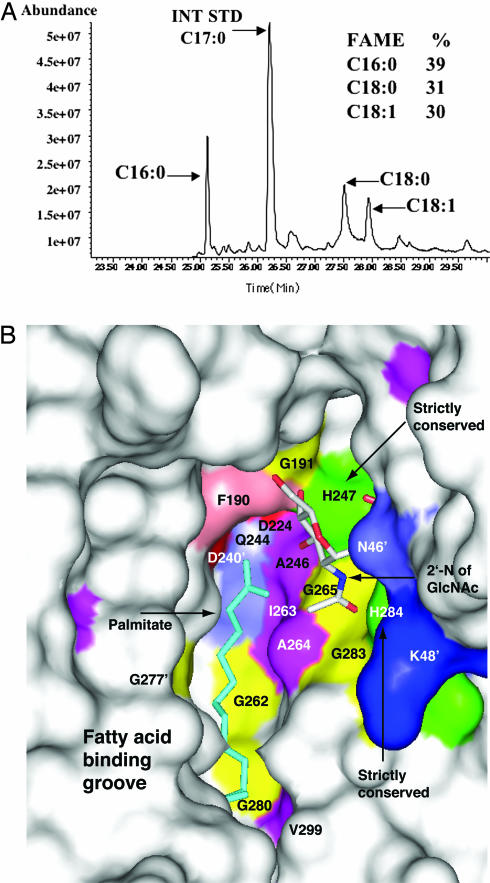
Fortuitously, strong electron density consistent with a FA, likely acquired from the E. coli expression system, was observed in grooves formed by LBD pairs (Fig. 3B and SI Fig. 8). It was necessary to characterize this ligand, and so LpxD was treated with ethanol and the extracted FA entities derivatized to the corresponding methyl esters (FAME) and hydroxy-FAMEs. Samples were analyzed by GC-MS, and the retention times and fragmentation patterns were compared with FAME standards. The ligands are indeed FAs and were identified as palmitic acid (C16:0), stearic acid (C18:0), and a monounsaturated C18:1 FA, observed in a ratio of ≈4:3:3 (Fig. 4A). Comparisons of the mean retention time differences between the internal standard (C17:0) and various C18:1 standards allowed the C18:1 unsaturated compound to be assigned as cis-vaccenic acid. No trimethylsilyl derivatized hydroxy-FAMEs were observed (SI Fig. 9). Palmitic acid, an abundant saturated FA in E. coli (23), is consistent with the observed electron density.
Fig. 4.
LpxD–FA complex. (A) Identification of bound FA. GC-MS analysis, chain length, and saturation states are indicated, and the key shows the relative percentages. (B) Surface view of conserved residues with ligands depicted as sticks. Palmitic acid is colored cyan, and UDP-GlcNAc is colored according to atom type: C, white; N, blue; O, red. Conserved residues are colored by type; basic residues are blue with the exception of His-247 and His-284, which are colored green. Acidic residues are red, aromatic residues are salmon, glycine residues are yellow, polar residues (Asn, Gln, Ser, Thr, and Cys) are slate blue, and aliphatic residues (Ala, Ile, Val, Leu, Met, and Pro) are magenta. Colored residues that form part of the FA and UDP-GlcNAc binding pockets include Gly-262, Gly-280, Gly-265, Ala-246, Ile-263, Ala-264, Asp-240, and Gln-244.
LpxD–FA Interactions.
A common theme in FA-binding proteins is the presence of a hydrophobic pocket or cavity that accommodates the FA tail (24). In LpxD, the FA binds in a hydrophobic groove created by residues on the first β-strands of coils 8–10 of one LBD (residues Ala-246, Gly-262, Ile-263, Ala-264, Gly-265, Gly-280, Ile-281, Thr-282, Gly-298, Val-299, and Thr-200) and the turns between coils six to nine of a second LBD (residues Val-258, Ala-259, Gly-277, Gln-278, Met-294, Ala-295, and Gln-296) (Figs. 2B and 4B). The ligand carboxylate, likely protonated, potentially forms hydrogen bonds with Asp-240 OD2 of one LBD and Gln-244 NE2 and the Gly-265 amide from the other LBD. In other FA-binding proteins, the size, shape, and hydrophobicity of FA-binding pockets contribute to ligand specificity: these characteristics influence the length and degree of saturation of FA bound (24). The FA binding site in LpxD suggests that chain length and degree of saturated FA bound is limited because the FA binding groove is narrow, linear, and ≈18 Å in length (measured from ND2 of His-247 of one subunit to OE1 of Gln-296 of an adjacent subunit).
We propose that this FA binding groove identifies the binding site for the myristic acid moiety of the substrate. The conditionally lethal point mutations of S271N of E. coli LpxD (25) and M288K, G289D and V291M of Salmonella typhimurium LpxD (26, 27) map to Ala-279, Thr-297, Gly-298, and Val-299 of C. trachomatis LpxD, respectively (Fig. 2C). These four residues are positioned in the FA binding groove of C. trachomatis LpxD and an increase in side chain size would have disruptive effects on both the structure and ligand binding. Residues that create the FA binding groove are highly conserved with >60% observed to be identical in 85 LpxD sequences (Fig. 2C). A similar groove, formed by two β-coils in Helicobacter pylori LpxA, binds 1-n-octyl-β-d-thioglucoside, which was an additive for crystallization (7) and a pentadecapeptide with antibacterial properties (10). The mutation of G173M in E. coli LpxA changes substrate preference from 14 to 10 carbon atoms (15). In a structural alignment of LpxA and LpxD, Gly-173 corresponds to Ala-295 (SI Fig. 5), and the alanine interacts with the FA tail in our LpxD structure (Fig. 3B).
Concluding Remarks.
We have determined the structure of C. trachomatis LpxD, the N-acyltransferase of lipid A biosynthesis. Complexes with UDP-GlcNAc and the fortuitous discovery of bound FA provide insights into specificity and mechanism at an early stage of lipid A biosynthesis. There is a strictly conserved π-stacking interaction for binding uracil and, within the catalytic center, two strictly conserved histidines implicated in the enzyme mechanism. The characterization of bound FA in a lipid A acyltransferase identifies residues which contribute to lipid binding and correlates to lethal point mutations in E. coli and S. typhimurium LpxD. In addition, this correlates to a groove implicated in substrate binding in LpxA by previous structural studies.
Materials and Methods
Cloning, Expression, and Purification of LpxD.
The lpxD gene was amplified from C. trachomatis (serovar B) genomic DNA, cloned into pET15b (Novagen, Madison, WI), and heat-shock-transformed into E. coli BL21(DE3) (Stratagene, La Jolla, CA). Gene expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside to cultures in Luria broth containing carbenicillin (50 mg·liter−1) when the A550 reached 0.6–0.8. Selenomethionine derivatization (Se-LpxD) used the methionine auxotrophic strain E. coli B834(DE3) (Novagen) cultured in M9 minimal media containing all amino acids (50 mg·liter−1) except methionine, which was replaced by selenomethionine (50 mg·liter−1). Cell pellets were resuspended in buffer [20 mM Tris (pH 8), 500 mM NaCl, 15 mM imidazole, and 3 mM 2-mercaptoethanol] containing lysozyme and DNase I (Roche Diagnostics, Burgess Hill, U.K.) and lysed in a French press. LpxD was purified on a 5-ml HiTrap Chelating HP column (GE Healthcare, Pittsburgh, PA) preloaded with Ni2+, using a gradient of 0–500 mM imidazole. Fractions of LpxD were pooled then dialyzed into 10 mM Tris (pH 8), 500 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine hydrochloride or DTT, and concentrated to 260–300 μM (theoretical ε0, 11,520 M−1·cm−1). Gel filtration (Superdex 200; GE Healthcare) was performed on the sample that led to the LpxD structure (Table 1). Sample purity was ascertained by SDS/PAGE and MALDI-TOF MS. The latter method also confirmed full incorporation of Se in Se-LpxD. Further details are given in SI Methods.
Crystallographic Methods.
Crystals of LpxD (Table 1) were grown in a matter of days at 20°C by hanging drop vapor diffusion using 1 μl of protein (5 or 10 mg·ml−1) and 1 μl of reservoir containing 1.2–1.3 M (NH4)2SO4, 0.1 M Mes (pH 6.5), 2% (vol/vol) PEG 400 or dioxane, and 1 mM tris(2-carboxyethyl)phosphine hydrochloride or DTT. To investigate LpxD–ligand interactions in the active site, crystals were grown in the presence of UDP-GlcNAc, which represents a fragment of the substrate. For complexes I and II, respectively, LpxD was incubated at 4°C for 30 min with 25 mM or 100 mM UDP-GlcNAc before crystallization. Tetragonal prisms (0.1 mm × 0.1 mm × 0.4 mm) were cryoprotected and flash-cooled to −170°C, and diffraction data were recorded in-house or at the European Synchrotron Radiation Facility (Table 1).
Data were integrated and scaled by using MOSFLM (28) and SCALA from the CCP4 suite of programs (29) or DENZO/SCALEPACK (30). The crystals, space group P41212, present a trimer in the asymmetric unit. A single-wavelength anomalous dispersion experiment identified 9 of 15 selenium positions in the asymmetric unit of Se-LpxD and provided phase estimates [SOLVE (31)] to 3.0-Å resolution. Density modification, phase extension, and construction of a model comprising ≈45% of the residues were performed with RESOLVE (32). The Se-LpxD model was completed in O (33) and refined with REFMAC5 (34). The structure of complex I was solved by molecular replacement (35) using the Se-LpxD model and refined using TLS (translation, libration, screw analysis) and maximum-likelihood restrained refinement without the use of noncrystallographic symmetry. The calculation of Rfree was performed on 5% of the data. Maps were inspected and the model was improved by using O and COOT (36). Waters were identified with the CCP4 program suite; PRODRG (37) provided ligand dictionaries used for refinement. Models of complex II and the native LpxD were obtained by rigid body refinement using complex I and then refined as described above. In these instances, noncrystallographic symmetry restraints were used during the initial stages of refinement. The occupancies for UDP-GlcNAc were based on consideration of refined thermal parameters and the appearance of electron and difference density maps.
Model geometry was analyzed by using PROCHECK (38). Residues are within allowed regions of a Ramachandran plot except for Asn-46 of subunit B in complex I; Asn-46 of subunits A and C; and Ala-50 of subunit B in LpxD. For complexes I, II, and LpxD, respectively, subunit A superposes onto B with an rmsd of 1.53 Å, 1.62 Å, and 1.57 Å over 345 Cα atoms; A superposes onto C with an rmsd of 2.27 Å, 2.21 Å, and 2.25 Å over 345 Cα atoms; and C superposes onto B with an rmsd of 1.27 Å, 1.09 Å, and 1.28 Å over 343 Cα atoms. The largest variation arises because of differences in the UBD and linker regions (residues 1–110), with the rmsd varying between 0.94 and 1.78 Å depending on the choice of model and chains. The rmsd is <0.65 Å for the corresponding superpositions of the LBD and HE (residues 110–345) in all of the models. Crystallographic contacts occur primarily through the UBDs of each subunit. Secondary structure was assigned by using a combination of DSSP (39), PROMOTIF (40), PROCHECK, and visual inspection. The trimer interface was analyzed with the Protein–Protein Interaction server (41). Figures were prepared in PyMOL (DeLano Scientific, San Carlos, CA), except for Fig. 1 (ChemDraw; CambridgeSoft, Cambridge, MA) and Fig. 2D and SI Fig. 5D (ALINE; C. S. Bond and A. W. Schüttelkopf, personal communication).
Extraction and Characterization of Bound FAs.
All glassware was sonicated and acid-washed before use during protein purification and sample handling for FA characterization. Ice-cold ethanol (800 μl) was slowly added to LpxD (200 μl of 227 μM) previously dialyzed in 500 mM NaCl/20 mM NH4HCO3. The mixture was vortexed and then incubated at −20°C for several days, after which precipitated protein was removed by centrifugation (800 × g for 15 min at 25°C). The supernatant containing extracted ligands was dried under N2. The sample was resuspended in methanol (200 μl) and spiked with an internal FA standard C17:0 (10 μl, 1 mM). Characterization and quantification of ligands were conducted by making the FAME derivatives followed by GC-MS analysis following established protocols (42). The presence of hydroxy-FAs was investigated by producing the trimethylsilyl derivatives of the FAME products. FAME identification was carried out by comparison of retention times with appropriate standards (Larodan Lipids, Malmö, Sweden) and trimethylsilyl derivatives of FAME of 2-hydroxy-myristate and 3-hydroxy-myristate standards (Sigma–Aldrich, St. Louis, MO). Further details are in SI Methods.
Supplementary Material
Acknowledgments
We thank D. Longbottom (Moredun Research Institute) for the gift of genomic DNA and C. Bond, A. Schüttelkopf, and the staff at the European Synchrotron Radiation Facility, in particular G. Leonard, for help and advice. This research was supported by The Wellcome Trust and the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets).
Abbreviations
- FA
fatty acid
- FAME
FA methyl ester
- LBD
lipid-binding domain
- UBD
uridine-binding domain
- HE
helical extension
- UDP-GlcNAc
UDP-N-acetylglucosamine
- ACP
acyl carrier protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2IU8, 2IU9, and 2IUA).
This article contains supporting information online at www.pnas.org/cgi/content/full/0606356104/DC1.
References
- 1.Cohen J. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Bojorquez LN, Dehesa AZ, Reyes-Teran G. Arch Med Res. 2004;35:465–479. doi: 10.1016/j.arcmed.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Galanos C, Luderitz O, Rietschel ET, Westphal O, Brade H, Brade L, Freudenberg M, Schade U, Imoto M, Yoshimura H, et al. Eur J Biochem. 1985;148:1–5. doi: 10.1111/j.1432-1033.1985.tb08798.x. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Whitfield C. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CR. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 6.Coggins BE, Li X, McClerren AL, Hindsgaul O, Raetz CR, Zhou P. Nat Struct Biol. 2003;10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee BI, Suh SW. Proteins. 2003;53:772–774. doi: 10.1002/prot.10436. [DOI] [PubMed] [Google Scholar]
- 8.Raetz CR, Roderick SL. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 9.Whittington DA, Rusche KM, Shin H, Fierke CA, Christianson DW. Proc Natl Acad Sci USA. 2003;100:8146–8150. doi: 10.1073/pnas.1432990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams AH, Immormino RM, Gewirth DT, Raetz CRH. Proc Natl Acad Sci USA. 2006;103:10877–10882. doi: 10.1073/pnas.0604465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly TM, Stachula SA, Raetz CR, Anderson MS. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 12.Rund S, Lindner B, Brade H, Holst O. J Biol Chem. 1999;274:16819–16824. doi: 10.1074/jbc.274.24.16819. [DOI] [PubMed] [Google Scholar]
- 13.Vaara M. FEMS Microbiol Lett. 1992;76:249–254. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- 14.Anderson MS, Raetz CR. J Biol Chem. 1987;262:5159–5169. [PubMed] [Google Scholar]
- 15.Wyckoff TJ, Lin S, Cotter RJ, Dotson GD, Raetz CR. J Biol Chem. 1998;273:32369–32372. doi: 10.1074/jbc.273.49.32369. [DOI] [PubMed] [Google Scholar]
- 16.Wyckoff TJ, Raetz CR. J Biol Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 17.Lee BI, Lee JY, Moon J, Han BW, Suh SW. Acta Crystallogr D. 2002;58:864–866. doi: 10.1107/s0907444902004845. [DOI] [PubMed] [Google Scholar]
- 18.Vuorio R, Harkonen T, Tolvanen M, Vaara M. FEBS Lett. 1994;337:289–292. doi: 10.1016/0014-5793(94)80211-4. [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y-M, Wu B, Zheng J, Rock CO. J Biol Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. J Biol Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 22.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure Fold Des. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 23.Ratledge C, Wilkinson SG. Microbial Lipids. London: Academic; 1988. [Google Scholar]
- 24.Hamilton JA. Prog Lipid Res. 2004;43:177–199. doi: 10.1016/j.plipres.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Vuorio R, Vaara M. J Bacteriol. 1992;174:7090–7097. doi: 10.1128/jb.174.22.7090-7097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirvas L, Koski P, Vaara M. EMBO J. 1991;10:1017–1023. doi: 10.1002/j.1460-2075.1991.tb08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirvas L, Vaara M. FEMS Microbiol Lett. 1992;90:289–294. doi: 10.1016/0378-1097(92)90662-8. [DOI] [PubMed] [Google Scholar]
- 28.Leslie AGW. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. Vol 26. Warrington, UK: Daresbury Lab; 1992. [Google Scholar]
- 29.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods in Enzymology. Vol 276. London: Academic; 1997. p. 307. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 36.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Schüttelkopf AW, van Aalten DM. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;283:283–291. [Google Scholar]
- 39.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 40.Hutchinson EG, Thornton JM. Protein Sci. 1996;15:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones S, Thornton JM. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fyffe SA, Alphey MS, Buetow L, Smith TK, Ferguson MA, Sorensen MD, Bjorkling F, Hunter WN. J Mol Biol. 2006;356:1005–1013. doi: 10.1016/j.jmb.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 43.Cruikshank DWJ. Acta Crystallogr D. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.