Abstract
Prp19/Pso4, a U-box containing E3 ligase, has a demonstrated role in pre-mRNA splicing, but has also been implicated in both yeast and mammalian cells as having a direct role in DNA damage processing. In this report we provide further evidence in support of this latter assertion. We show that hPrp19 forms an ubiquitylated oligomeric species that is resistant to disruption by SDS gel electrophoresis under nonreducing conditions suggesting that is mediated by a thiolester between ubiquitin and a cysteine residue in Prp19. The level of this species is significantly enhanced upon treatment of cells with DNA damaging agents, and its association with chromatin is increased. In addition, hPrp19 is known to form a stable core complex with Cdc5L, Plrg1, and Spf27; however, ubiquitylated hPrp19 fails to interact with either Cdc5L or Plrg1 indicating that DNA damage can induce profound alterations to the hPrp19 core complex. Finally, we show that overexpression of hPrp19 in human cells provides a pro-survival affect in that it reduces the levels of apoptosis observed after exposure of cells to DNA damage.
The pso4-1 mutant of S. cerevisiae was isolated in a genetic screen for strains sensitive to the cross-linking agent psoralen plus UVA (PUVA) [1, 2]. Characterization of this mutant indicated that it was particularly sensitive to bifunctional alkylating agents, but was also sensitive to a broad range of DNA damaging agents including IR, UV, and monofuntional alkylating compounds. Induced mutagenesis and induced and spontaneous mitotic recombination were all greatly reduced in this mutant suggesting that Pso4 is involved in a recombinational pathway of error-prone repair. Based on epistasis analysis PSO4 was assigned to both the yeast RAD6 and RAD52 groups indicating the pleiotropic nature of this mutation [3, 4]. Interestingly, cloning of PSO4 showed that it was allelic to PRP19 a previously characterized component of the pre-mRNA splicing complex in both yeast and human cells [5–10].
The complete function of Prp19 in mRNA splicing is not known, however, it has been shown that the Prp19p-associated complex is required for activation of the pre-mRNA splicesome and for the stable association of the small nuclear RNAs U5 and U6 with the spliceosome after U4 is dissociated [11, 12]. These findings suggest a possible structural role for the Prp19-associated complex in pre-mRNA splicing. Mammalian Prp19 associates with a large number of splicing factors, although, it appears to form a core complex with three other proteins including Cdc5L, Plrg1, and Spf27 [9]. The structure of the Prp19 core complex is not completely resolved, however, it has been shown that yeast Prp19 forms a tetramer via its coiled coil domains, and that the tetramer interacts with one copy of Cdc5L through the associated coiled coil domains [13]. In addition, a recent study has shown that within the tetramer the U-box domains of Prp19 interact to form two homodimers [14]. The stoichiometry of the other two members of the core complex are not known, however, both Plrg1 and Spf27 also directly interact with Prp19. The only known catalytic center in any of the four Pso4 complex members is a U-box domain located in the amino terminus of Prp19. U-box domains have been shown to contain an E3 ubiquitin (Ub) ligase activity [15, 16], and such activity has been demonstrated for Prp19 in vitro [14, 17, 18], and this function is required for pre-mRNA splicing in vivo [17, 18].
The discovery that PSO4 was allelic to PRP19 led to the possibility that the pleiotropic nature of this mutation in response to genotoxic agents may be due to a generalized splicing defect. However, an analysis of splicing of the intron containing RAD14 gene in the pso4-1 mutant showed that the contribution of Prp19/Pso4 in the repair of UV damage is independent of RAD14 pre-mRNA processing [19]. Furthermore, the human protein has also been shown to interact with terminal deoxynucleotidyl transferase, and to be involved in mediating cell survival after DNA damage [20]. In addition, we have recently demonstrated a biochemical role for the core complex in processing of ICLs in vitro, and showed a direct physical interaction between Cdc5L and WRN [21]. These findings indicate that the Prp19/Pso4 core complex has a direct role in mediating the cellular response to DNA damage in addition to its demonstrated role in pre-mRNA splicing. In this report, we present further evidence that mammalian Prp19/Pso4 is involved in the DNA damage response.
Materials and methods
Cell Culture and nucleic acid transfections
Hela cells were cultured in DMEM medium plus 10% fetal calf serum. Transfections of plasmids and siRNAs were performed using Lipofectamine 2000 and Oligofectamine (Invitrogen), respectively, following the manufacturer’s instructions. siRNAs were purchased from Dharmacon. The sequence of the hPrp19 siRNA was previously described [21]. For treatment with DNA damaging agents, methyl-methane sulfonate (MMS) or mitomycin C (MMC) was added to the medium 24 hours after transfection. Cells were subsequently harvested at indicated time points in Laemmli buffer (under non-reducing conditions) for immunoblot analysis, or cells were fixed for cell cycle analysis.
Cloning and Site-directed Mutagenesis
hPrp19 cDNA clone (Image clone number 5267341) was purchased from the American Type Culture Collection. Vectors for expression of GFP-Cdc5L and GFP-PLRG1 in mammalian cells were described previously [9]. hPrp19, Cdc5L, and Plrg1 cDNAs were subsequently cloned into the pEntr11 vector, and then moved to the pDest26, 27, 53 and pcDNA3.1/nV5 vectors of the Gateway cloning system (Invitrogen). A restriction fragment of Prp19 lacking the U-box domain was prepared by digestion with Nci I and Xho I and inserted into the pEntr11 vector to construct hPrp19-U plasmid. The hPrp19 point mutation clones, hPrp19-C5A and hPrp19-D40A, were prepared using a Site-Directed Mutagenesis kit (Stratagene), and were confirmed by DNA sequencing.
Phosphocellulose column fractionation and chromatin isolation
HeLa whole cell extracts were prepared and fractionated as described [21, 22]. For responses to DNA damage, HeLa cells were transfected with the indicated plasmid, and MMS was added to the medium 24 hours later. Chromatin fractionation was performed at the indicated time points.
Immunoblotting and pull down assays
hPrp19 and CDC5L antibodies were kindly provided by Paul Ajuh. V5 (Invitrogen), GST (Cell Signaling Technology), GFP and Ku70 (Santa Cruz Biotechnology ), Orc2 (BD Bioscience), and Flag (Sigma) antibodies were obtained from the indicated manufactures. Standard protocols were used for gel electrophoresis and immunoblotting with one modification. For electrophoresis of samples under reducing conditions, DTT was freshly added to the samples to a final concentration of 50 mM prior to adding SDS-loading buffer. For pull down assays, cells were lysed in NP40 buffer (0.5% NP40, 50 mM Tris (pH 8.0), 120 mM NaCl) and performed as described [21].
Cell cycle analysis
HeLa cells were transfected with the indicated plasmid. MMS was added to the medium 24 hours later, and cells were processed as described [23] at the indicated time points.
Results
hPrp19 forms ubiquitylated SDS-stable oligomers
During the course of purification of endogenous hPrp19 we noted the appearance in immunoblots of a higher molecular weight species that migrated at approximately the size of dimeric Prp19 (Fig. 1A). Although the oligomerization of Prp19 has been reported through interaction among its coiled-coil domains and between its U-box domains [13, 14], the species of hPrp19 observed here was resistant to SDS gel electrophoresis. To confirm that this band represented a hPrp19 species, we expressed a His-tagged version of hPrp19 (His-hPrp19) in HEK293 cells. An examination of the lysates by immunoblotting under nonreducing gel electrophoresis conditions indicated that hPrp19 does, in fact, form SDS-resistant species (Fig. 1B). Since Prp19 has been shown to autoubiquitylate itself in vitro [16] it was possible that these SDS-resistant species represented highly ubiquitylated forms. To address this possibility, we co-expressed HA tagged Ub (HA-Ub) and His-hPrp19 and again observed these higher molecular weight species (Fig. 1C, left panel). An anti-HA pull-down experiment showed that both monomeric and higher molecular weight forms of hPrp19 were ubiquitylated (Fig. 1C, right panel). Interestingly, when His-hPrp19 was electrophoresed in the presence of 50 mM DTT the higher molecular weight forms disappeared suggesting that they were unstable under reducing conditions (Fig. 1D). A similar scenario has been observed with the E2 Ub ligase Cdc34 in which self-association is mediated by an Ub thiolester attached to a cysteine residue [24]. The formation of the Ub thiolester and subsequent self-association were shown to be required for Cdc34’s catalytic activity.
Figure 1.
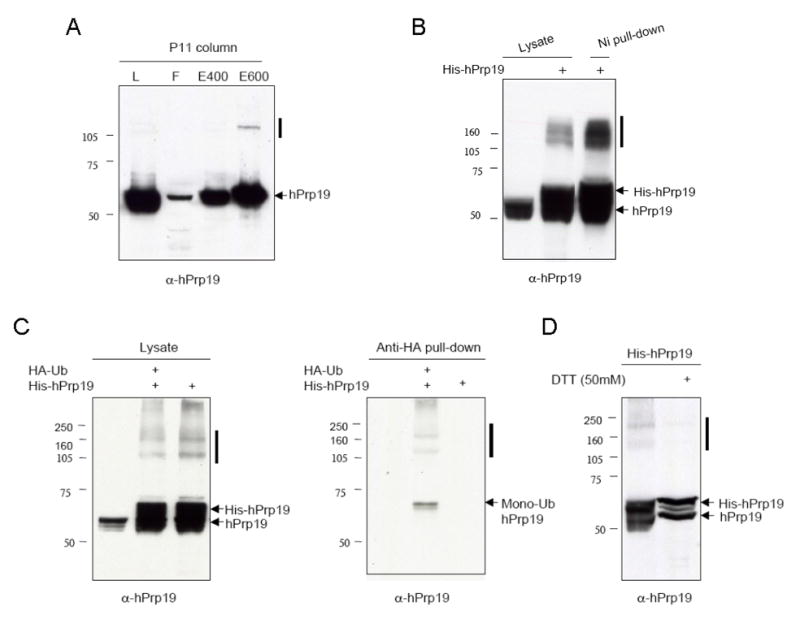
hPrp19 forms ubiquitylated SDS-resistant species. (A) Endogenous hPrp19 from HeLa cells was fractionated by phosphocellulose (P11) chromatography and analyzed by SDS-PAGE and immunoblotting. “L” indicates total lysate, “F” indicates flowthrough. (B) Immunoblot showing results of His-Prp19 overexpressed in HEK293 cells. (C) His-hPrp19 and HA-Ub were co-expressed in HEK 293 cells and resulting lysates were subjected to anti-HA pull down. (D) His-hPrp19 was expressed in HEK 293 cells and lysates were subjected to SDS-PAGE with or without DTT. Solid bars indicate the oligomerized forms of hPrp19.
To further confirm these initial conclusions about the structure of hPrp19, we expressed both GST and GFP tagged versions of the molecule. Again, higher molecular weight forms were detectable and these forms were sensitive to DTT (Fig. S1A). To determine if these higher molecular weight forms represented oligomerization of hPrp19, we co-expressed GST- and GFP-tagged hPrp19, performed a GST pull-down, and subsequently immunoblotted for GFP. This experiment showed that the higher molecular weight species contain both the GST and GFP tags indicating that these species are oligomers of hPrp19 (Fig. S1B, upper panesl). A similar experiment was performed using V5-tagged hPrp19 in place of GFP-hPrp19 and the same results were obtained (Fig. S1B, lower panels). In addition, we showed that these oligomeric species are sensitive to disruption by DTT, which again suggests the likelihood that they are mediated or stabilized by an Ub thiolester.
DNA damage affects chromatin association and the structure of the Prp19 core complex
Previous reported findings including ours have indicated that Prp19 plays a direct role in DNA repair processing [1–4, 20, 21], although the nature of this role remains undefined. To examine whether hPrp19 self-association is affected by exposure of cells to DNA damaging agents, we expressed GFP-hPrp19, and treated the transfected cells with methyl-methane sulfonate (MMS). As shown (Fig. 2A), with time the levels of oligomerized hPrp19 steadily increased. Similar results were also obtained upon examination of endogenous hPrp19, particularly after treatment with mitomycin C (MMC) (Fig. 2B,C). To determine if the localization of hPrp19 is altered by exposure of cells to DNA damage, we examined soluble (S) and chromatin (P) fractions after MMS treatment. Both monomeric and oligomerized forms of hPrp19 were shown with time to increase their association with the chromatin fraction (Fig. 2D).
Figure 2.

DNA damage induces oligomerization and enhanced chromatin association of hPrp19. (A) Immunoblot showing GFP-hPrp19 upon treatment of transfected cells with the indicated concentrations of MMS. (B,C) Immunoblot showing endogenous hPrp19 after exposure of HeLa cells to either MMS or MMC. (D) Immunoblot showing the presence of overexpressed V5-hPrp19 in soluble (S) or chromatin (P) fractions after exposure of cells to MMS. Orc2 is origin recognition complex protein 2. “NT” indicates not transfected.
The U-box domain of Prp19 is highly conserved from budding yeast to mammals (Fig. S2A). Mutation of conserved residues including C5 and D40 have been shown to inactivate the Ub E3 ligase function of the Prp19 U box domain [17]. We therefore prepared alanine mutants at these sites and examined the mutant proteins for oligomerization. Both the C5A and D40A mutants were able to form oligomers (Fig. S2B). In addition, GST and GFP tagged versions of the mutant protein still retained the ability to interact (Fig. S2C) indicating that the E3 Ub ligase function of hPrp19 is not required for self-association.
Since inactivation of the U-box by point mutations did not affect oligomerization, we examined whether deletion of the entire U-box would affect self-association (Fig. S3A). Interestingly, the U-box deletion mutant did not appear to be able to self-associate upon overexpression as shown by immunoblotting or a pull down assay (Fig. S3B). However, it retained the ability to interact with wild-type hPrp19 as shown by a pull-down assay (Fig. S3C). These findings coupled with those described above suggest that the U-box domain itself, but not its catalytic activity is required for self-association. We next addressed the question of whether the observed ubiquitylation of hPrp19 occurred within or outside of the U-box domain. For this experiment both wild-type and the U-box deletion mutant were co-expressed with HA-Ub, and pull-down assays were performed from the resulting lysates. Both forms of hPrp19 were pulled-down by anti-HA beads indicating that the site(s) of ubiquitylation lies outside of the U-box domain (Fig. S3D). The gel shown in Fig. S3D was run in the presence of DTT in order to collapse all the species into a single band. Since the wild-type hPrp19 expressed with or without co-expression of HA-Ub migrated at the same position in the gel, the HA-Ub has apparently been removed by exposure to DTT (the attachment of HA-Ub should have caused an approximate 10 kD increase in mass). This result agrees with our conclusion reached above that the ubiquitylation of hPrp19 is mediated by a DTT-sensitive thiolester.
For experiments shown in Fig. 3 the samples were electrophoresed in the presence of DTT to collapse all the oligomers into a single band. We next examined whether the ubiquitylation of hPrp19 affected its interaction with Cdc5L. For this experiment, we co-expressed V5-hPrp19, Flag-Cdc5L, and HA-Ub, and subsequently performed pull-down experiments. Surprisingly, while an anti-Flag pull-down confirmed the reported interaction between hPrp19 and Cdc5L (Fig. 3A, right panel), a pull-down with anti-HA indicated that the ubiquitylated form of hPrp19 did not interact with Cdc5L (Fig. 3A, middle panel). A similar result was also obtained with Plrg1 indicating that this component of the core complex also did not associate with the ubiquitylated form of hPrp19 (Fig. 3B). These experiments indicate that DNA damage and the ubiquitylation of hPrp19 that it elicits has profound affects upon the structure and localization of the Prp19 core complex.
Figure 3.

The ubiquitylated form of hPrp19 fails to interact with Cdc5L and Plrg1. (A) Immunoblots showing the occurrence of hPrp19 and Cdc5L in soluble (S) and chromatin (P) fractions after exposure of cells to MMS. (B) Immunoblots showing results of pull-down assays performed with cell extracts expressing V5-Prp19, Flag-Cdc5L, and HA-Ub. (C) Immunoblots showing results of pull-down assays performed with cell extracts expressing V5-Prp19, GST-Plrg1, and HA-Ub. All experiments shown were electrophoresed in the presence of the reducing agent DTT.
Prp19 Levels affect apoptotic responses in human cells
We have previously shown that siRNA-mediated depletion of hPrp19 and Cdc5L results in a dramatic reduction in clonal growth of HeLa cells [21]. To determine the basis for this observed reduction in proliferation, we again used siRNA to deplete hPrp19 and assayed for effects upon Cdc5L and Plrg1 levels. Both of these proteins were found to have reduced stability in the absence of hPrp19 (Fig. S3A). In addition, depletion of hPrp19 resulted in a significant increase in apoptosis compared to control cells at 48 hrs after transfection of siRNA (Fig. S3B,C). These findings appear to explain the reduction in growth caused by depletion of hPrp19.
The ubiquitylated form of hPrp19 is significantly increased upon overexpression of hPrp19 (see Fig. 1). Since this form is also increased in response to DNA damage, we asked whether overexpression of hPrp19 might modulate the affects of DNA damage-inducing agents. Either GFP- or GFP-hPrp19-expressing constructs were transfected into HeLa cells and 24 hrs later cells were exposed to MMS. At the indicated time points after MMS exposure, GFP positive cells were assayed for the degree of apoptosis (Fig. 4). GFP-hPrp19 positive cells exhibited an approximate two-fold decrease in apoptosis at each time point compared to the control cells suggesting that overexpression of Prp19 reduces the toxic affects of DNA damage introduced by MMS.
Figure 4.

Overexpression of hPrp19 reduces the levels of apoptosis induced by DNA damage. (A) Cell cycle analysis by FACS of cells transfected with GFP- or GFP-hPrp19-expressing constructs, and subsequently exposed to MMS. GFP positive cells are shown. (B) Quantitation of results shown in (A). Error bars represent the standard deviation of the mean.
Discussion
During the course of the purification of the hPrp19 complex [21] we noted a slower migrating SDS-resistant species that was recognized by hPrp19 antibody. Interestingly, exposure of cells to DNA damage caused an increase in the levels of this hPrp19 species. Characterization of this species showed that it is an oligomer (largely dimers and tetramers) of hPrp19 that is resistant to disruption by SDS gel electrophoresis, but that is disrupted upon exposure to reducing conditions. This species also contained Ub, which appeared to be involved in its formation. These characteristics were highly similar to that previously reported for the E2 Ub ligase Cdc34 [24]. Studies of this protein have shown that formation of a thiolester between Ub and a cysteine residue in Cdc34 is a prerequisite for self-association and the function of the ligase in multi-ubiquitylation of substrates. A catalytically functional U-box domain does not appear to be required for the self-association of hPrp19 since mutation of critical residues in the active site did not affect the attachment of Ub, although we cannot rule out the possibility that endogenous hPrp19 ubiquitylated the mutant protein in trans and thereby promoted oligomerization. Deletion of the entire U-box abolished oligomerization indicating that the U-box domain but not its catalytic activity is required for self-association. A somewhat surprising finding was that ubiquitylation of hPrp19 appeared to abrogate its interaction with both Cdc5L and Plrg1. A single molecule of Cdc5L is known to interact with a Prp19 tetramer via the associated coiled coil domains [13], and Cdc5L and Plrg1 have been shown to directly interact [9, 11]. Combined, these results suggest that the ubiquitylation of hPrp19 may affect the coiled coil domain, thus disrupting Cdc5L binding and promoting stronger interactions among the hPrp19 subunits. This finding is also consistent with our observation that Cdc5L did not exhibit increased chromatin association upon DNA damage. In fact, most of Cdc5L appeared to be constitutively bound to chromatin. Thus, in sum, our results indicate that in response to genotoxic damage a novel form of ubiquitylated hPrp19 is created that has enhanced self-association properties, binds to chromatin, and which no longer forms a core complex with Cdc5L and Plrg1. Although, the function of this novel hPrp19 form remains unresolved our findings strongly support the conclusion that this protein is involved directly in the DNA damage response.
Finally, our previous work showed that depletion of hPrp19 or Cdc5L in HeLa cells resulted in the failure of these cells to form colonies [21]. Here, we have shown that depletion of hPrp19 destabilizes Cdc5L and Plrg1, and ultimately induces apoptosis. Interestingly, we have also shown that overexpression of hPrp19 reduces the levels of apoptosis after exposure of cells to DNA damage. This pro-survival affect is likely due to a direct role of hPrp19 in the cellular response to DNA damage, and may be specifically related to the formation of ubiquitylated hPrp19 since this species is significantly increased upon overexpression of the ligase. Interestingly, in a similar context hPrp19 has been shown to be underexpressed in human senescent cells, and to increase the number of population doublings when overexpressed in human cells [25, 26].
Supplementary Material
Acknowledgments
We thank Paul Ajuh for gift of constructs and antibodies. This work was supported by NCI grants CA075160 and CA097175. DNA sequencing resources were supported by the Cancer Center Support (Core) Grant CA16672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Andrade HH, Marques EK, Schenberg AC, Henriques JA. The PSO4 gene is responsible for an error-prone recombinational DNA repair pathway in Saccharomyces cerevisiae. Mol Gen Genet. 1989;217:419–426. doi: 10.1007/BF02464912. [DOI] [PubMed] [Google Scholar]
- 2.Henriques JA, Vicente EJ, Leandro da Silva KV, Schenberg AC. PSO4: a novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat Res. 1989;218:111–124. doi: 10.1016/0921-8777(89)90017-7. [DOI] [PubMed] [Google Scholar]
- 3.Morais Junior MA, Brozmanova J, Benfato MS, Duraj J, Vlckova V, Henriques JA. The E. coli recA gene can restore the defect in mutagenesis of the pso4-1 mutant of S. cerevisiae. Mutat Res. 1994;314:209–220. doi: 10.1016/0921-8777(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 4.de Morais MA, Jr, Vicente EJ, Brozmanova J, Schenberg AC, Henriques JA. Further characterization of the yeast pso4-1 mutant: interaction with rad51 and rad52 mutants after photoinduced psoralen lesions. Curr Genet. 1996;29:211–218. doi: 10.1007/BF02221550. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SC, Tarn WY, Tsao TY, Abelson J. PRP19: a novel spliceosomal component. Mol Cell Biol. 1993;13:1876–1882. doi: 10.1128/mcb.13.3.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarn WY, Lee KR, Cheng SC. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grey M, Dusterhoft A, Henriques JA, Brendel M. Allelism of PSO4 and PRP19 links pre-mRNA processing with recombination and error-prone DNA repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4009–4014. doi: 10.1093/nar/24.20.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald WH, Ohi R, Smelkova N, Frendewey D, Gould KL. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. Embo J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. Rna. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajuh P, Sleeman J, Chusainow J, Lamond AI. A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J Biol Chem. 2001;276:42370–42381. doi: 10.1074/jbc.M105453200. [DOI] [PubMed] [Google Scholar]
- 12.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 13.Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, Walz T, Gould KL. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol Cell Biol. 2005;25:451–460. doi: 10.1128/MCB.25.1.451-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vander Kooi CW, Ohi MD, Rosenberg JA, Oldham ML, Newcomer ME, Gould KL, Chazin WJ. The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases(,) Biochemistry. 2006;45:121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 17.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loscher M, Fortschegger K, Ritter G, Wostry M, Voglauer R, Schmid JA, Watters S, Rivett AJ, Ajuh P, Lamond AI, Katinger H, Grillari J. The U-box E3 ligase SNEV interacts with PSMB4, the beta7 subunit of the 20S proteasome. Biochem J (2005) 2005;388:593–603. doi: 10.1042/BJ20041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revers LF, Cardone JM, Bonatto D, Saffi J, Grey M, Feldmann H, Brendel M, Henriques JA. Thermoconditional modulation of the pleiotropic sensitivity phenotype by the Saccharomyces cerevisiae PRP19 mutant allele pso4-1. Nucleic Acids Res. 2002;30:4993–5003. doi: 10.1093/nar/gkf632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan KN, Mitchell BS. Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc Natl Acad Sci U S A. 2003;100:10746–10751. doi: 10.1073/pnas.1631060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA Splicing and DNA Repair Complex Interacts with WRN for Processing of DNA Interstrand Cross-links. J Biol Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 22.Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varelas X, Ptak C, Ellison MJ. Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Mol Cell Biol. 2003;23:5388–5400. doi: 10.1128/MCB.23.15.5388-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grillari J, Hohenwarter O, Grabherr RM, Katinger H. Subtractive hybridization of mRNA from early passage and senescent endothelial cells. Exp Gerontol. 2000;35:187–197. doi: 10.1016/s0531-5565(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 26.Voglauer R, Chang MW, Dampier B, Wieser M, Baumann K, Sterovsky T, Schreiber M, Katinger H, Grillari J. SNEV overexpression extends the life span of human endothelial cells. Exp Cell Res (2005) 2005;312:746–759. doi: 10.1016/j.yexcr.2005.11.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


