SUMMARY
Transcript-specific translational control restricts macrophage inflammatory gene expression. The pro-inflammatory cytokine interferon-gamma induces phosphorylation of ribosomal protein L13a and translocation from the 60S ribosomal subunit to the interferon-gamma-activated inhibitor of translation (GAIT) complex. This complex binds the 3′ UTR of ceruloplasmin mRNA and blocks its translation. Here, we elucidate the molecular mechanism underlying repression by L13a. Translation of the GAIT element-containing reporter mRNA is sensitive to L13a-mediated silencing when driven by internal ribosome entry sites (IRES) that require initiation factor eIF4G, but is resistant to silencing when driven by eIF4F-independent IRESs, demonstrating a critical role for eIF4G. Interaction of L13a with eIF4G blocks 43S recruitment without suppressing eIF4F complex formation. Virus-, stress-, and caspase-directed attack of eIF4G is a well-known mechanism of global inhibition of cellular protein synthesis; however, our studies reveal a unique mechanism in which targeting of eIF4G by mRNA-bound L13a elicits transcript-specific translational repression.
INTRODUCTION
Translation initiation, the process of ribosome assembly on a mRNA, is the preferred target for translational control (Gebauer and Hentze, 2004; Gingras et al., 1999; Richter and Sonenberg, 2005). Initiation begins with assembly of the heterotrimeric eIF4F complex (containing eIF4A, eIF4G, and eIF4E) at the m7G cap structure of the message. eIF4G constitutes the backbone of the complex, and binds both cap-binding protein, eIF4E, and the RNA helicase, eIF4A. eIF4G also binds poly(A)-binding protein (PABP) which facilitates, by its interaction with the 3′-localized poly(A) tail, transcript circularization via end-to-end interaction (Mazumder et al., 2003b). The small ribosomal subunit contained in the 43S pre-initiation complex is then recruited to the mRNA by interaction with eIF4F. The 43S complex also contains the multi-subunit eIF3 complex, eIF1/eIF1A, and a pre-charged ternary complex of eIF2, GTP, and initiator Met-tRNA. Recruitment of 43S requires the interaction of eIF3 with an eIF4F component, eIF4G (Siridechadilok et al., 2005). The recruited 43S complex scans the 5′ UTR to identify the AUG initiation codon. Initiation codon recognition triggers hydrolysis of eIF2- and eIF5B-bound GTP, and permits joining of the 60S ribosomal subunit to form an elongation-competent 80S complex.
Disruption of any event in this highly orchestrated, multi-step process prevents formation of the 80S ribosomal complex and blocks synthesis of the encoded protein (Gebauer and Hentze, 2004; Richter and Sonenberg, 2005). Regulation can be global, affecting most cellular transcripts, generally by modification of a translation-initiation factor. Alternatively, regulation can be transcript-specific and directed by interaction of a regulatory protein or complex with the 5′ or 3′ UTR. For example, Drosophila cup and bicoid (Cho et al., 2005) and Xenopus maskin (Stebbins-Boaz et al., 1999) interact with the 3′ UTR of specific target mRNAs and block 43S complex recruitment. 43S recruitment is also inhibited by interaction of iron regulatory proteins (IRP)-1 and -2 to the iron-response element (IRE) in the 5′ UTR of ferritin mRNA (Gray and Hentze, 1994; Muckenthaler et al., 1998). Drosophila sex-lethal is unique in its adaptation of a dual inhibitory mechanism; sex-lethal inhibits stable association of 43S with msl-2 mRNA (Gebauer et al., 2003) and also blocks subsequent scanning (Beckmann et al., 2005). Further downstream in the initiation pathway, heterogeneous nuclear ribonucleoproteins (hnRNP) K and E1 block 60S ribosomal subunit joining at the initiation codon to silence translation of 15-lipoxygenase mRNA (Ostareck et al., 2001; Ostareck et al., 1997). Although the silencing mechanism has been elucidated in only a few cases, they expose a range of targets for repression, and suggest there is not a single, preferred regulatory step in the translation initiation pathway. In some cases, a ribosome function is targeted, e.g., 43S recruitment, scanning, or 60S joining. In other cases the target is eIF4E, and consequent disruption of eIF4F assembly. Interestingly, eIF4G has not been reported as a target of mRNA-specific translational control, despite the centrality of eIF4G in the initiation process, and its role as a common target by viruses and apoptosis-induced caspases for global inhibition of translation (Prevot et al., 2003a).
Recent studies suggest a critical role for mRNA-specific translational control in inflammation, particularly in processes related to limiting or resolving chronic leukocyte inflammation. In some cases, the expression of transcriptionally-induced leukocyte inflammatory proteins is subject to downstream fine-tuning by translational control mechanisms (Espel, 2005; Nathan, 2002). In studies of the inflammatory protein ceruloplasmin (Cp), we have shown interferon (IFN)-γ rapidly induces Cp protein and mRNA expression in U937 monocytic cells, but synthesis of the protein is stopped after about 16 h by a mechanism involving repression of translation (Mazumder and Fox, 1999; Mazumder et al., 1997). Translational silencing requires binding of an IFN-gamma-activated inhibitor of translation (GAIT) complex to a defined structural element (GAIT element) in the Cp mRNA 3′ UTR (Sampath et al., 2003). The GAIT complex contains four proteins: glutamyl-prolyl-tRNA synthetase (GluProRS), NS1-associated protein-1, glyceraldehyde 3-phosphate dehydrogenase, and ribosomal protein L13a (Mazumder et al., 2003a; Sampath et al., 2004). GluProRS is responsible for GAIT element recognition and binding, but the function of the other three GAIT complex proteins is unknown.
L13a is a candidate for the translation repressor since its phosphorylation and release from the 60S ribosomal subunit coincides with formation of the GAIT complex and inhibition of Cp mRNA translation. Inhibition requires the essential elements of mRNA circularization, i.e., poly(A) tail, poly(A)-binding protein (PABP), and eIF4G, suggesting the 3′-localized GAIT complex may interact with a 5′-localized translation factor. The specific mechanism of inhibition is unknown, but uncoupling of Cp mRNA from ribosomes suggests translation is blocked at an initiation step (Mazumder and Fox, 1999). In this report we show that L13a targets eIF4G to block Cp mRNA translation. We show that L13a phosphorylation is required for its activity, and that L13a binds eIF4G at the eIF3 binding site and blocks recruitment of the 43S ribosomal complex. The results show that eIF4G, a general translation initiation factor, can serve as a target in gene-specific translational silencing.
Results
Ribosomal Protein L13a Blocks Recruitment of 43S Ribosome Complex to GAIT Element-bearing RNA
The translational silencing activity of L13a, and the role of L13a phosphorylation, was tested by in vitro translation. A chimeric reporter mRNA containing an m7G cap, the 29-nt Cp mRNA GAIT element, and a 50-nt poly(A) tail (Luc-GAIT-poly(A)) was translated in a rabbit reticulocyte lysate (RRL) with T7 gene 10 mRNA as an internal specificity control (Fig. 1A). A similar construct containing an inactive, mutant GAIT element (U87C replacement in the 3′ UTR) was used as an additional control (Luc-GAIT (mut.)-poly(A)). We took advantage of our finding that essentially all L13a produced by baculovirus-infected insect cells is phosphorylated (phospho-L13a), whereas L13a made by E. coli is unmodified (Mazumder et al., 2003a). Phospho-L13a markedly inhibited Luc-GAIT-poly(A) translation, but not translation of the reporter containing the mutant GAIT element or T7 gene 10 (Fig. 1B). Unphosphorylated L13a, either the E. coli protein or alkaline phosphatase-treated L13a from insect cells, was inactive. A 3-fold excess of unphosphorylated L13a failed to reverse the silencing activity of phospho-L13a further confirming its specificity of action (Supplement, Fig. 1). These results establish the requirement for L13a phosphorylation for GAIT element-mediated translational silencing activity, confirming previous results using a reporter with an extended Cp 3′ UTR segment (Mazumder et al., 2003a).
Figure 1. Translational Silencing by Phospho-L13a Blocks 80S Recruitment to Reporter mRNA Containing a GAIT Element.
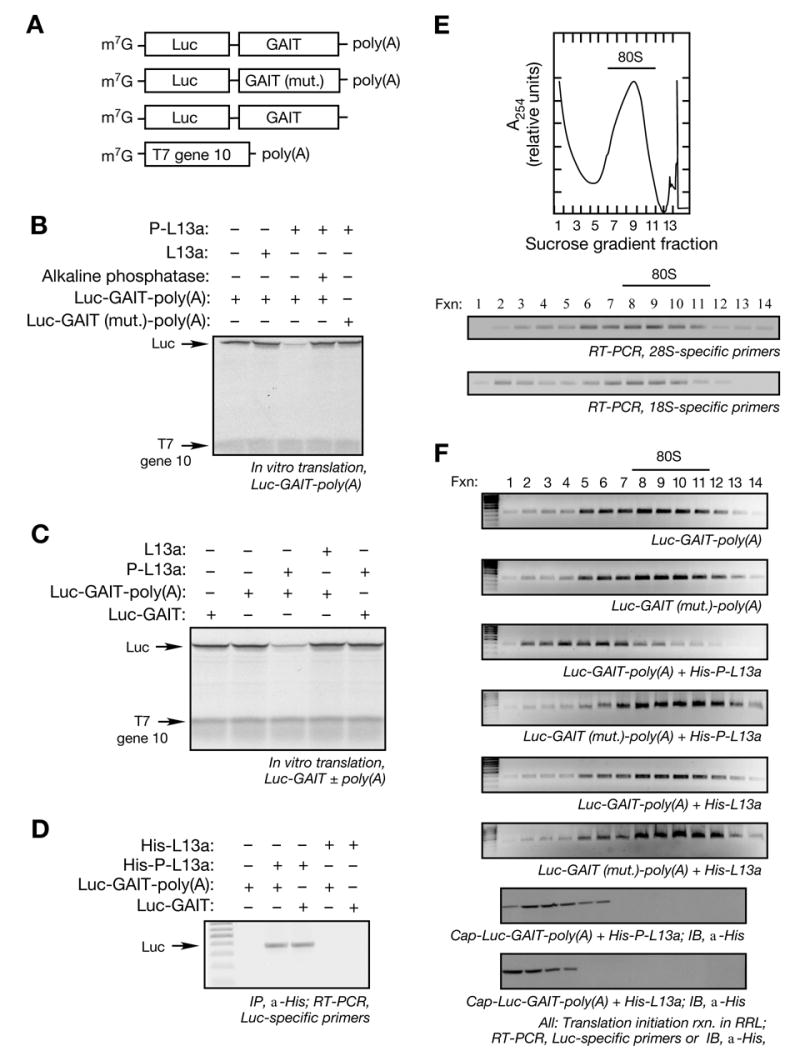
(A) Schematic of Luc reporter RNAs used in translation initiation reactions. m7G-capped, chimeric reporter Luc RNAs with wild-type and mutant (mut.) GAIT element, and with and without a poly(A) tail. T7 gene 10 RNA was used as a specificity control.
(B) Translational silencing of GAIT element-containing reporter RNA by phospho-L13a. Luc RNAs (200 ng) were subjected to in vitro translation using [35S]Met and a RRL. T7 gene 10-poly(A) RNA (100 ng) was co-translated in the same reaction as control. Recombinant phospho-L13a (5 μg) made by baculovirus-infected insect cells was treated with shrimp alkaline phosphatase (3 U) for 90 min. Newly translated, [35S]Met-labeled proteins were resolved by 7% SDS-PAGE and detected by fluorography.
(C) Poly(A) tail is required for translational silencing by phospho-L13a. Luc RNA with and T7 gene 10 RNAs were subjected to in vitro translation by a RRL. Partially-purified recombinant phosphorylated or unmodified L13a (5 μg) was added to the in vitro translation reaction. [35S]Met-labeled proteins were resolved by SDS-PAGE and detected by fluorography.
(D) Poly(A) tail is not required for L13a recruitment to the GAIT element. Translation initiation reactions were reconstituted in RRL, using Luc reporters (500 ng) with or without a poly(A) tail, in the presence of His-tagged, phosphorylated or unmodified L13a (10 μg). L13 was immunoprecipitated (IP) with monoclonal anti-His antibody, and bound RNA detected by RT-PCR with the Luc-specific primers.
(E) RNA detection in sucrose density gradient fractions. Initiation reaction containing Luc RNAs (500 ng) and partially purified, His-tagged phosphorylated or unmodified L13a (10 μg) was reconstituted in RRL in the presence of 0.5 mM cycloheximide. The reaction was subjected to sucrose density gradient centrifugation using 10 to 25% gradient and A254 was monitored (top panel). The 80S peak is marked by a horizontal bar. The authenticity of the 80S fractions was confirmed by RT-PCR amplification using 28S- (middle panel) and 18S- (bottom panel) specific rRNA primers.
(F) Assembly of 80S ribosome complex. Initiation reactions of a Luc reporter RNA carrying a wild-type or mutant GAIT element (500 ng), and phosphorylated or unmodified L13a (5 μg), were reconstituted in a RRL and subjected to sucrose density gradient centrifugation. Fractions (1 ml) were collected from the top. Total RNA was isolated and subjected to RT-PCR using Luc-specific primers (top 6 panels). The 80S peak is marked by a horizontal bar. Migration of phospho-L13a in the gradient was determined by precipitation of the fractions with trichloroacetic acid (TCA) followed by immunoblot (IB) analysis with anti-His antibody (bottom 2 panels).
GAIT element-mediated translational silencing by a U937 cell lysate requires the essential components of transcript circularization (Mazumder et al., 2001). To show that inhibition by recombinant phospho-L13a likewise requires end-to-end transcript interactions, the requirement for a poly(A) tail on the target mRNA was examined. The absence of a poly(A) tail did not influence basal translation of the Luc-GAIT reporter; however, translation of the Luc-GAIT reporter containing a poly(A) tail was selectively inhibited by phospho-L13a (Fig. 1C). L13a binding to a GAIT element-bearing Luc reporter RNA was investigated in a RRL by immunoprecipitation of His-tagged L13a followed by RT-PCR using Luc-specific primers. Phospho-L13a bound the Luc reporter irrespective of the presence of a poly(A) tail; however, unphosphorylated L13a bound neither RNA (Fig. 1D). Efficient binding of phospho-L13a to a poly(A)-minus reporter indicates that binding is not sufficient for translational silencing, but the poly(A) tail may contribute to effective presentation of phospho-L13a. For example, the poly(A) tail, by end-to-end interaction with eIF4G via the PABP intermediary, may deliver 3′ UTR-bound L13a to the vicinity of the initiation site in the Cp mRNA 3′ UTR.
To determine if phospho-L13a blocks translation at the initiation step, we first inspected the terminal initiation event, i.e., joining of the 60S ribosome subunit to mRNA-bound 43S complex (which contains the 40S ribosome subunit) to form an elongation-competent 80S ribosome complex. Initiation was determined in a RRL in the presence of cycloheximide, which allows 80S formation on mRNA but blocks elongation and increases steady-state amount of 80S complex. Ribosomes were permitted to assemble on test RNAs, and initiation complexes resolved by sucrose density-gradient fractionation. A representative 80S peak from Luc-GAIT-poly(A) RNA fractionation is shown (Fig. 1E, top panel). The identity of the 80S peak was authenticated by RT-PCR amplification of 28S and 18S rRNA (Fig. 1E, lower two panels). Reporter RNA was determined by RT-PCR amplification using Luc-specific primers. Rapid formation of 80S complex on the Luc reporter containing either wild-type or mutant GAIT element was observed (Fig. 1F, top two panels). Phospho-L13a prevented formation of 80S complex only on a Luc reporter bearing the wild-type GAIT element; unphosphorylated L13a was ineffective (Fig. 1F, 3rd to 6th panels). Migration of phosphorylated and unmodified L13a was examined in initiation reactions of the reporter carrying wild-type GAIT element. Phospho-L13a was in the light, pre-80S fractions consistent with its binding to GAIT element-containing RNA and shifting with the RNA to pre-80S fractions (Fig. 1F, 2nd panel from bottom). Unmodified L13a does not bind the GAIT element reporter and it is not part of a ribonucleoprotein complex and thus it also migrates in the light fractions (Fig. 1F, bottom panel). Thus, formation of the 80S ribosomal complex on GAIT element-containing transcripts is impaired in the presence of phospho-L13a, and translation must be blocked at this, or an earlier initiation step.
Upstream of 80S complex formation, the 43S complex is recruited to the trimeric eIF4F complex assembled at the mRNA 5′ cap to form the 48S pre-initiation complex. To determine whether L13a blocks 43S recruitment we used sucrose gradient analysis of initiation complexes assembled in RRL in the presence of a non-hydrolysable GTP analog, GMP-PNP. The analog stalls 43S ribosomal pre-initiation complex at the initiator codon and increases the steady-state amount of 48S complex (Anthony and Merrick, 1992). Luc reporter RNAs were replaced with shorter, 32P-labeled GAIT element-bearing reporters based on the 400-nt rabbit α-globin RNA to improve detection of 48S complex formation (Fig. 2A) (Ostareck et al., 2001). In a preliminary characterization, 48S and 80S peaks were detected in the presence of GMP-PNP and cycloheximide, respectively (Fig. 2B). In the presence of GMP-PNP, phospho-L13a markedly reduced 48S complex formation on a reporter containing wild-type GAIT element (Fig. 2C). The inhibition was specific since 48S formation was not reduced after substitution with unmodified L13a or mutated GAIT element. We tested whether inhibition required a poly(A) tail on the silenced transcript. Phospho-L13a failed to inhibit 48S complex formation on an α-globin reporter RNA carrying the GAIT element, but without a poly(A) tail (Fig. 2C). This result confirms that the inhibition of 48S formation by phospho-L13a is poly(A)-dependent and suggests an important role for transcript closure in the inhibitory process.
Figure 2. Phospho-L13a Blocks Assembly of the 48S Complex.
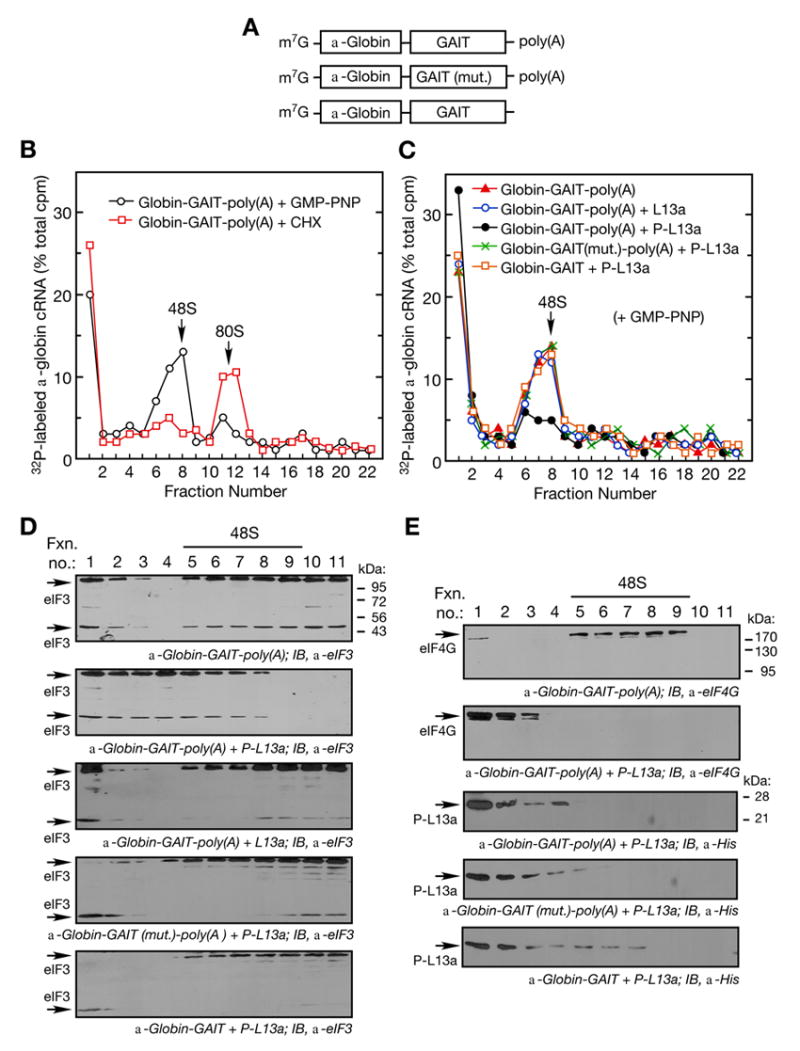
(A) Schematic of α-globin RNA constructs used in translation initiation reactions. Shown are α-globin RNA reporters carrying the wild-type (top) or mutant (middle) GAIT element with a poly(A) tail, and α-globin RNA carrying the wild-type GAIT element without a poly(A) tail (bottom).
(B) Ribosomal complex formation on GAIT element-containing RNA. Translation initiation reactions containing 32P-labeled, α-globin-GAIT-poly(A) reporters (300,000 cpm) were reconstituted in RRL in presence of GMP-PNP (black ○, 2 mM) or cycloheximide (red □, CHX, 0.5 mM) and resolved by sucrose density gradient centrifugation. Radioactivity in fractions was determined and peak positions of 48S and 80S complexes are indicated.
(C) Phospho-L13a blocks 48S complex formation on a GAIT element-containing RNA in a poly(A)-dependent way. Translation initiation reactions were reconstituted in a RRL with GMP-PNP (2 mM) as in Fig. 2B, but in the presence of His-tagged, phospho-L13a (5 μg). Conditions tested were: α-globin-GAIT-poly(A) (red ▲), α-globin-GAIT-poly(A) + L13a (blue ○), α-globin-GAIT-poly(A) + phospho-L13a (black ●), α-globin-GAIT (mut.)-poly(A) + phospho-L13a (green ×), α-globin-GAIT (no poly(A) tail) + phospho-L13a (orange □). The reaction was subjected to sucrose density gradient centrifugation, fractions (0.75 ml) were collected, and radioactivity determined.
(D) Phospho-L13a prevents formation of eIF3-containing 48S complex. Gradient fractions from experiments with α-globin-GAIT-poly(A) reporter and buffer control (top), phospho-L13a (middle), and unmodified L13a (bottom) were subjected to TCA precipitation followed by IB analysis with anti-eIF3 antibody. Two eIF3 subunits (44 and 110 kDa) are indicated.
(E) Migration of eIF4G and phospho-L13a in the sucrose gradient. Gradient fractions from Fig. 2C containing α-globin-GAIT-poly(A) reporter and buffer control (top panel) or phospho-L13a (2nd panel) were subjected to TCA-precipitation followed by IB analysis with anti-eIF4G antibody. Migration of phospho-L13a in the presence of α-globin-GAIT-poly(A) (3rd panel), α-globin-GAIT (mut.)-poly(A) (4th panel), or α-globin-GAIT (bottom panel) was determined by IB analysis with anti-His antibody.
To verify the inhibition of 48S complex formation by phospho-L13a, 43S recruitment to target RNAs was investigated. We measured association of eIF3, the 43S component that binds cap-bound eIF4G, with GAIT element-containing α-globin reporter RNAs by sucrose gradient fractionation and immunoblot analysis (Fig. 2D). Addition of phospho-L13a to the RRL shifted eIF3 from the 48S fractions to the lower density RNP fractions when reporter RNA carrying the GAIT element was used (Fig. 2D, 2nd panel). In three control experiments using unmodified L13a with reporter carrying wild-type GAIT element, or phospho-L13a with reporter carrying mutant GAIT element or without the poly(A) tail, eIF3 was not shifted from the 48S fractions (Fig. 2D, bottom three panels). The small differences in sedimentation characteristics of 110 and 44 kDa eIF3 subunits may be due to the experimental variability in subunit degradation. Because eIF4G is the eIF4F component required for 48S formation, co-migration of eIF4G and phospho-L13a in the sucrose gradients was examined by immunoblot analysis. In the presence of GAIT element-containing RNA, but in the absence of phospho-L13a, eIF4G co-migrated with the 48S complex (Fig. 2E, top panel). However, addition of phospho-L13a shifted eIF4G from the 48S fractions to the low density, RNP-containing fractions (Fig. 2E, 2nd panel). In the presence of RNA containing either wild-type or mutant GAIT element, phospho-L13a also was found in the same RNP fractions as eIF4G (Fig. 2E, 3rd and 4th panels). After removal of the poly(A) tail some association of phospho-L13a with the 48S fractions was seen (Fig. 2E, bottom panel), consistent with formation of 48S on poly(A)-minus RNA in the presence of phospho-L13a (Fig. 2C) and with poly(A)-independent recruitment of phospho-L13a to the GAIT element (Fig. 1D). Together these results show that phospho-L13a inhibits 43S recruitment to a transcript containing a 3′ UTR-located GAIT element.
L13a Binding to Target RNA does not Hinder Assembly of Cap-binding, eIF4F Complex
43S recruitment to a target mRNA for cap-dependent translation requires prior assembly of the trimeric eIF4F complex on the m7G cap. This event is the earliest step in initiation that involves the template mRNA, and eIF4F complex assembly is blocked by several translation suppressors (Pestova et al., 2001; Richter and Sonenberg, 2005). We considered the possibility that L13a binding similarly interfered with eIF4F assembly on the target transcript. Initiation of GAIT-containing reporter RNA was reconstituted in RRL in the presence of phospho-L13a and with the non-hydrolysable GTP analog, GMP-PNP (Fig. 3A). GMP-PNP prevents the joining of 60S and increases the steady-state amount of the 48S complex, consisting of the 43S ribosome complex, mRNA, and eIFs (Anthony and Merrick, 1992; Ji et al., 2004; Muckenthaler et al., 1998). An initial experiment was done to verify cap-independent binding of His-tagged phospho-L13a to the GAIT element-bearing reporter. Binding was examined by immunoprecipitation with anti-His, followed by RT-PCR with Luc-specific primers. Phospho-L13a bound the RNA target containing the GAIT element but not the mutant GAIT element (Fig. 3B). Also, as expected for a protein that binds a 3′ UTR element, phospho-L13a binding to RNA was not disrupted by interference with assembly at the 5′ cap using exogenous m7G or human rhinovirus (HRV)-2A protease.
Figure 3. Formation of Cap-binding Complex, eIF4F, is not Disrupted by Phospho-L13a.
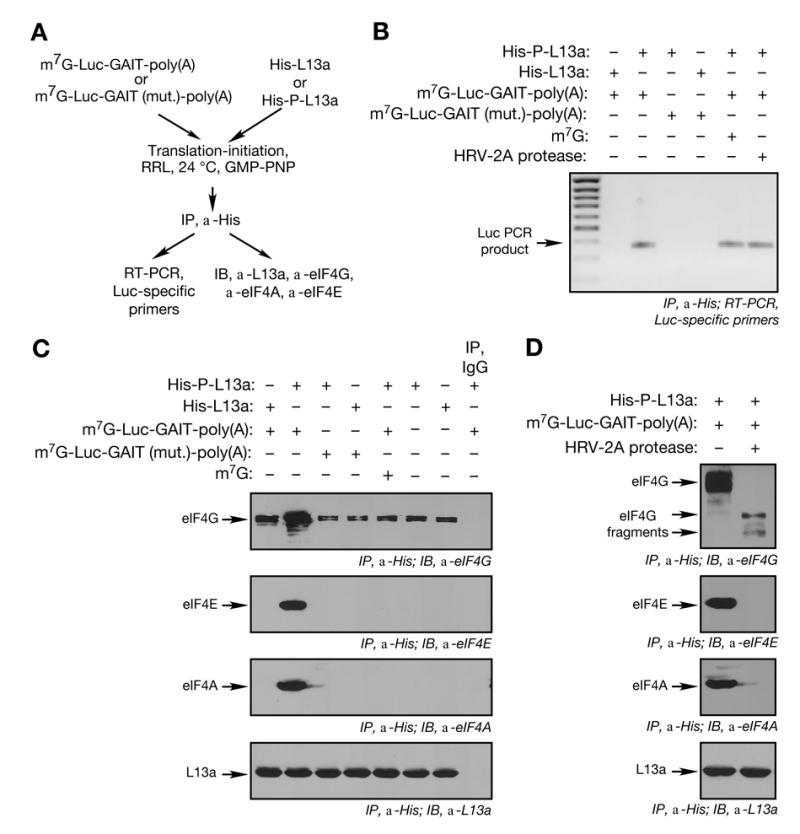
(A) Schematic of initiation reactions reconstituted on GAIT element-bearing RNA with His-tagged, phospho-L13a, and immunoprecipitation and detection procedures.
(B) Binding of phospho-L13a to GAIT element-bearing RNA in RRL. Translation initiation reactions were reconstituted in RRL using Luc reporters (500 ng) in presence of His-tagged, phosphorylated or unmodified L13a (10 μg). L13a was immunopre-cipitated (IP) with anti-His antibody, followed by RT-PCR with Luc-specific primers.
(C) Analysis of eIF4F components interacting with RNA-bound phospho-L13a. Translation initiation reaction was reconstituted in RRL. L13a was immunoprecipitated (IP) with anti-His antibody and precipitates subjected to immunoblot analysis with anti-eIF4G (upper panel), anti-eIF4E (2nd panel), anti-eIF4A (3rd panel) and anti-L13a (bottom panel) antibodies. A control lane shows immunoprecipitation with IgG.
(D) HRV-2A protease-sensitivity of eIF4F complex assembly. Translation initiation reaction was reconstituted in RRL with or without HRV-2A protease (2 μg). Immunoprecipitation (IP) and immunoblot (IB) procedures were as in Fig. 3C.
We investigated whether the eIF4F complex remains bound to an L13a-silenced reporter RNA. Bound eIF4F components were identified by immunoprecipitation with anti-His antibody, and immunoblot analysis using anti-eIF4G, -eIF4E, and -eIF4A antibodies. All three eIF4F components remained bound to a GAIT element-containing RNA in the presence of phospho-L13a (Fig. 3C, lane 2). The interaction between phospho-L13a and the intact eIF4F complex was RNA-dependent since it was not seen in the absence of reporter RNA or with a reporter containing an inactive GAIT element. Treatment with RNase A/T1 abrogated eIF4F complex recovery indicating the interaction required the GAIT-containing RNA (Supplement, Fig. 2). Weak binding of L13a to eIF4G, but not to eIF4A or eIF4E, occurred in the absence of RNA, possibly due to direct interaction between free L13a and free eIF4G. The authenticity of the eIF4F complex was examined by assessing cap-dependent interaction with the phospho-L13a-silenced reporter. Cap dependence was shown by the loss of interaction in the presence of exogenous m7G, which blocks eIF4E binding to the cap (Fig. 3C). Furthermore, cleavage of eIF4G with HRV-2A protease, prevents cap-dependent assembly of eIF4F, blocked the interaction of phospho-L13a with eIF4F components eIF4A and eIF4E, while permitting a weak interaction with eIF4G (Fig. 3D, top panel). Thus, phospho-L13a blocks 43S recruitment to GAIT element-bearing transcripts without disrupting eIF4F complex assembly, and an intact eIF4G is necessary for interaction of phospho-L13a with eIF4F.
eIF4G is the Target of L13a-mediated Translational Silencing Activity
Our results indicate that L13a blocks initiation at a step after eIF4F assembly on the mRNA 5′-cap, but before recruitment of 43S complex. Moreover, the presence of eIF4F on the silenced transcript suggests that an eIF4F constituent may be the target of L13a. To more precisely define the site of inhibition, we took advantage of 5′-situated internal ribosome entry sites (IRES) that initiate translation in the absence of specific eIF4F components. Translation initiation driven by the cricket paralysis virus (CrPV) IRES (Jan and Sarnow, 2002; Pestova and Hellen, 2003; Pestova et al., 2004; Wilson et al., 2000) and hepatitis C virus (HCV) IRES (Pestova et al., 1998) support 43S and 60S recruitment independent of any eIF4F component. In contrast, initiation by encephalomyocarditis virus (EMCV) IRES requires eIF4A and the central domain of eIF4G, but not cap-binding protein eIF4E (Kolupaeva et al., 2003; Pestova et al., 1996a). RNA constructs were prepared containing one of the three IRESs upstream of Luc, the GAIT element, and a 50-nt poly(A) tail (Fig. 4A). The 5′-end was blocked with an unmethylated cap, ApppG, to prevent cap-dependent translation. Excess, exogenous m7G cap was added to the RRL to maximize cap-independent translation of IRES-driven transcripts. As expected, translation of capped Luc RNA and T7 gene 10 RNA was sensitive to exogenous cap, but translation of RNAs driven by CrPV, HCV, and EMCV IRESs was not (Fig. 4B). This result shows IRES-driven translation of GAIT element-bearing reporter RNAs is efficient in a RRL. Phospho-L13a did not inhibit CrPV IRES- and HCV IRES-mediated translation of reporter RNAs indicating that one or more eIF4F components were required for silencing (Fig. 4C). In contrast, EMCV IRES-driven translation of the reporter was blocked by phospho-L13a (but not by non-phosphorylated L13a). The effect of phospho-L13a on 80S assembly on IRES-driven reporters was examined by sucrose density gradient fractionation as above (Fig. 4D, representative gradient shows EMCV IRES-containing RNA). Phospho-L13a blocked association of 80S ribosomes with transcripts driven by EMCV-IRES, but not by CrPV or HCV IRES (Fig. 4E). These results suggest that an eIF4F component required for EMCV IRES-mediated translation, e.g., eIF4G, is required for silencing by phospho-L13a, and may be its target.
Figure 4. Virus IRES-driven RNAs show an eIF4F Component is Required for Translational Silencing by Phospho-L13a.
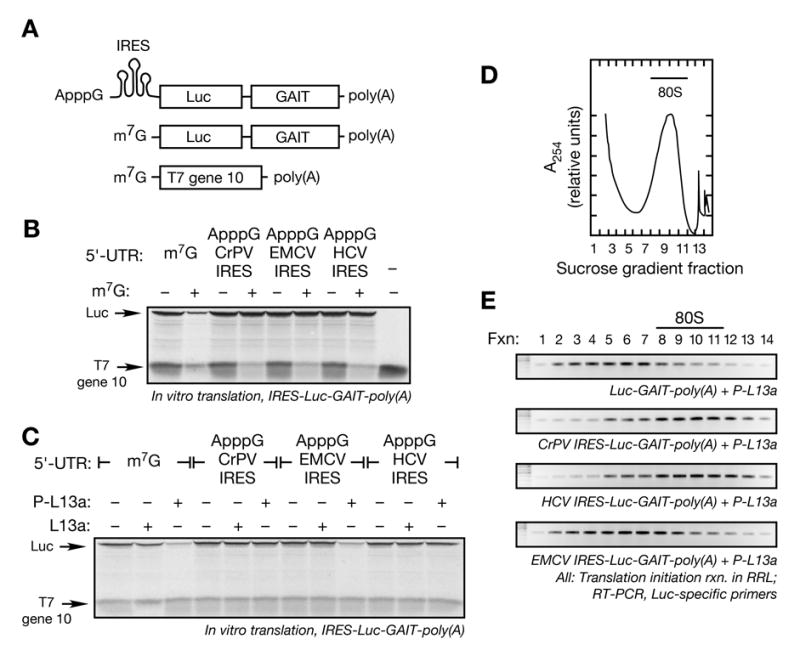
(A) Schematic of ApppG-capped, Luc-GAIT element-poly(A) RNAs with CrP, HCV, or EMCV IRES sequences in the 5′ UTR (top), m7G-Luc-GAIT element-poly(A) (middle), and m7G-T7 gene 10-poly(A) (bottom).
(B) In vitro translation of IRES-driven reporters. IRES-driven and m7G-capped reporter RNAs (200 ng) were subjected to in vitro translation in RRL in presence or absence of exogenous cap analog, m7G (200 μM). Newly translated, [35S]Met-labeled Luc and T7 gene 10 were resolved by SDS-PAGE (7%) and detected by fluorography.
(C) Sensitivity of IRES-driven reporters to repression by phospho-L13a. ApppG-IRES-Luc-GAIT-poly(A) RNAs (200 ng) were subjected to in vitro translation in RRL with capped, T7 gene 10-poly(A) RNA (100 ng) in presence of recombinant phosphorylated or unmodified L13a (5 μg). Newly synthesized [35S]Met-labeled proteins were resolved by SDS-PAGE and detected by fluorography.
(D) Ribosomal RNA detection in sucrose density gradient fractions. Initiation reaction of IRES-driven Luc RNA reporters (500 ng) were reconstituted in RRL in the presence of cycloheximide (0.5 mM). The reaction was subjected to sucrose density gradient centrifugation and A254 was monitored. The 80S peak is indicated (bar). A representative 80S peak obtained from the initiation reaction of EMCV IRES containing RNA in presence of phospho-L13a is shown.
(E) Effect of phospho-L13a on 80S assembly on IRES-driven RNAs. Initiation reactions of Luc RNA-GAIT element reporters (500 ng) driven by an m7G cap (top panel), CrPV IRES (2nd panel), HCV IRES (3rd panel), or EMCV IRES (bottom panel) were reconstituted in RRL in the presence of phospho-L13a (10 μg) and cycloheximide (0.5 mM), and resolved by sucrose density gradient centrifugation. Total RNA isolated from 1 ml fractions was subjected to RT-PCR analysis using Luc-specific primers.
We investigated whether a specific eIF4G domain is required for translational silencing by phospho-L13a. We took advantage of the human rhinovirus (HRV)-2A protease which cleaves eIF4G in the C-terminus near the binding site for eIF3 of the 43S small ribosomal complex (Skern, 2002). The central eIF4G domain that remains, permits EMCV IRES-driven translation, but not cap-dependent translation (Pestova et al., 1996b; Prevot et al., 2003b). EMCV IRES-containing Luc reporter constructs are shown in Fig. 5A. Immunoblot analysis showed eIF4G in the RRL was cleaved entirely by HRV-2A protease into 110 kDa C-terminal and 72 kDa N-terminal fragments (Fig. 5B). The identity of the eIF4G cleavage products was verified by antibodies raised against N-and C-terminus peptides (Supplement, Fig. 3). EMCV IRES-driven translation of the Luc-GAIT-poly(A) reporter was unaffected by eIF4G cleavage, but translation of cap-driven T7 gene 10 was blocked (Fig. 5C, lane 3). Phospho-L13a blocked translation of EMCV IRES-driven reporter even after protease cleavage of eIF4G (Fig. 5C, lane 6). In a control experiment, phospho-L13a did not alter the ability of HRV-2A protease to cleave eIF4G (Supplement, Fig. 4). These results implicate the C-terminal domain of eIF4G as the L13a target in translational silencing. The inhibition of EMCV IRES-driven translation by phospho-L13a was shown to be poly(A) tail-dependent (Fig. 5D, lanes 3 and 9). Interestingly, the inhibition of translation was poly(A)-independent when eIF4G was cleaved with HRV-2A protease (Fig. 5D, lanes 6 and 8). Taken together these results suggest the importance of an intact eIF4G for poly(A)-dependent inhibition of translation by phospho-L13a.
Figure 5. eIF4G C-terminal Domain is Necessary and Sufficient for Translational Silencing by Phospho-L13a.
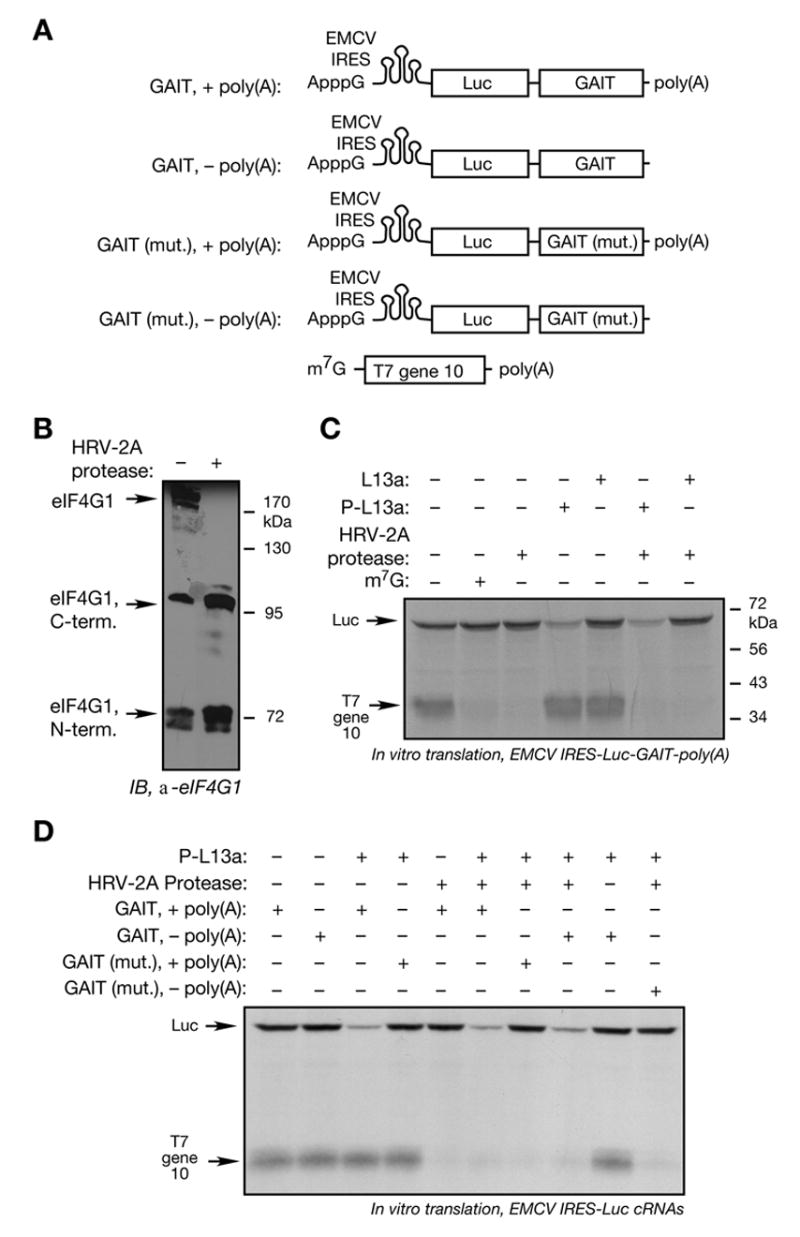
(A) Schematic of ApppG-capped, Luc-GAIT element RNAs with upstream EMCV IRES sequences containing wild-type or mutant GAIT element with and without poly(A) tail (top 4 panels). m7G-T7 gene 10-poly(A) (bottom panel).
(B) Cleavage of RRL eIF4G by HRV-2A protease. Recombinant HRV-2A protease (2 μg) was incubated with RRL (100 μg) for 30 min at 37 °C, and subjected to SDS-PAGE and immunoblot analysis with anti-eIF4G antibody. The antibody is a 1:1 mixture of antisera raised against peptide sequences from the N- (amino acids 175-200) and C-(amino acids 1179-1206) termini of eIF4G1.
(C) Inhibition of EMCV IRES-driven translation by phospho-L13a in the presence of HRV-2A protease. ApppG-capped Luc RNA (200 ng) containing GAIT element with EMCV IRES and m7G capped T7 gene 10 RNA (above) were translated in RRL with phosphorylated or unmodified L13a, and in presence or absence of exogenous m7G cap and HRV-2A protease. Newly translated, [35S]Met-labeled proteins were resolved by SDS-PAGE and detected by fluorography.
(D) Inhibition of EMCV IRES-driven translation by phospho-L13a is poly(A)-independent when eIF4G is cleaved. ApppG-capped Luc RNA (200 ng) with EMCV IRES containing GAIT element with or without the poly(A) tail, and m7G capped T7 gene 10 RNA, were translated in RRL in presence or absence of phosphorylated L13a, and HRV-2A protease. Newly translated, [35S]Met-labeled proteins were resolved by SDS-PAGE and detected by fluorography.
L13a Blocks 43S Recruitment by Interaction with eIF4G at its eIF3-binding Site
Our co-immunoprecipitation suggested that L13a interacts with eIF4G in a RRL (Fig. 3C). To verify this interaction in vivo, human embryonic 293T kidney cells were transfected with a vector driving expression of Flag-tagged L13a. Immunoprecipitation with anti-Flag antibody, followed by immunoblot with anti-eIF4G antibody showed specific interaction of L13a with eIF4G (Fig. 6A). The eIF4G domain that binds L13a was investigated using HRV-2A protease to cleave eIF4G. His-tagged phospho-L13a was added to a RRL pretreated with HRV-2A, and L13a immunoprecipitated with anti-His antibody and probed with anti-eIF4G. Phospho-L13a interaction with the C terminus of eIF4G was shown by co-immunoprecipitation of the 110 kDa, C-terminal fragment (Fig. 6B). The 72 kDa, N terminus was not detected in the bound or supernatant fractions suggesting the fragment degrades under our recovery conditions.
Figure 6. L13a and eIF3 Share a Common eIF4G Binding Site.
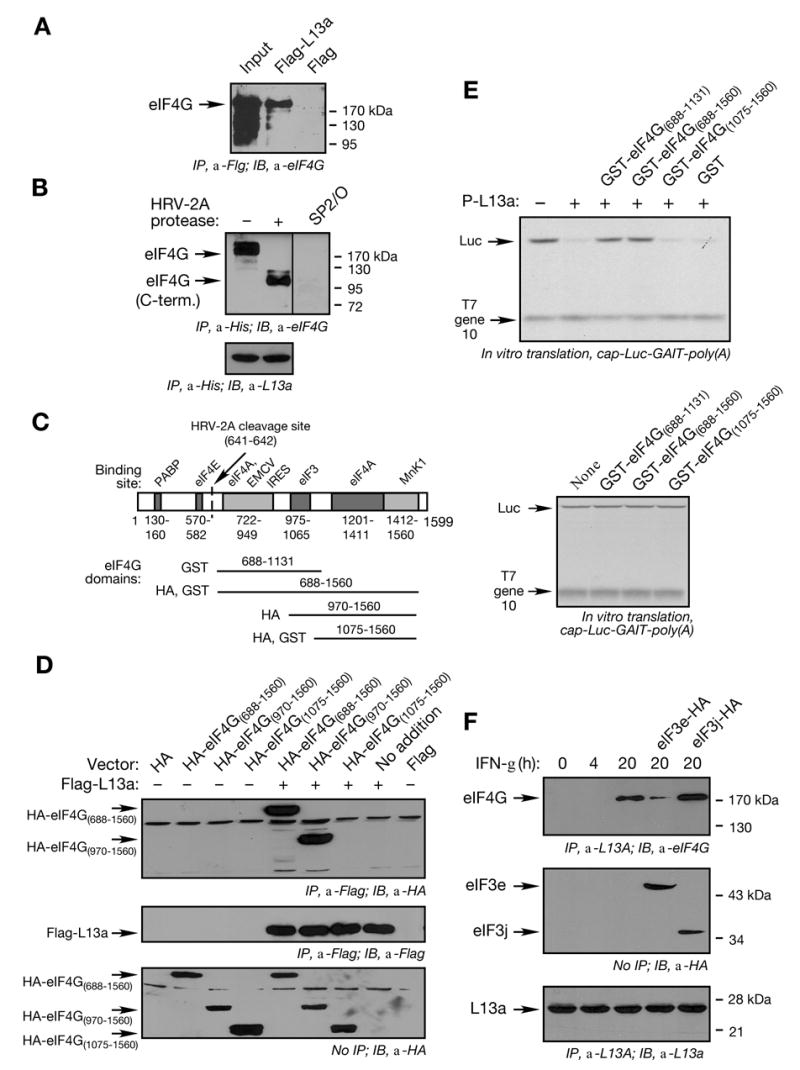
(A) L13a binds eIF4G in 293 T cells. 293T cells were transfected with pcDNA3-Flag-L13a or pcDNA3-Flag constructs. After 20 h, cell extracts were prepared and subjected to immunoprecipitation (IP) with anti-Flag antibody. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblot (IB) with anti-eIF4G antibody.
(B) L13a binds the C-terminal fragment of eIF4G. eIF4G in RRL was cleaved by HRV-2A protease. His-tagged phospho-L13a was incubated with RRL in presence or absence of HRV-2A protease followed by immunoprecipitation with anti-His antibody and immunoblot with anti-eIF4G antibody, or SP2/O as control antibody (upper panel). Immunoprecipitation efficiency was shown by re-probing the blot with anti-L13a antibody (lower panel).
(C) Domain organization of eIF4G. Domains of eIF4G responsible for binding of eIF4E, eIF4A, eIF3, PABP, MnK1, and EMCV IRES, and the cleavage site of HRV-2A protease are shown. Schematic showing eIF4G deletion fragments made as GST-fusion proteins or HA-tagged protein is indicated below.
(D) Mapping the interacting domain of eIF4G with L13a. 293T cells were co-transfected with Flag-L13a and HA-tagged eIF4G(688–1560), eIF4G(970–1560), or eIF4G(1075–1560). Cell lysates were immunoprecipitated with anti-Flag antibody. Immunoprecipitates were resolved by SDS-PAGE and subjected to immunoblot analysis with anti-HA (upper panel) or with anti-Flag (middle panel) antibodies. Aliquots of lysates without immunoprecipitation were probed by immunoblot with anti-HA antibody (lower panel).
(E) Exogenous eIF4G C-terminal fragment overcomes repression by phospho-L13a. Phospho-L13a (5 μg) was pre-incubated with partially-purified GST fusion proteins (50 μg) eIF4G(688–1131), eIF4G(688–1560), and eIF4G(1075–1560), and GST control protein. Pre-incubated solutions were added to RRL and cap-Luc-GAIT element-poly(A) (200 ng) and T7 gene 10-poly(A) were subjected to in vitro translation (upper panel). As a control, partially-purified GST fusion proteins containing eIF4G domains were added to the in vitro translation reaction in the absence of phospho-L13a (lower panel). Newly synthesized [35S]Met-labeled proteins were resolved by SDS-PAGE and detected by fluorography.
(F) Overexpression of eIF3e blocks binding of L13a to eIF4G. U937 cells were transfected with HA-tagged eIF3e or eIF3j by electroporation using Amaxa method. 5 x 106 U937 cells were treated with IFN-γ for 4 or 20 h. Cell lysates were subjected to immunoprecipitation (IP) with anti-L13a antibody and immunoblot (IB) with anti-eIF4G (top panel), anti-HA (middle panel), or anti-L13a (bottom panel) antibodies. IP with anti-L13a antibody was done using Sieze-X immunoprecipitation kit (Pierce) to avoid contamination from the antibody light and heavy chains.
The eIF4G C-terminus contains binding sites for multiple initiation factors, including eIF3, eIF4A, and Mnk1 (Prevot et al., 2003a). The L13a interaction site was investigated by construction of vectors expressing HA-tagged eIF4G domains overlapping the initiation factor-binding sites (Fig. 6C). 293T cells were co-transfected with these vectors and Flag-L13a. L13a co-immunoprecipitated with eIF4G(688–1560) and eIF4G(970–1560), but not with eIF4G(1075–1560) (Fig. 6D, upper panel). Control experiments showed uniform expression of transfected Flag-L13a (Fig. 6D, middle panel) and all eIF4G deletion fragments (Fig. 6D, lower panel). The eIF3 binding site in eIF4G, between amino acids 975 to 1065, is present in eIF4G(970–1560) but not in eIF4G(1075–1560), indicating this domain is critical for L13a binding to eIF4G. To verify the importance of this interaction in translational silencing, dominant-negative activity of eIF4G deletion fragments was determined. C-terminal eIF4G fragments were expressed as GST-fusion proteins in E. coli (Fig. 6C), pre-incubated with recombinant phospho-L13a, and then added to an in vitro translation reaction driven by m7G-Luc-GAIT-poly(A) RNA. eIF4G(688–1131) and eIF4G(688–1560) blocked translation repression by phospho-L13a, but eIF4G(1075–1560) was ineffective (Fig. 6E, upper panel). The GST fusion proteins eIF4G added in the absence of phospho-L13a were inactive (Fig. 6E, lower panel). Thus, only eIF4G fragments containing the eIF3 binding site inhibited phospho-L13a activity. Together these results indicate that eIF3 and L13a share the same or neighboring binding sites on eIF4G.
To test whether phospho-L13a competes with eIF3 for binding to eIF4G, we took advantage of the recent finding that the eIF3e subunit of eIF3 binds with high affinity to eIF4G and is required for 43S recruitment (LeFebvre et al., 2006). U937 cells were transfected with plasmids expressing HA-tagged eIF3e and eIF3j (as negative control), and then treated with IFN-γ for 20 h. Over-expression of eIF3e, but not eIF3j, substantially blocks the inducible binding between L13a and eIF4G as determined by co-immunoprecipitation (Fig. 6F, upper panel). Control studies showed nearly equivalent expression of transfected eIF3 subunits (Fig. 6F, middle panel), and consistent immunoprecipitation of L13a under all conditions (Fig. 6F, lower panel). Together these results suggest that direct competition of phospho-L13a with eIF3e for binding to eIF4G is the mechanism by which L13a blocks 43S recruitment and silences translation.
DISCUSSION
We have determined the molecular mechanism by which the inflammatory protein Cp is translationally silenced in cells in response to prolonged treatment with IFN-γ. This repression is critical because uncontrolled expression of Cp may have injurious consequences due to the oxidative properties of the copper protein (Fox et al., 2000; Mukhopadhyay et al., 1997). Previously we have shown that IFN-γ induces Cp mRNA and protein expression in U937 cells within 4 h, but that translation of the transcript halts after about 16 h (Mazumder and Fox, 1999; Mazumder et al., 1997). Also, we have reported that translational silencing of Cp mRNA requires binding of the 4-component GAIT complex to the 3′ UTR GAIT element (Sampath et al., 2004). Little is known about the function of each of the four GAIT proteins in the silencing mechanism, except that GluProRS is the GAIT complex protein that interacts directly with the GAIT element, and is responsible for mRNA target specificity. Here we show that L13a is the GAIT protein directly responsible for translational silencing of target transcripts. This finding is consistent with our previous result showing that the entire cell complement of L13a is phosphorylated and released from the 60S ribosomal subunit at about the same time that translation is halted, about 16 h after IFN-γ treatment (Mazumder et al., 2003a).
Transcript Circularization and other Requirements for L13a Silencing Activity
We have determined the requirements for silencing activity by recombinant L13a, and its specific mechanism of action. We took advantage of our finding that recombinant L13a produced by baculovirus infected insect cells is phosphorylated and active in an in vitro translation assay, whereas recombinant L13a from E. coli is unmodified and inactive (Mazumder et al., 2003a). Translational silencing of a reporter RNA in a RRL requires phosphorylated L13a, and both a wild-type Cp GAIT element and a poly(A) tail in the 3′ UTR. Identical requirements are observed for L13a inhibition of the recruitment of 43S and 60S complexes to reporter transcripts during initiation. The requirement for a functional GAIT element indicates the inhibition of initiation is mediated by the GAIT complex, and the RRL itself likely contributes the other 3 GAIT proteins (all constitutive products of “housekeeping” genes).
The requirement for a poly(A) tail on the target transcript is of substantial interest. In addition to its role in cap-dependent translation, the poly(A) tail enhances translation of EMCV IRES indicating possible communication between 5′ and 3′ termini during IRES-mediated translation (Bergamini et al., 2000; Michel et al., 2001; Svitkin et al., 2001). Therefore, it is not unexpected that phospho-L13a silences EMCV IRES-driven translation of GAIT element-containing RNA. However, our finding that phospho-L13a silences EMCV IRES-mediated translation even after cleavage of eIF4G was surprising since the remaining C terminus fragment of eIF4G does not bind PABP or facilitate end-to-end interaction. A possible explanation is that phospho-L13a has a very high affinity for the C-terminus of the cleaved eIF4G-fragment that remains bound to the EMCV IRES, and this high affinity binding may compensate for the loss of mRNA circularization in the absence of a poly(A) tail. This possibility is consistent with a previous report showing equal efficiency of translation of EMCV IRES-driven mRNAs with and without poly(A) tails under the conditions of eIF4G cleavage by HRV-2A protease (Svitkin et al., 2001). In both cases, eIF4G cleavage causes translation (or repression) of EMCV IRES-driven mRNAs to be poly(A)-independent. The efficient translation they observed may be due to increased affinity of eIF3e for the cleaved eIF4G fragment, which overcomes the need for the poly(A) tail. We speculate that in our studies, phospho-L13a (which shares with eIF3e the same eIF4G binding site) may also have an increased affinity for cleaved eIF4G, which similarly overcomes the requirement for a poly(A) tail.
Our results show that the poly(A) tail is not necessary for binding phospho-L13a to the GAIT element (Fig. 1D), but rather it is necessary for end-to-end transcript closure by a succession of interactions with PABP and eIF4G (Mazumder et al., 2001). Similarly, a recent report on translational repression by miRNA shows activity of the inhibitory, 3′ UTR-binding miRNP complex requires target mRNA polyadenylation (Wang et al., 2006). Transcript closure may be necessary to carry phospho-L13a from the distal 3′ UTR into the vicinity of the 5′-localized translation initiation complex where it can exert its inhibitory activity. This mechanism is consistent with our previous report that translational silencing activity of Cp mRNA by a lysate from U937 cells requires the essential elements of mRNA circularization, i.e., poly(A) tail, eIF4G, and PABP (Mazumder et al., 2001; Varani, 2001). Our new result showing the eIF4F complex remains intact on a silenced transcript is consistent with this mechanism.
eIF4G is the L13a Target for Transcript-specific Translation Inhibition
eIF4G is essential for translation initiation. It is a component of the cap-binding complex, facilitates end-to-end transcript interactions, is a scaffold for RNA-unwinding proteins and other initiation factors, and makes initial contact with the ribosome via its interaction with eIF3 of the 43S complex. The central position of eIF4G and its manifold interactions makes it an obvious target for translation regulation. In fact, eIF4G is a target common to multiple pathways that inhibit global protein synthesis, e.g., cell responses to virus infection, apoptosis, and stress. However, these pathways do not inhibit eIF4G by a single mechanism, rather a diversity of inhibitory mechanisms have evolved (Fig. 7A). For example, eIF4G is proteolytically cleaved upon virus infection by viral proteases, e.g., 2A protease encoded by polio-, rhino- and coxsackie-virus, and L protease encoded by foot-and-mouth disease virus (Prevot et al., 2003a). Cleavage of eIF4G prevents assembly of the eIF4F complex resulting in global inhibition of cap-dependent translation, while permitting translation of cap-independent, IRES-driven transcripts. Similarly, cleavage of eIF4G by caspase-3 occurs in apoptotic cells (Clemens et al., 1998; Marissen and Lloyd, 1998). Other viral and cellular proteins bind eIF4G and inactivate it without cleavage. For example, adenovirus 100-kDa protein binds eIF4G and displaces Mnk1, an eIF4E kinase that activates translation (Cuesta et al., 2004). NSP3 protein of rotavirus (Piron et al., 1998) and NS1 protein of influenza virus (Aragon et al., 2000) bind the eIF4G N-terminus, thereby displacing PABP. Upon heat shock or other cell stress, the Hsp27 chaperone protein binds eIF4G and traps it in insoluble complexes (Cuesta et al., 2000). In all of these examples, eIF4G is targeted by an RNA-independent mechanism to cause global inhibition of translation.
Figure 7. Global and Transcript-specific Translational Silencing by Targeting eIF4G.
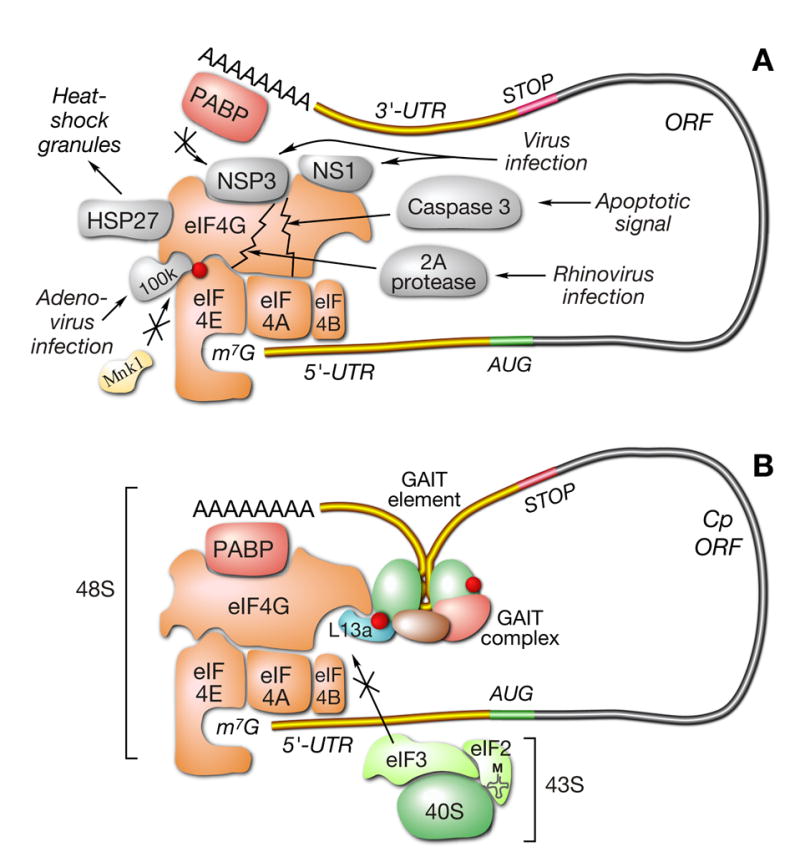
(A) Global inhibition of translation by targeting eIF4G. Virus infection, apoptosis, and cell stress target eIF4G by different mechanisms.
(B) L13a targets eIF4G for transcript-specific inhibition of translation. L13a in the 3′ UTR-bound GAIT complex targets eIF4G without cleavage of eIF4G or disruption of cap-binding eIF4F complex. Binding of phospho-L13a to eIF3 binding site of eIF4G blocks interaction of 43S ribosomal complex and represses translation initiation.
Our results reveal eIF4G is not only involved in global inhibition of protein synthesis, but it is also the principal target for transcript-specific translational repression by L13a (Fig. 7B). However, there are several aspects of the inhibition by L13a that differentiate it from the global mechanisms. Foremost is the finding that inhibition by phospho-L13a is RNA-dependent, and in fact, depends on the presence of the GAIT element in the transcript 3′ UTR. The absolute requirement for a specific structural element renders this inhibitory mechanism transcript-specific rather than global. Moreover, unlike most global inhibitors of translation, phospho-L13a does not alter initiation factors by proteolysis or by disruption of assembled complexes; translation inhibition by phospho-L13a occurs in the presence of an intact eIF4F complex.
Binding studies with HRV-2A protease fragments show that L13a binds the eIF4G C-terminus. Detailed analysis of the binding site using recombinant deletion fragments shows that L13a binds eIF4G(970–1560) but not eIF4G(1075–1560) thereby showing a requirement for eIF4G(970–1074). This domain coincides with the eIF3 binding site (eIF4G(975–1065)). Thus, L13a and eIF3 share the same binding site on eIF4G. In addition, our results show that phospho-L13a decreases eIF3 recruitment to a GAIT-containing reporter RNA and blocks 48S complex formation. These results are consistent with a model in which mRNA-bound L13a inhibits 43S recruitment by blocking eIF3 binding to eIF4G. The decreased recruitment of eIF3 to a GAIT-containing reporter RNA and the absence of 48S complex are consistent with this inhibitory mechanism. The kinetics of IFN-γ-induced binding of phospho-L13a and eIF4G (Fig. 6F) matches that of L13a phosphorylation and translational silencing of GAIT element-containing RNA (Mazumder et al., 2003a). Over-expression of eIF3e, which exhibits high-affinity binding to eIF4G (LeFebvre et al., 2006), blocks this interaction supporting a model in which GAIT element-bound phospho-L13a and 43S-bound eIF3 compete for binding to cap-bound eIF4G. Unexpectedly, non-phosphorylated L13a binds weakly to eIF4G in a RRL even in the absence of RNA (Fig. 3C). Under this condition, L13a interacts with eIF4G, but not eIF4E or eIF4A, suggesting L13a binds free eIF4G not associated with the eIF4F cap-binding complex. We surmise that L13a binds free eIF4G with low affinity, but binding is enhanced when eIF4G is associated with cap-bound eIF4F. This concept is supported by the decrease in RNA-dependent binding between L13a and eIF4G when eIF4F assembly is inhibited by exogenous cap or treatment with HRV-2A protease. Binding to eIF4G may be increased further when L13a is phosphorylated and bound to the transcript 3′ UTR as part of the GAIT complex; enhanced binding could result from conformational change in complex-bound L13a, or from more efficient presentation of L13a to the target due to end-to-end transcript interactions, or both.
Diversity of Mechanisms Directing Transcript-specific Translational Control
Only a few systems of mRNA-specific translational control have been elucidated at the molecular level. However, it is already apparent that diverse mechanisms have evolved that inhibit initiation at multiple steps (Gebauer and Hentze, 2004). The specific inhibitory mechanism driven by L13a has unique characteristics that differentiate it from previously described translation-initiation regulators. The inhibition of 43S recruitment by phospho-L13a distinguishes it from mechanisms in which initiation is blocked after 43S recruitment to the mRNA. For example, SXL binds to a poly(U)-rich region in the 5′ UTR of msl-2 mRNA in D. melanogaster, and inhibits 43S ribosome scanning (Beckmann et al., 2005). In a second example, binding of hnRNPs K and E1 to the 3′ UTR DICE element of reticulocyte 15-lipoxygenase inhibits 60S ribosomal subunit joining to the 43S subunit (Ostareck et al., 2001; Ostareck et al., 1997). Inhibition of 43S recruitment has been reported for several translation regulators; however, their inhibitory mechanisms are distinct from that used by L13a. A complex containing cytoplasmic-polyadenylation-element-binding protein (CPEB) and maskin binds the cytoplasmic polyadenylation element (CPE) in the 3′ UTR of specific target mRNAs during vertebrate oocyte development and silences their translation (Stebbins-Boaz et al., 1999). The CPEB-maskin complex binds eIF4E and prevents its interaction with eIF4G, and thus blocks eIF4F assembly. Analogous inhibitory mechanisms are used by D. Melanogaster cup and bicoid, which bind the 3′ UTRs of nanos and caudal mRNAs, respectively, and repress translation by acting as transcript-specific eIF4E-binding proteins (Cho et al., 2005; Niessing et al., 2002). The inhibition of ferritin mRNA translation by IRPs resembles the inhibition by L13a as it also blocks 43S complex recruitment without interfering with eIF4F assembly (Gray and Hentze, 1994; Muckenthaler et al., 1998). In contrast to GAIT complex binding to the 3′ UTR, IRPs bind a 5′ UTR element. More importantly, IRP binding to the IRE does not bind a specific general translation factor, since replacement of the IRP/IRE couple with an irrelevant protein/RNA element recapitulates translation repression (Stripecke et al., 1994). IRP binding to the 5′ UTR most likely blocks 43S recruitment by non-specific steric interference. A highly divergent mechanism has been described in which modification of a general initiation factor regulates mRNA-specific translation (Dever, 2002). For example, amino acid starvation causes phosphorylation of eIF2α and inhibition of global protein synthesis; however, translation of GCN4, ATF4, and C/EBP mRNAs are increased. Translation activation results from an enhanced rate of re-initiation from upstream open reading frames in these transcripts (Dever, 2002). In these cases, the basis for mRNA-specific translational control by general initiation factors relies primarily on the transcript-selective discrimination of the translation machinery.
Our finding that eIF4G is targeted by a 3′ UTR-bound inhibitory complex that blocks 43S recruitment provides a new mechanism of mRNA-specific translational repression. The presence of an intact eIF4F cap-binding complex, despite the interaction of L13a and eIF4G, is a notable feature that differentiates this mechanism from those in which attack on eIF4E prevents eIF4F assembly. The preservation of eIF4F is unusual, but not unexpected, given that the eIF4G component facilitates end-to-end transcript closure that is required for translation inhibition by L13a. In other cases in which eIF4E is targeted, 5′ to 3′ transcript interactions are mediated by the trans-acting factors themselves, not by the poly(A) tails. Thus, the requirement for an intact eIF4F complex for L13a-mediated repression may be related to the poly(A)-dependency of the mechanism. It will be of interest to learn if the requirement for an intact eIF4F is a common feature in as-yet undiscovered translational control mechanisms that, like L13a, are poly(A)-dependent and target general initiation factors.
EXPERIMENTAL PROCEDURES
Reagents
RRL, methionine-free amino acids and RNasin were from Promega (Madison, WI). Shrimp alkaline phosphatase, reverse transcriptase, Taq polymerase, and restriction enzymes were from Fermentas (Hanover, MD). SP6 Message Machine kit for in vitro transcription was from Ambion (Austin, TX). Translation grade [35S]methionine and transcription grade α-[32P]UTP were from Amersham (Piscataway, NJ). Cycloheximide, GMP-PNP, and all other reagents were from Sigma (St. Louis, MO). Rabbit anti-peptide polyclonal antibody against human L13a was purified on a peptide column (Mazumder et al., 2003a). Anti-eIF4G antibodies were from Simon Morley and Mathias Hentze. Anti-eIF4A antibody was provided by Hans Trachsel and Michael Altmann. Anti-eIF3 antibody was a gift from John Hershey. Anti-eIF4E was purchased from Abcam (Cambridge, MA). HRV-2A protease was from Tim Skern. Anti-Flag and anti-HA antibodies were from Sigma and Roche, respectively.
Plasmid Construction
Construction of the plasmids PSP64-Luc-GAIT-poly(A) and PSP64-Luc-GAIT (mut.)-poly(A) were described previously (Sampath et al., 2003). These plasmids were modified and used to generate capped, poly(A)-tailed (A50) RNAs m7G-Luc-GAIT-poly(A) and m7G-Luc-GAIT (mut.)-poly(A). IRES-containing plasmids were made by PCR amplification of nt 265–828 of EMCV genome using pGEM-Cat-EMCV-Luv (from Graham Belsham) (Drew and Belsham, 1994), nt 40–373 of HCV genome using pHCV(40–373).NS from Tatyana Pestova (Pestova et al., 1998), and nt 6029–6219 of CrPV genome using pCup1 LEU2 IGR URA3 from Peter Sarnow (Thompson et al., 2001). PCR products containing IRES sequences were cloned into Pst1 and BamH1 sites of PSP64-Luc-GAIT-poly(A) and PSP64-Luc-GAIT (mut.)-poly(A) (Sampath et al., 2003) to give plasmids EMCV(or HCV or CrPV)-Luc-GAIT-poly(A) and EMCV(or HCV or CrPV)-Luc-GAIT (mut.)-poly(A). These plasmids were used to generate RNAs containing ApppG at the 5′ cap.
Plasmids PSP64-α-globin-GAIT-poly(A)50 and PSP64-α-globin-GAIT (mut.)-poly(A)50 were made by PCR amplification of the 429-nt α-globin fragment from PHST101 (Komar et al., 1997), excision of Luc, and cloning into the BamH1 and Stu1 sites of PSP64-Luc-GAIT-poly(A) and PSP64-Luc-GAIT (mut.)-poly(A) (Sampath et al., 2003). These plasmids were used to generate m7G-α-globin-GAIT-poly(A)50 and m7G-α-globin-GAIT (mut.)-poly(A)50 RNAs. GST-fusion proteins containing specific eIF4G1 domains were produced from plasmids pGEX-4G-GST-eIF4G1(688–1133), pGEX-4G-GST-eIF4G1(688–1560), and pGEX-4G-GST-eIF4G1(1075–1560), made by cloning the appropriate eIF4G1 sequences into the Sal1 and Not1 sites of pGEX-5x-1. HA-tagged plasmids containing the three eIF4G1 domains were made by cloning the appropriate DNA sequences into BamH1 and Not1 sites of pcDNA3.1-HA. pGEX4G1404 was used as a template for PCR amplification of individual eIF4G1 domains (Hundsdoerfer et al., 2005). A plasmid expressing HA-tagged eIF3e was generated by inserting the eIF3e open reading frame from the vector pNOp48Nde (from John Hershey) into the BamH1 and Not1 sites of pcDNA3.1-HA. A plasmid expressing HA-tagged eIF3j in pcDNA5/FRT/HA-p35 was a gift from John Hershey. pFlag-L13a was made by cloning full-length L13a ORF into the EcoR1 and BamH1 sites of pFlag-CMV-5 (Sigma, MO).
In Vitro Transcription
m7G-Luc-GAIT-poly(A) RNA was produced by in vitro transcription using Sp6 RNA polymerase, using as template PSP64-Luc-GAIT-poly(A) after linearization by PvuII. m7G-α-globin-GAIT-poly(A) RNA was made by in vitro transcription of PSP64-α-globin-GAIT-poly(A) after linearization by EcoR1. The same RNA, but without the poly(A) tail, was made by linearization of the same plasmid by Sac1 prior to addition of Sp6 RNA polymerase. Sp6 Message Machine kit was used to make m7G-capped transcripts. The IRES-containing RNA ApppG-EMCV (or HCV or CrPV)-Luc-GAIT-poly(A) was made by in vitro transcription using Sp6 RNA polymerase from PvuII-linearized EMCV(or HCV or CrPV)-Luc-GAIT-poly(A) plasmid. Sp6 Maxiscript kit (Ambion) was used with ApppG (unmethylated cap, Ambion) with limiting GTP concentration to perform the in vitro transcription reaction. The RNAs were purified by LiCl precipitation and washed with 75% ethanol. m7G-T7 gene 10 RNA was made by in vitro transcription of pGEMEX-2 (Promega) by T7 RNA polymerase using T7 Message Machine kit. PSP64-α-globin-GAIT-poly(A) and PSP64-α-globin-GAIT (mut.)-poly(A) plasmids were linearized by either EcoR1 or Sac1. The linearized plasmids were transcribed by SP6 RNA polymerase using SP6 Message Machine kit in the presence of [α-32P]UTP to generate radiolabeled of m7G-α-globin-GAIT and m7G-α-globin-GAIT (mut.) RNA with and without the poly(A) tail.
Preparation of Recombinant L13a
pET-17b-L13a and pET-15b-L13a were used to express recombinant, unphosphorylated L13a and His-tagged L13a in E. coli (Mazumder et al., 2003a). pFASTBAC1-L13a and pFASTBAC HTb-L13a were used to express phosphorylated, recombinant L13a and His-tagged L13a in baculovirus-infected insect cells. The construction of these plasmids and protein purification was described previously (Mazumder et al., 2003a). His-tagged L13a was purified by Ni-affinity chromatography.
In Vitro Translation of RNA by Reticulocyte Lysate
Purified RNA (200 ng) produced by in vitro transcription was added to RRL (35 μl), 20 μM methionine-free amino acid mixture, 40 U of RNAsin, 20 μCi of translation grade [35S]methionine, and 5 μg of recombinant L13a in a total volume of 50 μl for 60 min, 30 °C. An aliquot (5 μl) was resolved by 7% SDS-PAGE. The gel was fixed, treated with Amplify (Amersham), dried, and radiolabeled bands detected by autoradiography.
Determination of eIF4F assembly on GAIT element-containing RNA
m7G-Luc-GAIT-poly(A) or m7G-Luc-GAIT (mut.)-poly(A) RNA (500 ng) was incubated with RRL (25 μl) and His-tagged L13a (10 μg) in presence of 2 mM GMP-PNP at 30 °C. After 5 min, the reaction was terminated with ice-cold buffer containing 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, 0.05% Triton X-100, 50 mM Hepes, (pH 7.5). Immunoprecipitation was done using 10 μl of monoclonal anti-His antibody (Novagen). From one aliquot the GAIT element containing Luc RNA was extracted by Trizol (Invitrogen) and subjected to reverse transcription using Luc-specific primers. Another aliquot was subjected to SDS-PAGE, followed by immunoblot analysis with anti-eIF4G, anti-eIF4E, anti-eIF4A, and anti-L13a antibodies.
Sucrose Gradient Analysis of Complex Formation during Translation Initiation
In vitro-synthesized RNA (500 ng) was incubated in RRL (100 μl) in the presence of 0.5 mM of cycloheximide for 5 min at 30 °C. The initiation reaction was stopped by placement on ice and layering with a 10 to 25% linear sucrose gradient in buffer containing 100 mM KCl, 5 mM MgCl2, 20 mM Hepes (pH 7.4), 2 mM dithiothreitol, and 0.5 mM cycloheximide. After centrifugation for 16 h, 20,000 rpm, the gradients were unloaded by upward displacement using a programmable density gradient system with a UA-6 detector (ISCO).
To detect the 48S ribosomal complex, 32P-labeled α-globin reporter RNAs (300,000 cpm) were incubated with RRL (100 μl) in presence of GMP-PNP (2 mM) or cycloheximide (0.5 mM). Reaction products were resolved on a 5 to 25% linear sucrose gradient by centrifugation for 18 h at 20,000 RPM. Fractions (0.75 ml) were collected and radioactivity determined by scintillation counting. For eIF3 detection, fractions were subjected to TCA-precipitation, followed by SDS-PAGE and immunoblot analysis with anti-eIF3 antibody.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant HL79164 from NIH, and by Cleveland State University (to B.M) and HL29582 (to P.L.F.). We acknowledge Simon Morley for anti-eIF4G, Christian Thoma and Matthias Hentze for anti-eIF4G and eIF4G plasmid pGEX4G1404, Hans Trachsel and Michael Altmann for anti-eIF4A, John Hershey for anti-eIF3 and eIF3 subunit constructs, Gideon Dreyfuss for SP2/O antibody, Tim Skern and Carla Sousa for HRV-2A protease, Graham Belsham for pGEM-CAT-EMCV-LUC plasmid, Tatyana Pestova for pHCV(40-373).NS plasmid, Peter Sarnow for pCup1 LEU2 IGR URA3 plasmid and Rupak Mukhopadhyay for assistance in monocyte transfection studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J, Nieto A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol. 2000;20:6259–6268. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann K, Grskovic M, Gebauer F, Hentze MW. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell. 2005;122:529–540. doi: 10.1016/j.cell.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. Rna. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Clemens MJ, Bushell M, Morley SJ. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol. 2004;78:7707–7716. doi: 10.1128/JVI.78.14.7707-7716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Drew J, Belsham GJ. trans complementation by RNA of defective foot-and-mouth disease virus internal ribosome entry site elements. J Virol. 1994;68:697–703. doi: 10.1128/jvi.68.2.697-703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fox PL, Mazumder B, Ehrenwald E, Mukhopadhyay CK. Ceruloplasmin and cardiovascular disease. Free Radic Biol Med. 2000;28:1735–1744. doi: 10.1016/s0891-5849(00)00231-8. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Grskovic M, Hentze MW. Drosophila sex-lethal inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol Cell. 2003;11:1397–1404. doi: 10.1016/s1097-2765(03)00176-x. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. Embo J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci U S A. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Kommer A, Krasheninnikov IA, Spirin AS. Cotranslational folding of globin. J Biol Chem. 1997;272:10646–10651. doi: 10.1074/jbc.272.16.10646. [DOI] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Fox PL. Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3′ untranslated region. Mol Cell Biol. 1999;19:6898–6905. doi: 10.1128/mcb.19.10.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Mukhopadhyay CK, Prok A, Cathcart MK, Fox PL. Induction of ceruloplasmin synthesis by IFN-gamma in human monocytic cells. J Immunol. 1997;159:1938–1944. [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003a;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003b;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Imataka H, Sonenberg N, Fox PL. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol Cell Biol. 2001;21:6440–6449. doi: 10.1128/MCB.21.19.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel YM, Borman AM, Paulous S, Kean KM. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol Cell Biol. 2001;21:4097–4109. doi: 10.1128/MCB.21.13.4097-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay CK, Mazumder B, Lindley PF, Fox PL. Identification of the prooxidant site of human ceruloplasmin: a model for oxidative damage by copper bound to protein surfaces. Proc Natl Acad Sci U S A. 1997;94:11546–11551. doi: 10.1073/pnas.94.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- Niessing D, Blanke S, Jackle H. Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev. 2002;16:2576–2582. doi: 10.1101/gad.240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. Embo J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell. 2003a;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Prevot D, Decimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. Embo J. 2003b;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Fox PL. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol Cell Biol. 2003;23:1509–1519. doi: 10.1128/MCB.23.5.1509-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Skern T, Hampolz B, Guarne A, Fita I, Bergmann E, Peterson J, James MNG. Structure and function of picornavirus proteinases. In: Wimmer BLS a E., editor. Molecular Biology of Picornavirus. Washington, D.C: ASM Press; 2002. pp. 199–212. [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Stripecke R, Oliveira CC, McCarthy JE, Hentze MW. Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. Rna. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci U S A. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G. Delivering messages from the 3′ end. Proc Natl Acad Sci U S A. 2001;98:4288–4289. doi: 10.1073/pnas.091108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


