Abstract
We developed an assay for the detection and quantitation of porcine circovirus type 2 (PCV2) with the SYBR Green I-based real-time PCR. The real-time PCR provides a broad dynamic range, detecting from 103 to 1011 copies of DNA per reaction. No cross-reactions were found in specimens containing PCV1. Because of the high sensitivity and specificity of the assay with a relatively rapid and simple procedure, real-time PCR can be used as a routine assay for the clinical diagnosis of PCV2 infection. In this study we applied real-time PCR assay to 80 clinical samples, collected from 40 pigs with postweaning multisystemic wasting syndrome (PMWS) and 40 healthy pigs in comparison with conventional PCR assay. In 56 of 80 samples, PCV2 DNA was detected by conventional PCR assay. All samples positive for PCV2 DNA in conventional PCR assay were also positive in real-time assay, and 12 of 24 samples that tested negative for PCV2 DNA in the conventional assay were tested positive in real-time PCR assay. Real-time PCR assay increased the number of samples in which PCV2 was detected by 15%. It is, therefore, considered to be a useful tool for the detection of PCV2.
Keywords: Porcine circovirus type 2 (PCV2), Real-time PCR, Sensitivity
INTRODUCTION
Porcine circovirus (PCV) is a small, naked DNA virus with diameter of 17 nm (Tischer et al., 1982), and is a member of the Circoviridae family. PCV isolated as a persistent contaminant from a porcine kidney cell line is nonpathogenic and is designated as PCV1 (Allan et al., 1995; 2000). Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in pigs and was first described in 1991 (Harding, 1997). The causative agent of PMWS is believed to be a pathogenic strain of PCV, named PCV2 (Allan et al., 1999). PMWS is a disease that can affect nursery and fattening pigs, and is being reported in pigs throughout the world (Allan et al., 1998; Kennedy et al., 1998; Segales and Domingo, 2002; Wen et al., 2005). The disease is characterized by weight loss, dyspnoea and jaundice as well as the pathological findings of interstitial pneumonia, generalized, enlarged lymph nodes, hepatitis and nephritis (Ladekjær-Mikkelsen et al., 2002). In addition, PCV2 was found to be associated with porcine dermatitis and nephropathy syndrome (PDNS) (Wellenberg et al., 2004), respiratory disease complex (Kim and Chae, 2003) and reproduction failure (Sanchez et al., 2001). Several qualitative PCR methods have been described for the detection of PCV2 in fresh and fixed material (Allan et al., 1999; Hamel et al., 2000; Kim and Chae, 2003; Larochelle et al., 1999; Ouardani et al., 1999). However, as PCV2 is so common within the swine population that conventional PCR can only be used as reference for PMWS diagnosis (Brunborg et al., 2004). Conventional PCR is time-consuming and prone to sample contamination by DNA amplified previously. This increases the potential for false-positive results. Furthermore the load of PCV2 is closely related to PMWS (Brunborg et al., 2004; Liu et al., 2000; Olvera et al., 2004; Rovira et al., 2002), so quantifying the number is important for diagnosing PCV2 in suspected pig.
Real-time PCR is an excellent diagnostic tool with high sensitivity and specificity and fast turnaround time. This system is so-called because the accumulated amplicons can be monitored directly during the DNA amplification process. The quantitation of DNA is based on the determination of the threshold cycle when the amplified PCR product is first detected.
The higher the initial DNA copy number input, the sooner the product of amplification is detected. Reports on virus detection based on real-time PCR technology have been described for various human and animal viruses. There are only five published studies of real-time PCR assays for the detection of PCV2 (Brunborg et al., 2004; Chung et al., 2005; Ladekjær-Mikkelsen et al., 2002; Olvera et al., 2004; Rovira et al., 2002). But all of them used a hybridization probe method in the TaqMan system and are mostly applied on serum samples and the primers were mostly based on their local PCV2 strains. Although some papers (Gilpin et al., 2003; Fenaux et al., 2004) used an SYBR Green I to quantify the PCV2 DNA, but a regression lines between the C T values and the input concentrations were not given.
In this study, the development of a quantitative real-time PCR for the detection of PCV2 based on SYBR Green I dye using LightCycler (LC) system is reported. To evaluate the usefulness of this real-time PCR for diagnosing and monitoring ill pigs with PCV2 infection, we compared results of real-time PCR and conventional PCR using clinical samples from 40 PMWS affected and healthy pigs, respectively.
MATERIALS AND METHODS
Samples
Tissue samples (liver, mesenteric lymph node) were collected from 40 living or dead pigs from 21 to 90 d of age suffering from PMWS. Thirty-three of them originated from five different farrow-to-finish large operations located in eastern and western Hangzhou. The rest of them were obtained from southern Hangzhou. The samples were collected from Jan. 2003 through Oct. 2005. Gross lesions consisted of lymph node and spleen enlargement and non-collapsed, rubbery Samples were stored at −70 °C until DNA isolation.
Liver and mesenteric lymph node samples were collected from 40 healthy randomly selected pigs aged 5 to 6 months from an abattoir in Oct. 2005 and stored at −70 °C until use.
PCV2 DNA isolation
Tissue samples were minced into small pieces and homogenized in phosphate buffered saline (PBS) and centrifuged at 10000×g for 5 min. The virus DNA was extracted from the supernatant by column chromatography using mini DNA extraction kit (Shanghai Shenergy Biocolor Bioscience & Technology Company, Shanghai, China). Cell lysis and precipitation of DNA was performed as recommended by the manufacturer. Briefly, 200 µl of supernatant was incubated with digestion buffer and a final proteinase K concentration of 0.2 mg/ml for 30 min at 55 °C. Then DNA was bound to spin columns and washed twice. Finally, the genomic DNA dissolved in 50 µl of distilled water was stored at −20 °C until use.
Primer designing for SYBR Green I-based quantitative PCR
The primer design for PCV2 quantitation using real-time PCR with SYBR Green I was based on nucleotide sequences of ORF2 (open reading frame 2) retrieved from GenBank (accession Nos. DQ231511~DQ231519 and DQ235696), which were isolated from Hangzhou during 2003~2005. Nucleotide sequences were aligned to identify conserved regions using DNAStar software (DNASTAR, Madison, WI, USA). The primers were as follows: P1, 5′-ATAACCCAGCCCTTCTCCTACC-3′; P2, 5′-GGCCTACGTGGTCTACATTTCC-3′. The length of the amplified products was 145 bp.
Construction of standard plasmids for SYBR quantitative PCR
The plasmid (pPCV2) used as the standard DNA was constructed by ligation of a PCR fragment in pMD18-T simple vector according to the instructions of the manufacturer (TaKaRa, Dalian, China). The cloned fragment was comprised of a 702 bp region of the PCV2 ORF2 (capsid gene) amplified by primers P3: GCCGAGGTGCTGCCGCT and P4: CAGTTCGTCACCCTTTCCCC. Plasmid was propagated in Escherichia coli DH5α cells and was purified by using a Bio-Dev gel extraction kit (Bio-Dev, Beijing, China) and quantified by measuring OD 260 using spectrophotometer (Thermo Electron, USA). Ten-fold dilutions were made in order to get 1012~101 plasmids per 2 µl sample for the real-time PCR. The dilutions were stored at −20 °C, while stock plasmid was stored at −70 °C.
Real-time PCR
Real-time PCR was done using the LC system with SYBR Green I detection and T m analysis. SYBR® Premix Ex Taq™ (perfect real time) PCR kit was purchased from TaKaRa (Dalian, China). Five hundred nanograms DNA in an aliquot of 2 µl was used as template. Viral load was expressed as copies per 500 ng DNA. The procedure was optimized with regard to concentrations of primers, and denature/extension temperature.
The optimized reaction was carried out in a 20 µl final reaction volume containing 0.4 µmol/L concentration of each forward and reverse primer, 2 µl DNA solution, 6.4 µl distilled water, 10 µl of kit-supplied SYBR® PCR master mix (including HotStart Ex Taq HS DNA polymerase, reaction buffer, dNTP mix, and SYBR Green I). Prior to cycling, the glass capillaries were sealed, and placed into the LC sample carousel, and centrifuged at 400×g for 10 s in the LC carousel centrifuge (Eppendorf, Germany). The thermal profile for the real-time PCR was 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 20 s. Three concentrations of pPCV2-ORF2 (109, 105, and 103 copies per sample) were included in each run, and served as positive controls as well as to derive the standard curve used for quantitation of PCV2 DNA in tissue samples. Positive and negative reference samples were tested along with the unknown samples in each run.
Melting curve analysis of the PCR product
Melting curve analysis was performed to measure the specificity of PCR product. After PCR cycling, samples were heated to 95 °C for 0 s and 65 °C for 15 s and then heated to 95 °C at a linear transition rate of 0.1 °C/s. Fluorescence of the samples was monitored continuously while the temperature was increasing. SYBR Green I is released upon denaturation, which results in a decreasing fluorescence of the signal. The LC software calculates the T m. All samples were analyzed once.
Conventional PCR
Viral DNA sequences were amplified by conventional PCR as previously described (Huang et al., 2004) using the oligonucleotide primers PCV2-1 (5′-TAGGTTAGGGCTGTGGCCTT-3′) and PCV2-2 (5′-CCGCACCTTCGGATATACTG-3′). Amplicons of 264 bp were detected by electrophoresing 8 µl aliquots through 1% agarose gel containing 0.5 µg/ml ethidium bromide. To ensure quality of data, negative and positive reference samples were always applied in each PCR reaction. DNA extraction and amplification were performed in different rooms.
Statistics
The viral concentrations were expressed as the viral DNA copies per 500 ng DNA of tested tissues. The data obtained from PMWS and healthy pigs were analyzed using t test, considering a value of P<0.05 as statistically significant.
RESULTS
Primer design
The current assay leads to a PCR product of 145 bp, with primers covering 44 bp of the 145 bp. Alignment of the 10 Hangzhou PCV2 strains with the chosen primers showed that there were up to two mismatches with some PCV2 strains of Hangzhou. However, the present real-time PCR can efficiently amplify various PCV2 field strains of Hangzhou even if PCV2 nucleotide sequence showed up to two mismatches in the primers.
Optimization strategies
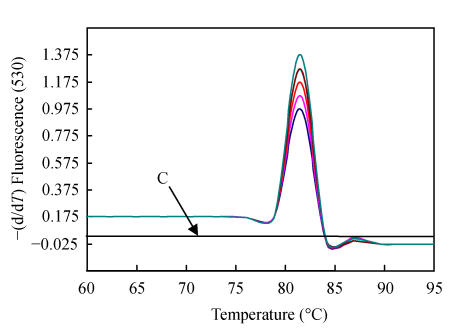
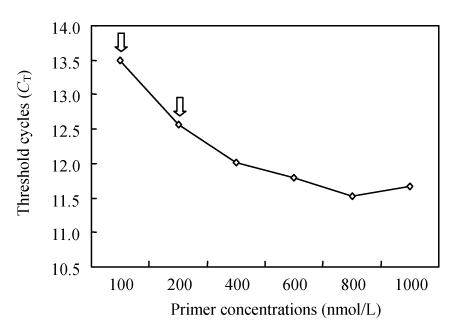
Real-time PCR offers continuous monitoring of the PCR reaction cycle by cycle by using fluorescence. The detection in our assay is based on the binding of the fluorescent dye SYBR Green I, which binds all double stranded DNA molecules. Since this process is sequence independent, the characteristic T m of the target is determined by performing a melting curve analysis on the PCR products to test the formation of primer-dimers and non-specific products, which compete with formation of specific PCR products. T m analysis of the real-time PCR product revealed that the denaturation/elongation temperature in the range of 55~65 °C in cycles had no effect. The distinct temperature at which the double stranded amplicon was denatured was 82.1 °C (range: 81.9~82.4 °C). No increase in fluorescence signal from primer-dimers or other non-specific amplification products (distinguished from the specific product through their lower T m) was monitored (Fig.1). In addition, concentrations of primers were essential for optimal working conditions. As shown in Fig.2, when the PCV2 primer concentrations were lowered from 1000 nmol/L to 100 nmol/L, the PCV2 C T increased markedly by 1.8 cycles. The C T only increased by 0.9 cycles if the forward and reverse primers were adjusted to 200 nmol/L. To maintain PCV2 amplification at a moderate and reproducible level, we chose the combination of 200 nmol/L for the forward and reverse primers.
Fig. 1.
Melting curve analysis of PCR products obtained following 40 cycles of amplification
Melting peaks were produced by plotting the change in the relative intensity of fluorescence for 11 different Hangzhou PCV2 strains over temperature. Note that the T m of the PCR products of PCV2 isolates was 82.1 °C. No peaks of amplification products from negative control (arrow C) were observed
Fig. 2.
Amplifications of PCV2 DNA. The effect of the PCV2 forward and reverse primer concentrations on PCV2 amplification
The experiment was carried out with forward and reverse primer concentrations varying from 100 to 1000 nmol/L, The C T value obtained from each set of amplification conditions was plotted against the concentrations of the forward or reverse primer. Left open arrow, when the reverse primer concentration was lowered from 1000 nmol/L to 100 nmol/L, the PCV2 C T increased markedly by 1.8 cycles; the C T only increased by 0.9 cycles if the reverse primer was adjusted to 200 nmol/L (right open arrow)
Detection and quantitation limit of the assay
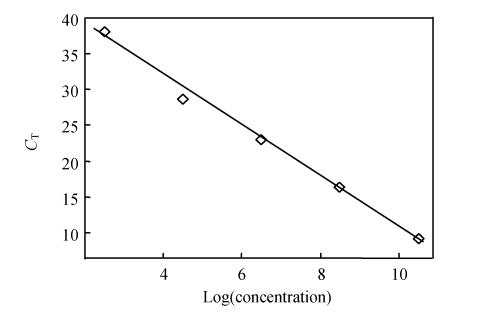
Ten-fold serial plasmid dilutions were tested and used to construct the standard curve by plotting the plasmid copy number logarithm against the measured C T values. Samples were repeated three times. The generated standard curve covered a linear range of 1×103 to 1×1011 copies per 2 µl for PCV2. The efficiency was 2.02 (A perfect amplification reaction would produce a standard curve with an efficiency of “2”, because the amount of target DNA would double). The amount of 1×1012 copies per 2 µl was outside the linear range of the standard curve, amounts between 10~100 copies per 2 µl were not detected (Fig.3).
Fig. 3.
Regression lines between the C T values and the input concentrations of PCV2 plasmid DNA in real-time PCR detected by using SYBR Green I
Specificity of the assay
The specificity of the assay was examined with regard to PCV1. Duplicates of PCV1 DNA, isolated from 200 µl of a suspension of clinical samples infected with PCV1, were run under the optimal conditions of the assay, and no increase in fluorescence being observed.
Reproducibility of the real-time PCR for PCV2
When the standard PCV2 plasmid DNA was used for the evaluation of the coefficients of variation (CVs) of the real-time PCR, the intra- and inter-assay CVs for C T values ranged between 0.29% and 0.82%, and 0.29% and 1.24%, respectively (Table 1).
Table 1.
Variance analysis of C T values quantified by real-time PCR in serially diluted standard plasmid solutions
| Concentration of standard plasmid (copies/ml) | n | Intra-assay variability |
Inter-assay variability |
||||
|
CT |
CV (%) |
CT |
CV (%) | ||||
| Mean | SD | Mean | SD | ||||
| 109 | 10 | 14.2 | 0.08 | 0.56 | 14.5 | 0.18 | 1.24 |
| 107 | 10 | 20.8 | 0.06 | 0.29 | 20.7 | 0.06 | 0.29 |
| 105 | 10 | 26.7 | 0.22 | 0.82 | 26.8 | 0.16 | 0.59 |
Quantitation of viral load in tissue samples
The assay was tested on lymph nodes and livers from both healthy and PMWS affected pigs. There was a significant difference between PCV2 load in healthy and PMWS affected pigs (P<0.01) with a cut-off between healthy and PMWS pigs at 106 (liver) or 107 (lymph node samples) PCV2 genomes per 500 ng DNA. The virus load of specimens that were tested positive for PMWS by the real-time PCR was 1×106 to 1×1010 (liver) and more than 1×107 per 500 ng DNA (lymph node), while the virus load of specimens that were tested positive for the healthy pigs was 103~106 per 500 ng DNA (liver or lymph node samples).
Comparison of results obtained by real-time PCR and conventional PCR
Results of the real-time PCR and conventional PCR assays are shown in Table 2. All liver and lymph node samples positive for PCV2 DNA in conventional PCR were also positive in real-time PCR assay, whereas half of the samples negative for PCV2 DNA in the conventional PCR assays were positive in the real-time PCR assays. The copy number of PCV2 DNA in tissue samples which were positive in both methods ranged from 1×105 to 1×1012 copies per 500 ng DNA. The copy number of PCV2 DNA of twelve lymph node samples, which were positive by the real-time PCR assay but negative by the conventional PCR assay, was 1×103 to 1×105 copies per 500 ng DNA.
Table 2.
Comparison of real-time PCR and conventional PCR
| Conventional PCR assay result (n) | Real-time PCR assay result (n) |
||
| Positive | Negative | Total | |
| Positive | 56 | 0 | 56 |
| Negative | 12 | 12 | 24 |
| Total | 68 | 12 | 80 |
DISCUSSION
The established real-time PCR assay for PCV2 DNA quantitation was found to be in the range of 103 to 1011 with excellent linearity. The new assay increased the number of samples in which PCV2 were detected by 15% more than conventional PCR.
Several PCV2 specific serological assays were established, but they could not differentiate between maternal antibodies and antibodies due to infection. Furthermore, presence of PCV2 antibodies is not relevant to the disease symptoms. Also seroconversion is commonly detected around 20 d after infection, so these methods cannot detect latent infection or early infection. PCV2 infection is common in pig herds and its shedding has been reported in PCV2-infected pigs without any clinical symptoms. The ability to quantify viral loads with the quantitative PCR assay is a valuable tool to gain further understand of mechanisms of PCV2 infection.
For precise estimation of viral loads in samples, a competitive quantitative PCR was applied that can detect and quantify target DNA simultaneously (Liu et al., 2000), but it requires an exogenous competitor as a control that needs to be constructed and characterized, and requires post-PCR electrophoresis. Real-time PCR is performed in a closed tube and requires no post-PCR electrophoresis. Therefore, contamination with amplicons can be avoided. This assay is more reproducible and convenient than competitive quantitative PCR tests. TaqMan real-time PCR requires an efficient probe with double-labelled fluorescent reporter dyes to be incorporated in each reaction. And sometimes the probe cannot bind with target sequence when gene sequence variation occurs.
The primers used in this study were positioned in ORF2 that were conserved between 11 different Hangzhou strains on which sequence data were available. Our system has advantages in having wider range than the fluorogenic probe and competitive quantitative PCR system. The established real-time PCR for PCV2 DNA quantitation were found to be in the (3~11)-log10 dynamic range with excellent linearity (Fig.3). This compares favorably with other methods described previously: a competitive PCR (Liu et al., 2000) or real-time PCR based on SYBR Green I (Fenaux et al., 2004; Gilpin et al., 2003). The former have given linear ranges of 4- to 6-log10, whereas the latter have not presented the range. The detection of PCV2 over a wide range of concentrations allows us to measure viral loads in animals with various levels of infection without further sample concentration or dilution. Furthermore, our method assay was reproducible, as indicated by the low intra- and inter-assay CVs for C T values obtained with the standard plasmids. The intra-assay CVs were equal or less than 0.82%. The limitation of the comparison between conventional and real-time PCR is the absence of a gold standard. There were 12 samples that tested negative by conventional PCR but positive by real-time PCR. Another approach to increase the sensitivity of conventional PCR is to perform nested PCR (Kiatipattanasakul-Banlunara et al., 2002), which has been reported to be more sensitive than single-round PCR. However, a major disadvantage of nested PCR is the high risk of carryover and cross-contamination. Since the real-time PCR uses a completely sealed capillary system, the risk of contamination is much lower.
Brunborg et al.(2004) presented encouraging results with PCV2 Norwegian isolates by using TaqMan system, our study has the unique advantage of having the real-time assay validated with various PCV2 strains from Hangzhou. With these primer sets, our PCR assay detected all PCV2 identified by the conventional PCR method. Moreover, SYBR Green I can bind to any double-strand DNA, so the dye can also be used in diagnosis of other viruses [e.g. porcine reproductive and respiratory syndrome virus (PRRSV)], and most of real-time machines can detect the fluorescence emitted by SYBR Green I. These will lower the diagnosis costs and make the method more applicable and practicable than probe. The LightCycler 2.0 is very quick. It can reach 20 °C/s during cycles and the real-time PCR can finish within 30 min.
We cloned a plasmid containing viral ORF2 for the generation of standard PCV2 for quantitative PCR. The detection limit of this assay was 1000 copies of PCV2 DNA. Steps were taken to avoid false positives in quantitative PCR, because SYBR Green I dye can bind to any double stranded DNA, even nonspecific amplicons. A “hot start” Ex Taq HS polymerase, which requires incubation at 95 °C to activate just before the first denaturation step of thermocycling, was applied. Any nonspecific amplicon would be eliminated before the amplification, with no subsequent contamination detected on further runs. The specificity of PCR was verified by the performance of melting curve analysis for the PCR product, which depends on its GC content, length, and sequence. All the Hangzhou PCV2 strains were amplified and examined by melting curve analysis. The melting temperatures of PCR products were 82.1 °C indicating an amplification of a specific product. The specificity of the real-time PCR with SYBR for PCV2 was also determined by testing samples infected with PCV1. As expected, the PCV1 was not detected using the established real-time PCR. Changing the annealing temperature to 55 °C or 65 °C did not affect the efficiency and did not lead to detection of PCV1 in the assay. Also, this proved our primer was specific. Furthermore, this makes it easier to design other primer sets to detect two or more viruses simultaneously, which is our next goal, because Chung et al.(2005) have established a real-time PCR based on SYBR Green I to detect PRRSV. Finally, according to the instructions provided by TaKaRa we did routine analyses with an annealing/elongation temperature of 60 °C.
A high PCV2 load in tissues in an animal is a good indicator of its PMWS status. Significant differences in viral load of PCV2 in serum from sick and healthy animals have been described by other authors (Brunborg et al., 2004; Liu et al., 2000; Olvera et al., 2004; Rovira et al., 2002). Brunborg et al.(2004) reported a cut-off of 1×107 PCV2 genomes per 500 ng DNA for PMWS. We also found that animals with mild, moderate and severe symptom had PCV2 loads of 1×106~1×1010, 1×109~1×1010 and 1×1010~1×1012 copies per 500 ng DNA, respectively, while healthy pigs had only 1×103~1×105 copies. There was a slight difference between our and Brunborg’s results. One of the reasons for this difference may be the time of sample collection, if samples were collected from the dead animals, or pigs were dead prior to sending to the laboratory and the collected tissue samples were in various states of degradation, they may have influenced the recovery of nonpackaged viral DNA, and an underestimation of viral load in tissues (Brunborg et al., 2004). The other reason may be the inherent nature of different method, e.g. TaqMan, SYBR have different sensitivity and efficiency.
References
- 1.Allan GM, McNeilly F, Cassidy JP, Reilly GA, Adair B, Ellis WA, McNulty MS. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44(1):49–64. doi: 10.1016/0378-1135(94)00136-K. [DOI] [PubMed] [Google Scholar]
- 2.Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10(1):3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 3.Allan GM, McNeilly F, Meehan BM, Kennedy S, Mackie DP, Ellis JA, Clark EG, Espuna E, Saubi N, Riera P, et al. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol. 1999;66(2):115–123. doi: 10.1016/S0378-1135(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 4.Allan GM, McNeilly F, Ellis J, Krakowka S, Meehan B, McNair I, Walker I, Kennedy S. Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol. 2000;145(11):2421–2429. doi: 10.1007/s007050070031. [DOI] [PubMed] [Google Scholar]
- 5.Brunborg IM, Moldal T, Jonassen CM. Quantitation of porcine circovirus type 2 isolated from serum/plasma and tissue samples of healthy pigs and pigs with postweaning multisystemic wasting syndrome using a TaqMan-based real-time PCR. J Virol Methods. 2004;122(2):171–178. doi: 10.1016/j.jviromet.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Chung WB, Chan WH, Chaung HC, Lien Y, Wu CC, Huang YL. Real-time PCR for quantitation of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 in naturally-infected and challenged pigs. J Virol Methods. 2005;124(1-2):11–19. doi: 10.1016/j.jviromet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J Virol. 2004;78(24):13440–13446. doi: 10.1128/JVI.78.24.13440-13446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilpin DF, McCullough K, Meehan BM, McNeilly F, McNair I, Stevenson LS, Foster JC, Ellis JA, Krakowka S, Adair BM, et al. In vitro studies on the infection and replication of porcine circovirus type 2 in cells of the porcine immune system. Vet Immunol Immunopathol. 2003;94(3-4):149–161. doi: 10.1016/S0165-2427(03)00087-4. [DOI] [PubMed] [Google Scholar]
- 9.Hamel AL, Lin LL, Sachvie C, Grudeski E, Nayar GP. PCR detection and characterization of type-2 porcine circovirus. Can J Vet Res. 2000;64(1):44–52. [PMC free article] [PubMed] [Google Scholar]
- 10.Harding J. Post-weaning multisystemic wasting syndrome (PMWS): preliminary epidemiology and clinical presentation. Proc Am Assoc Swine Pract. 1997;28:503. [Google Scholar]
- 11.Huang C, Hung JJ, Wu CY, Chien MS. Multiplex PCR for rapid detection of pseudorabies virus, porcine parvovirus and porcine circoviruses. Vet Microbiol. 2004;101(3):209–214. doi: 10.1016/j.vetmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy S, Allan G, McNeilly F, Adair BM, Hughes A, Spillane P. Porcine circovirus infection in Northern Ireland. Vet Rec. 1998;142(18):495–496. [PubMed] [Google Scholar]
- 13.Kiatipattanasakul-Banlunara WTR, Suzuki K, Albarenque SM, Thanawongnuwech R, Nakayama H, Doi K. Detection of porcine circovirus 2 (PCV-2) DNA by nested PCR from formalin-fixed tissues of post-weaning multisystemic wasting syndrome (PMWS) pigs in Thailand. J Vet Med Sci. 2002;64(5):449–452. doi: 10.1292/jvms.64.449. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Chae C. Multiplex nested PCR compared with in situ hybridization for the differentiation of porcine circoviruses and porcine parvovirus from pigs with postweaning multisystemic wasting syndrome. Can J Vet Res. 2003;67(2):133–137. [PMC free article] [PubMed] [Google Scholar]
- 15.Ladekjær-Mikkelsen AS, Nielsen J, Stadejek T, Storgaard T, Krakowka S, Ellis J, McNeilly F, Allan G, Botner A. Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2) Vet Microbiol. 2002;89(2-3):97–114. doi: 10.1016/S0378-1135(02)00174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods. 1999;80(1):69–75. doi: 10.1016/S0166-0934(99)00032-4. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Wang L, Willson P, Babiuk LA. Quantitative, competitive PCR analysis of porcine circovirus DNA in serum from pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 2000;38(9):3474–3477. doi: 10.1128/jcm.38.9.3474-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olvera A, Sibila M, Calsamiglia M, Segales J, Domingo M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J Virol Methods. 2004;117(1):75–80. doi: 10.1016/j.jviromet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol. 1999;37(12):3917–3924. doi: 10.1128/jcm.37.12.3917-3924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rovira A, Balasch M, Segales J, Garcia L, Plana-Duran J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. Experimental inoculation of conventional pigs conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol. 2002;76(7):3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez REJr, Nauwynck HJ, McNeilly F, Allan GM, Pensaert MB. Porcine circovirus 2 infection in swine foetuses inoculated at different stages of gestation. Vet Microbiol. 2001;83(2):169–176. doi: 10.1016/S0378-1135(01)00425-4. [DOI] [PubMed] [Google Scholar]
- 22.Segales J, Domingo M. Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet Q. 2002;24(3):109–124. doi: 10.1080/01652176.2002.9695132. [DOI] [PubMed] [Google Scholar]
- 23.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295(5844):64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 24.Wellenberg GJ, Stockhofe-Zurwieden N, de Jong MF, Boersma WJ, Elbers AR. Excessive porcine circovirus type 2 antibody titers may trigger the development of porcine dermatitis and nephropathy syndrome: a case-control study. Vet Microbiol. 2004;99(3-4):203–214. doi: 10.1016/j.vetmic.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Wen L, Guo X, Yang H. Genotyping of porcine circovirus type 2 from a variety of clinical conditions in China. Vet Microbiol. 2005;110(1-2):141–146. doi: 10.1016/j.vetmic.2005.07.003. [DOI] [PubMed] [Google Scholar]