Abstract
The RNA chaperone, Hfq, plays a diverse role in bacterial physiology beyond its original role as a host factor required for replication of Qβ RNA bacteriophage. In this study, we show that Hfq is involved in the expression and secretion of virulence factors in the facultative intracellular pathogen, Salmonella typhimurium. A Salmonella hfq deletion strain is highly attenuated in mice after both oral and intraperitoneal infection, and shows a severe defect in invasion of epithelial cells and a growth defect in both epithelial cells and macrophages in vitro. Surprisingly, we find that these phenotypes are largely independent of the previously reported requirement of Hfq for expression of the stationary phase sigma factor, RpoS. Our results implicate Hfq as a key regulator of multiple aspects of virulence including regulation of motility and outer membrane protein (OmpD) expression in addition to invasion and intracellular growth. These pleiotropic effects are suggested to involve a network of regulatory small non-coding RNAs, placing Hfq at the centre of post-transcriptional regulation of virulence gene expression in Salmonella. In addition, the hfq mutation appears to cause a chronic activation of the RpoE-mediated envelope stress response which is likely due to a misregulation of membrane protein expression.
Introduction
The bacterial Sm-like protein, Hfq, has been increasingly recognized as a post-transcriptional regulator of global gene expression (Valentin-Hansen et al., 2004). Hfq was first identified in Escherichia coli as a host factor required for replication of Qβ RNA bacteriophage (Franze de Fernandez et al., 1968), and shown to be an RNA-binding protein that forms homohexamers of ∼12 kDa subunits (Franze de Fernandez et al., 1972). Hfq was early observed to be an abundant protein (Carmichael et al., 1975), but its importance in uninfected bacteria remained unclear until it was shown that an hfq insertion mutant of E. coli exhibited broad, pleiotropic phenotypes affecting growth rate, cell morphology and tolerance of stress conditions (Tsui et al., 1994). Independently, genetic analysis of Azorhizobium caulinodans and Yersinia enterocolitica mutants, showing defects in nitrogen fixation or toxin production respectively, found that these phenotypes were due to mutations in hfq (Kaminski et al., 1994; Nakao et al., 1995). Subsequently, Hfq was shown to promote efficient translation of rpoS mRNA in E. coli and Salmonella (Brown and Elliott, 1996; Muffler et al., 1996), and to alter the stability of several other mRNAs (e.g. Vytvytska et al., 1998; Hajnsdorf and Regnier, 2000), indicating that this protein acts to regulate gene expression at the post-transcriptional level. Hfq has also emerged as a key player in mRNA translational control by small non-coding RNAs (sRNAs). Here, Hfq was first observed to be involved in translational repression of rpoS mRNA by OxyS, a small regulatory RNA that is part of the oxidative stress response in E. coli (Zhang et al., 1998). Since then, numerous E. coli sRNAs have been shown to associate with Hfq and to require this protein for their own stability and/or for interactions with their target mRNAs (reviewed in Valentin-Hansen et al., 2004; Majdalani et al., 2005; Romby et al., 2006). These include two E. coli sRNAs, DsrA and RprA, which activate rpoS translation in response to stress conditions (reviewed in Repoila et al., 2003); note, however, that the RpoS regulatory function of these sRNAs may not be conserved in Salmonella (Jones et al., 2006).
Several recent studies addressed a potential role of Hfq in the virulence of pathogenic bacteria. A Brucella abortus hfq mutant displayed significantly reduced survival in cultured murine macrophages, and attenuated virulence in a mouse model (Robertson and Roop, 1999). Similarly, Hfq was reported to be essential for the virulence of Vibrio cholerae (Ding et al., 2004). An hfq mutant of this bacterium fails to colonize the suckling mouse intestine, a model of cholera pathogenesis. Hfq also contributes to the pathogenesis of Listeria monocytogenes in mice (Christiansen et al., 2004), and to Legionella pneumophila virulence in amoeba and macrophage infection models (McNealy et al., 2005). Furthermore, the hfq mutation reduces the virulence of the opportunistic human pathogen Pseudomonas aeruginosa by affecting both cell-associated (flagellum, adhesion factors) as well as extracellular virulence factors, e.g. elastases and pyocyanin (Sonnleitner et al., 2003). In most of these cases, the observed virulence defects were accompanied by reduced stress tolerance, likely reflecting a compromised ability to cope with the harsh environment in the host cell (Robertson and Roop, 1999; Christiansen et al., 2004; McNealy et al., 2005).
A role for Hfq in bacterial virulence was first indicated by its requirement for efficient expression of the major stress sigma factor, σS (also known as RpoS, KatF or σ38) in the enteric bacteria, E. coli and Salmonella. Here, hfq mutants display greatly reduced RpoS levels in stationary phase, due to inefficient translation of the rpoS mRNA (Brown and Elliott, 1996; Muffler et al., 1996). In Salmonella, σS is an important virulence factor as it mediates the expression of the Salmonella plasmid virulence (spv) genes, which are required for systemic infection, and enables bacteria to cope with diverse stresses (nutrient deprivation, oxidative and acid stress, DNA damage) relevant to the environments faced in their mammalian hosts (Fang et al., 1992; Bang et al., 2005). A Salmonella rpoS mutant exhibits significantly reduced virulence in mice (Fang et al., 1992), and mutated rpoS alleles are often found in attenuated Salmonella strains (Robbe-Saule et al., 1995; Wilmes-Riesenberg et al., 1997).
Based on the importance of Hfq for σS expression and the many phenotypes shared by hfq and rpoS mutants in E. coli and Salmonella (Fang et al. 1992; Muffler et al., 1997), it has generally been assumed that Hfq would be important for Salmonella virulence. However, experimental evidence for a more general role of Hfq, i.e. beyond promoting rpoS mRNA translation, has so far been lacking. To address these questions, we constructed and characterized a set of hfq mutants and control strains in Salmonella enterica serovar Typhimurium (S. typhimurium). We find that loss of Hfq results in drastically reduced virulence in vitro and in vivo. These phenotypes, which are largely σS-independent, are associated with loss of cell motility, altered membrane composition, reduced adhesion and abrogated effector protein secretion. The results indicate that Hfq plays a much more dominant role in Salmonella virulence than previously believed.
Results
Construction of Salmonella hfq mutant and control strains
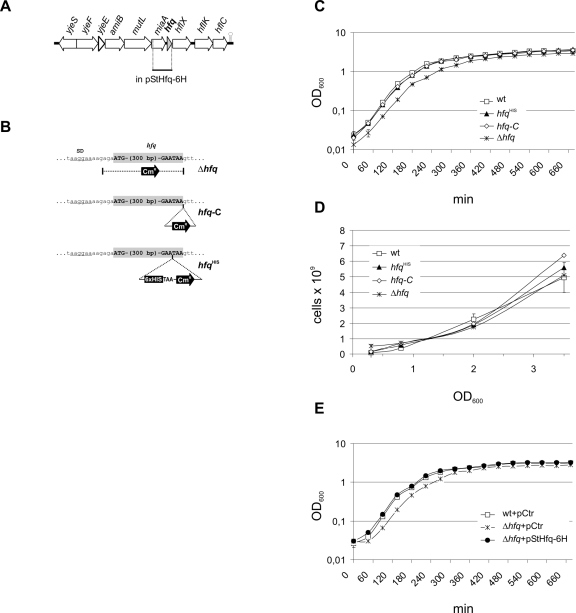
The hfq gene is located in clockwise orientation at bps 4604575–4604883 in the genome of S. typhimurium strain LT2 (McClelland et al., 2001). As in E. coli, it is located in the yjeF-yjeE-amiB-mutL-miaA-hfq-hflX-hflK-hflC cluster of genes (Fig. 1A), part of which may form an operon (Tsui and Winkler, 1994). The Salmonella and E. coli hfq genes are 93% and 94% identical at the nucleotide and amino acid level respectively, with all amino acid deviations being located in the Hfq C-terminal region (Brown and Elliott, 1996). The sequence of the hfq region taken from the unfinished genome of the virulent Salmonella strain used in this study, SL1344 (http://www.sanger.ac.uk/Projects/Salmonella), was compared with that of strain LT2 and found to be identical.
Fig. 1.
Details of Salmonella hfq mutants and their growth characteristics. A. Genomic location of hfq in SL1344. The region cloned on complementation plasmid, pStHfq-6H, is indicated. B. Schematic representation of the insertion sites of the cat resistance cassette in the deletion mutant Δhfq, the control strain hfq-C, and the chromosomally HIS-tagged strain, hfqHIS. C and D. Growth and cell viability of hfq mutant strains (open squares: wild-type; filled triangles: hfqHIS; open diamonds: hfq-C; stars: Δhfq). (C) OD600 values of triplicate cultures in LB medium were determined in 45 min intervals. (D) Bacteria were plated to determine viable counts (from triplicate cultures) at an OD of 0.3 and of 2, and 6 h after cultures had reached an OD of 2. E. Complementation of the slight growth defect of the Δhfq strain by plasmid pStHfq-6H (open squares: wild-type strain carrying control plasmid pVP012; stars: Δhfq carrying a control plasmid; filled circles: Δhfq complemented with pStHfq-6H).
Based on the sequence data, three hfq mutant or control strains were constructed in SL1344 to study Hfq functions in vivo (Fig. 1B). In the Δhfq mutant, the entire hfq coding region is replaced by a cat (chloramphenicol resistance) marker. As the cat gene used here does not carry a transcriptional terminator, transcription of the polycistron should be unaffected. hfq-C is a control strain in which the cat gene is inserted after the hfq stop codon. In control strain hfqHIS, the cat gene is inserted before the UAA stop codon. In addition, this latter insertion adds six histidine codons to the last hfq codon, thus producing a chromosomally encoded His-tagged Hfq protein.
Growth characteristics of the hfq mutant and control strains
All three hfq strains formed normal colonies when grown on standard Luria–Bertani (LB) plates at 37°C, although the Δhfq strain exhibited slightly slower growth. At room temperature (22°C) however, the Δhfq mutant grew much more slowly than the wild type, seen as a smaller colony size, whereas the hfq-C and hfqHIS derivatives showed normal growth (data not shown). When we compared the growth of all strains in LB liquid medium with aeration at 37°C, no differences were observed among the wild type, and the two control strains, hfq-C and hfqHIS (Fig. 1C). The deletion mutant, Δhfq, showed a longer lag phase after inoculation into fresh medium and reached stationary phase at a lower optical density as compared with the other three strains. However, parallel determination of viable counts at three different growth phases showed that cell viability of Δhfq was uncompromised (Fig. 1D).
The observation that the hfq-C and hfqHIS strains showed growth rates identical to the wild-type strain supported the suggestion that the slightly altered growth of the Δhfq mutant was due to the lack of Hfq protein rather than to polar effects caused by the insertion of the cat cassette. To corroborate this, the hfqHIS allele including 1014 bp of the upstream miaA coding sequence was cloned in a low-copy vector (pSC101* origin), resulting in plasmid pStHfq-6H. This plasmid fully complemented the reduced growth of the Δhfq strain (Fig. 1E), also indicating that the major hfq promoter is located within the miaA coding region.
Certain growth conditions, e.g. oxygen limitation and high osmolarity, are known to activate Salmonella invasion gene expression in vitro (e.g. Lee and Falkow, 1990; Song et al., 2004). As these so-called Salmonella pathogenicity island 1 (SPI1)-inducing conditions were used extensively in this study (see below), we also determined the growth behaviour of all aforementioned strains under these conditions. As seen with aerobic growth, the Δhfq mutant strain exhibited a slightly extended lag phase but reached the same optical density as the wild type while the two control strains hfqHIS and hfq-C show growth indistinguishable from the wild-type strain (Fig. S1).
The hfq mutation attenuates virulence in mice
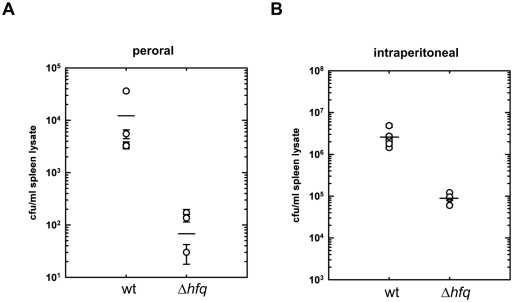
To address the role of Hfq in Salmonella pathogenesis, we first examined the effect of the hfq deletion in a typhoid fever mouse model of Salmonella infection. Groups of 4- to 5-week-old, female Balb/c mice (five mice per strain) were infected perorally with 108 cfu of either the wild-type or Δhfq strains. Mice infected with the wild-type strain showed typical symptoms of infection beginning the following day, whereas mice infected with the Δhfq mutant showed no signs of illness during the course of the experiment. The infected animals were sacrificed 72 h post infection, and organ colonization was determined by plating dilutions of homogenized spleen lysates to agar plates. As shown in Fig. 2A, the hfq mutant was recovered at > 100-fold reduced levels relative to the wild-type strain after peroral infection, and for at least two of the mice, no bacteria were recovered. These observations suggested that the hfq mutation resulted in defects in either invasion of intestinal epithelial cells, macrophage survival, or both.
Fig. 2.
The Δhfq mutant is severely attenuated in mice. A. Groups of five Balb/c mice were infected perorally with suspensions of ∼108 bacteria of either the wild-type or Δhfq strains. Bacterial loads in spleen homogenates were determined 72 h post infection. For intraperitoneal infections (B) 1:1 mixtures of both, wild-type and Δhfq strain, each strain at ∼105 bacteria, were used for infections. Forty-eight hours post infection, spleens were removed and the cfu ml−1 for each strain was determined in spleen homogenates by plating to selective plates for calculation of the relative ratios of the two, co-infecting strains (competitive index, CI, see text).
To determine whether the virulence defect of the hfq mutant extended beyond invasion-related defects, mice were also co-infected intraperitoneally with a mixture of the wild-type and Δhfq strains, where uptake by resident macrophages should circumvent the need for invasion. Two, independent experiments indicated that the hfq mutant showed at least a 30- to 100-fold reduced uptake and/or survival in macrophages and subsequent carriage to the spleen compared with the wild-type strain (Fig. 2B), leading to calculated competitive indices (CI; Shea et al., 1999) of 0.01–0.03. This is consistent with the idea that both uptake and intracellular survival/proliferation in macrophages were affected. It should be noted that the post-infection time points for determination of bacterial counts shown were chosen to avoid premature death of the infected animals. In preliminary experiments, in animals still surviving 1 week post infection in the mixed infection experiments, the Δhfq strain showed a > 1000-fold reduction in cfu relative to the wild-type strain (CI of 0.0005−0.001; data not shown).
The hfq mutant is impaired in the invasion of non-phagocytic cells
Oral infection by Salmonella results in active invasion of non-phagocytic epithelial cells of the host intestine. To determine the effect of the hfq mutation on the invasion rate of non-phagocytic cells in vitro, cultured HeLa cells were infected with the wild-type and several hfq mutant and control strain strains. A Salmonella SL1344 Δspi1 mutant, which lacks the entire SPI1, served as a negative control in these experiments. SPI1 encodes a type three secretion system (TTSS) and several effector proteins that mediate the uptake of Salmonella by non-phagocytic eukaryotic cells (Galan and Curtiss, 1989; Mills et al., 1995; Collazo and Galan, 1996). HeLa cells were infected with a multiplicity of infection (moi) of 10 with bacteria grown aerobically to early stationary phase (OD600 of 2). Following gentamicin treatment to kill remaining extracellular bacteria, the number of intracellular bacteria was determined 2 and 6 h post infection (Table 1).
Table 1.
Invasion and intracellular replication (% of the bacterial input).
| Aerobic growth to early stationary phase (OD600 of 2), gentamicin protection assay (HeLa cells) | SPI1-inducing growth conditions, gentamicin protection assay (HeLa cells) | Aerobic growth to early stationary phase (OD600 of 2), macrophage survival assay (RawB) | |||||
|---|---|---|---|---|---|---|---|
| Strain/infection time | 2 h | 6 h | 2 h | 6 h | 1 h | 4 h | 24 h |
| wt | 14.16 | 30.38 | 29.4 | 82.11 | 16.53 | 29.79 | 47.48 |
| hfqHIS | 7.92 | 14.53 | 13.59 | 41.58 | 5.54 | 10.31 | 18.08 |
| hfq-C | 4.76 | 19.66 | 15.74 | 38.59 | 4.90 | 12.37 | 15.74 |
| Δhfq | 0.13 | 0.13 | 3.25 | 6.58 | 0.39 | 0.40 | 3.25 |
| Δspi1 | 0.00 | 0.01 | 0.05 | 0.07 | ND | ND | ND |
| ΔrpoS | 8.79 | 22.61 | 22.88 | 65.89 | ND | ND | ND |
| wt + pCtr | ND | ND | 22.19 | 70.74 | 9.98 | 26.55 | 28.63 |
| Δhfq + pCtr | ND | ND | 2.35 | 6.29 | 0.54 | 0.57 | 0.67 |
| Δhfq + pStHfq-6H | ND | ND | 40.87 | 118.38 | 15.16 | 41.21 | 42.24 |
The hfq deletion mutant showed a 100-fold reduced initial rate of invasion at 2 h post infection compared with the wild-type strain. We also compared the number of intracellular bacteria present after an additional 4 h. Within these 4 h, the number of wild-type bacteria doubled, whereas the number of hfq mutant bacteria remained unchanged, suggesting an intracellular growth defect in addition to an invasion defect. Despite its drastic invasion defect, the invasion rate of the hfq mutant remains above that of a non-invasive Δspi1 mutant for which only single cells could be recovered (Table 1 and Fig. S2A).
To determine whether the hfq mutant was still impaired in invasion when grown under SPI1-inducing conditions, the invasion assays were repeated with bacterial cultures grown for 12 h under high-salt, oxygen-limiting conditions (Table 1 and Fig. S2B). These growth conditions increased the invasion rate of both the wild type and the Δhfq strain to 30% and 3% respectively (as calculated for the 2 h time point). However, the Δhfq strain remained 10-fold less invasive than the wild type, and intermediate with respect to the non-invasive Δspi1 mutant. While the wild-type strain showed more than one replication in additional 4 h, the Δhfq strain only doubled in the 4 h period.
Three other strains included as controls in all of these experiments, ΔrpoS, hfq-C and hfqHIS, all displayed only slightly reduced invasion rates in the range of 1.3- to threefold in comparison with the wild type, and none of these strains were affected in intracellular growth (Table 1 and Fig. S2A and B). To corroborate that the lack of Hfq protein was the main cause of the invasion defect of the Δhfq mutant, we tested whether it could be complemented by a plasmid-borne hfq allele. Providing Hfq in trans with plasmid pStHfq-6H not only fully restored invasion to the hfq deletion strain, but enhanced invasion relative to the wild type (Table 1 and Fig. S2B). Taken together, these data suggest that Hfq is required for efficient invasion of non-phagocytic cells, which is likely to underlie the strong attenuation of virulence seen in oral mouse infections.
We also examined both the invasion and long-term intracellular growth phenotypes of the Δhfq mutant in an intestinal epithelial cell line (Fig. S3A). Consistent with the results using the HeLa cell line, the initial invasion rate of LoVo cells was 10- to 100-fold reduced at either a 10-fold higher or equivalent infective dose as the wild-type strain respectively. In addition, whereas the wild-type strain showed an approximately 10- to 20-fold increase in intracellular cfu over a 24 h period, the Δhfq strain showed either no change or a slight reduction in viable bacteria over the same period. These results were consistent with a requirement for Hfq for both invasion as well as intracellular replication in non-phagocytic cells.
The hfq strain survives but shows an intracellular growth defect in macrophages
Salmonella survival in the host is also dependent on the ability to survive and replicate in macrophages. To test a possible role for Hfq in macrophage survival, we infected in vitro cultured murine macrophages (RawB) with equal numbers of wild-type and hfq mutant bacteria (Table 1 and Fig. S2C). At 1 h post infection, we noted 30-fold fewer intracellular bacteria in macrophages infected with the hfq mutant, likely reflecting the reduced invasion rate of this strain. However, complementation with plasmid pStHfq-6H fully restored macrophage invasion, comparable to levels observed with wild-type bacteria. Intracellular replication as determined 4 and 24 h after infection also revealed drastic differences between the wild-type strain and the hfq deletion mutant. While the wild-type and the hfq-C and hfqHIS control strains at least doubled within the 4 h post infection, the hfq deletion mutant showed no significant increase in intracellular bacteria per macrophage. At 24 h post infection the number of intracellular bacteria had increased to > threefold as compared with the 1 h time point for the wild-type, the control strains and the complemented deletion mutant (Table 1).
In other experiments, infection of the J774A.1 murine macrophage cell line showed a similar reduction in initial uptake, but no significant increase in intracellular cfu for up to 24 h (Fig. S3B). Thus, Hfq appeared to have little or no effect on the expression of genes required for macrophage survival, although the lack of significant intracellular growth in both epithelial and macrophage cell lines suggested an effect on expression of the second, major pathogenicity island, SPI2, which is required for intracellular proliferation (Shea et al., 1996; Cirillo et al., 1998; Hensel et al., 1998).
Lack of Hfq results in global changes of protein expression and loss of protein secretion
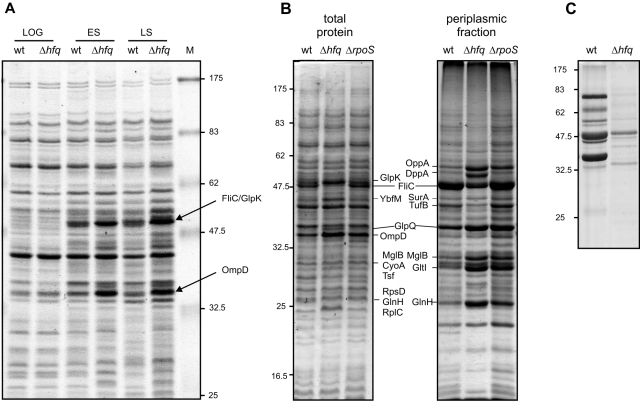
Considering the pleiotropic effect of Hfq on mRNA stability and translational regulation in other bacteria, we sought to determine Hfq-dependent changes in protein expression. We first compared the whole-cell protein patterns in one-dimensional gels of wild-type and Δhfq cells from cultures grown aerobically in l-broth in three different growth phases: exponential growth, early and late stationary phase. As shown in Fig. 3A, Δhfq cells exhibit no significant difference to the wild type in exponential phase. In contrast, in stationary phase the Δhfq mutation showed a markedly different protein pattern, with the most prominent and reproducible changes being two abundant protein bands of ∼40 and ∼55 kDa (Fig. 3A). Mass spectrometry (MALDI-TOF) identified the 40 kDa band as the major outer membrane protein (OMP), OmpD. Analysis of the 55 kDa band proved more complex, because MALDI-TOF analysis indicated the presence of two proteins, GlpK (glycerol kinase) and FliC (major phase-1 flagellin). This band was further resolved with longer gel runs (Fig. 3B, left panel) and revealed that in the Δhfq mutant, FliC levels were strongly reduced whereas GlpK accumulated to higher levels. Parallel analysis of the protein profile of an rpoS deletion strain showed that the Hfq-dependent regulation of OmpD, FliC and GlpK, was not related to lower σS levels in Δhfq cells. Additional analyses revealed an increase in the levels of HtrA, YbfM, OmpF, CyoA and Tsf, and a decrease of the ribosomal proteins RpsD and RplC in the Δhfq strain. To obtain a preliminary picture of global changes in the expression profiles of less abundant proteins, we also analysed early stationary phase samples of wild-type and Δhfq cells resolved on two-dimensional gels (Fig. S4). Of the 69 protein candidates analysed by MALDI-TOF, 32 were upregulated in Δhfq cells, whereas 37 showed downregulation. These results are summarized in Table 2 (further details are given in Table S1).
Fig. 3.
Altered protein expression in Salmonella Δhfq. SDS-PAGE (10–12% gels) of protein samples of SL1344 wild-type and Δhfq prepared from different growth phases (LOG: logarithmic phase, OD600 of 0.3; ES: early stationary phase, OD600 of 2; LS: late stationary phase, 6 h after cells had reached an OD600 of 2). A. Total protein samples. B. Total protein and periplasmic fractions; samples of a ΔrpoS strain were included as an additional control. C. Secreted protein fractions of early stationary phase bacteria.
Table 2.
Results of 1D and 2D gel analysis of protein patterns of SL1344 wild-type and Δhfq cultures grown to early stationary phase (OD600 = 2).
| Candidate proteina | Regulationb | Localizationc | Functiond | Analysise |
|---|---|---|---|---|
| CarA | – | CP | Carbamoyl-phosphate synthetase, glutamine-hydrolysing small subunit | 2D |
| SurA | + | CP | Peptidyl-prolyl cis-trans isomerase, survival protein | 1D, 2D |
| HtrA | + | PP | Periplasmic serine protease Do, heat shock protein | 1D, 2D |
| PyrH | – | CP | Uridine 5′-monophosphate kinase | 2D |
| Upp | – | CP | uracil phosphoribosyltransferase | 2D |
| YaeT | + | (OM) | Putative outer membrane antigen | 2D |
| GltI | + | PP | ABC transporter periplasmic binding protein; ABC superfamily, glutamate/aspartate transporter | 1D, 2D |
| SucD | – | CP | Succinyl-CoA synthetase, alpha subunit | 2D |
| Pal | + | PP | Tol protein required for outer membrane integrity, uptake of group A colicins, and translocation of phage DNA to cytoplasm | 2D |
| YbgF | – | (PP) | Putative periplasmic protein | 2D |
| Dps | – | CP | Stress response DNA-binding protein; starvation induced resistance to H2O2; DNA protection during starvation protein | 2D |
| CspD | + | CP | Cold shock-like protein CspD; similar to CspA but not cold shock induced | 2D |
| TrxB | – | CP | Thioredoxin reductase; thioredoxin reductase | 2D |
| FabF | – | CP | 3-oxoacyl-[acyl-carrier-protein] synthase II | 2D |
| IcdA | + | CP | Isocitrate dehydrogenase in e14 prophage, specific for NADP+ | 2D |
| PagC | + | OM | PhoP regulated: reduced macrophage survival; virulence membrane protein PagC precursor | 2D |
| STM1254 | – | (OM) | Putative outer membrane lipoprotein | 2D |
| STM1328 | – | (OM) | Putative OMP | 2D |
| AroD | – | CP | 3-Dehydroquinate dehydratase | 2D |
| LppB | – | OM | Putative methyl-accepting chemotaxis protein; major outer membrane lipoprotein | 2D |
| LppA | – | OM | Murein lipoprotein, links outer and inner membranes; major outer membrane lipoprotein | 2D |
| YnaF | – | CP | Putative universal stress protein | 2D |
| Tpx | + | CP | Thiol peroxidase | 2D |
| TrpB | – | CP | Tryptophan synthase beta chain | 2D |
| OppA | + | PP | ABC superfamily, oligopeptide transport protein with chaperone properties | 1D, 2D |
| KdsA | – | CP | 3-deoxy-D-manno-octulosonic acid 8-P synthetase | 2D |
| PrsA | – | CP | Phosphoribosylpyrophosphate synthetase | 2D |
| FliC | – | OM/SUP | Flagellin, filament structural protein | 2D |
| Gnd | – | CP | Gluconate 6-phosphate dehydrogenase, decarboxylating | 2D |
| GlpQ | + | PP | Glycerophosphodiester phosphodiesterase, periplasmic | 1D, 2D |
| AckA | – | CP | Acetate kinase A (propionate kinase 2) | 2D |
| HisJ | – | PP | ABC superfamily, histidine-binding periplasmic protein | 2D |
| CysP | + | PP | ABC superfamily, thiosulphate transport protein | 2D |
| MaeB | + | CP | Paral putative transferase; phosphate acetyltransferase | 2D |
| NlpB | + | OM | Lipoprotein-34 | 2D |
| STM2494 | + | (IM) | Putative inner membrane or exported | 2D |
| NifU | – | CP | NifU homologue involved in Fe-S cluster formation | 2D |
| YfiA | – | CP | ribosome associated factor, stabilizes ribosomes against dissociation; putative sigma(54) modulation protein | 2D |
| LuxS | – | CP | Quorum sensing protein, produces autoinducer – acyl-homoserine lactone-signalling molecules | 2D |
| SipA | – | SUP | Cell invasion protein | 2D |
| SipC | – | SUP | Cell invasion protein | 2D |
| GudD | – | CP | d-Glucarate dehydratase | 2D |
| Ptr | + | PP | Protease III | 2D |
| OmpX | –/+ | OM | Ail and ompX homologue; outer membrane protein X precursor | 2D |
| YraP | + | (PP) | Paral putative periplasmic protein; possible lipoprotein | 2D |
| RbfA | – | CP | Ribosome-binding factor, role in processing of 10S rRNA | 2D |
| GreA | + | CP | Transcription elongation factor, cleaves 3′ nucleotide of paused mRNA | 2D |
| Mdh | –/+ | CP | Malate dehydrogenase | 2D |
| AccB | + | CP | acetyl-CoA carboxylase, BCCP subunit, biotin carboxyl carrier protein | 2D |
| FkpA | + | CP | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | 2D |
| DppA | + | PP | ABC superfamily, dipeptide transport protein | 1D, 2D |
| YiaD | + | (OM) | Putative outer membrane lipoprotein | 2D |
| Kbl | – | CP | 2-amino-3-ketobutyrate CoA ligase (glycine acetyltransferase) | 2D |
| PstS | + | PP | ABC superfamily, high-affinity phosphate transporter | 2D |
| RbsB | + | PP | ABC superfamily, d-ribose transport protein; d-ribose-binding periplasmic protein | 2D |
| FadA | – | CP | 3-ketoacyl-CoA thiolase (thiolase I, acetyl-CoA transferase), small (beta) subunit of the fatty acid-oxidizing multienzyme complex | 2D |
| RplL | – | CP | 50S ribosomal subunit protein L7/L12 | 2D |
| MalE | – | PP | ABC superfamily maltose transport protein, substrate recognition for transport and chemotaxis | 2D |
| AphA | + | PP | Non-specific acid phosphatase/phosphotransferase, class B | 2D |
| OsmY | – | PP | Hyperosmotically inducible periplasmic protein, RpoS-dependent stationary phase gene | 2D |
| Tsf | + | CP | Protein chain elongation factor EF-Ts | 1D |
| CyoA | + | IM | Cytochrome o ubiquinol oxidase subunit II | 1D |
| YbfM | + | (OM) | Putative OMP | 1D |
| GlnH | + | PP | ABC superfamily (bind_prot), glutamine high-affinity transporter | 1D |
| OmpF | + | OM | OMP 1a (ia; b; f), porin | 1D |
| MglB | + | PP | ABC superfamily (peri_perm), galactose transport protein | 1D |
| STM2786 | + | PP | Tricarboxylic transport | 1D |
| RpsD | – | CP | 30S ribosomal subunit protein S4 | 1D |
| RplC | – | CP | 50S ribosomal subunit protein L3 | 1D |
| GlpK | + | CP | Glycerol kinase | 1D |
| TufB | – | CP | Protein chain elongation factor EF-Tu (duplicate of tufA) | 1D |
Nomenclature according to coliBASE (http://colibase.bham.ac.uk/; Chaudhuri et al., 2004).
Up- or downregulation in hfq strain as compared with SL1344.
Predicted subcellular protein localization: CP, cytoplasmic; PP, periplasmic; OM, outer membrane; IM, inner membrane; SUP, secreted.
Functional classification according to KEGG (http://www.genome.jp/kegg/; Goto et al., 1997).
Protein identified on one-dimensional (1D) or two-dimensional (2D) gel.
See Table S1 for further details.
Loss of Hfq also affected the composition of the periplasmic protein population (Fig. 3B, right panel). While some of the changes in protein expression seen in Δhfq cells are shared with the rpoS deletion strain (e.g. OppA and GltI), loss of Hfq leads to a specific increase in DppA, a decrease in TufB levels, and higher levels of OppA, MglB, GltI and GlnH as compared with the ΔrpoS strain (Table 2).
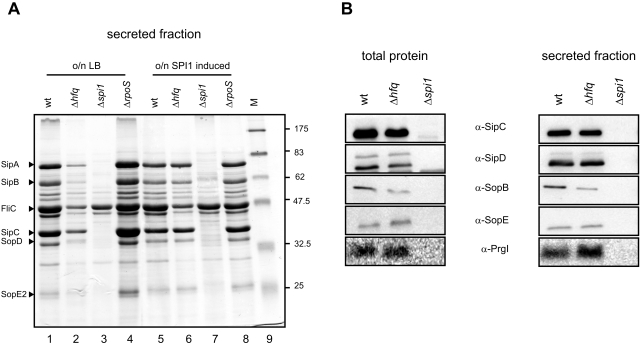
The most drastic effects of the hfq deletion, however, were observed with the secreted protein fraction (Fig. 3C). FliC, the most prominent protein found in Salmonella supernatants (Komoriya et al., 1999) and other secreted proteins typically seen in SL1344 supernatants, e.g. effector proteins that are translocated by the SPI1 TTSS (Ehrbar et al., 2002), were either strongly reduced or undetectable. The loss of secreted SPI1 effectors was consistent with the reduced invasion phenotype of the Δhfq strain. None of these reductions were observed with the ΔrpoS strain (Fig. 5A).
Fig. 5.
SPI1-inducing conditions restore effector levels and their secretion in the Δhfq strain. A. Comparison of secreted proteins of wild-type, Δhfq, Δspi1 and ΔrpoS grown for 12 h under standard conditions (lanes 1–4) or SPI1-inducing conditions (lanes 5–8) by SDS-PAGE analysis. B. Western blot detection of effector and needle proteins in total protein samples and secreted fractions of bacteria grown for 12 h under SPI1-inducing conditions. Bacterial strains from left to right: wild-type, Δhfq, Δspi1.
Overexpression of HilA in Δhfq rescues SPI1 effector protein expression but not secretion
Consequently, we sought to determine if the Hfq-dependent loss of secreted SPI1 effectors was due to a more general defect on SPI1 gene expression. The activation of SPI1 genes is mediated by a transcription factor cascade. On top of this cascade, the transcription factors, HilC and HilD, along with RtsA (encoded outside SPI1) cooperate to transmit environmental signals that lead to derepression of hilA (Bajaj et al., 1996; Lucas and Lee, 2001; Schechter and Lee, 2001; Ellermeier et al., 2005). HilA is the SPI1 major transcriptional activator responsible for most of the SPI1 TTSS and effector gene expression, both directly and indirectly through its activation of InvF (Darwin and Miller, 1999; Eichelberg and Galan, 1999; Lostroh and Lee, 2001). In addition, HilA also activates expression of secreted effector proteins encoded outside SPI1, e.g. SopB encoded within SPI5 (Ahmer et al., 1999).
To quantify the amount of HilA protein, we constructed a chromosomal FLAG epitope-tagged derivative of the hilA gene. Quantification of Western blot signals obtained for HilAFLAG revealed a > sixfold reduction of the protein in the Δhfq mutant as compared with the wild type (Fig. 4A, left panel). In addition, Northern blot quantification showed that in Δhfq cells hilA mRNA was reduced to ∼8% of wild-type levels (Fig. 4B). Several transcriptional reporter fusions were also used to determine if the changes in hilA expression resulted from a reduced hilA promoter activity (Fig. 4B). Depending on the fusion used, hilA transcription in Δhfq was found to be reduced to between 30% and 70% of wild-type levels. Collectively, this suggested that Hfq regulates HilA synthesis at both the transcriptional and the post-transcriptional level.
Fig. 4.
The hfq deletion mutant is impaired in HilA expression and shows reduced effector levels. A. HilA levels in wild-type and Δhfq Salmonella grown to early stationary phase. Shown are Western blots probed for chromosomally encoded HilAFLAG protein (left panel), or HilAmyc protein as expressed from pBAD-HilA expression plasmid (right panel). Bacteria carrying the empty pBAD vector were included as control. B. hilA promoter activity determined with a transcriptional hilA-gfp fusion in early stationary phase (PhilA), and hilA mRNA levels as determined by Northern analysis. Given are relative values obtained for Δhfq, with the levels determined for the wild-type strain set to 100%. C. Western blot detection of effector and needle proteins in total protein samples and secreted fractions of bacteria grown to early stationary phase. Bacterial strains from left to right: wild-type, Δspi1, wild-type strain carrying a pBAD control vector, wild-type strain carrying a pBAD-HilA expression plasmid, Δhfq carrying a pBAD control vector, Δhfq with pBAD-HilA expression plasmid. All strains were grown in LB medium complemented with 0.05% l-arabinose to facilitate HilA expression from plasmid pBAD-HilA.
To verify that the lower HilA levels in the Δhfq strain cause a reduction of SPI1 effector protein synthesis, we first determined the intracellular levels of SipC, SipD, SopB and SopE on Western blots, all of which were readily detected in wild-type cells (Fig. 4C, lanes 1 and 3). In stark contrast, no (SipC, SopB, SopE) or drastically reduced (SipD) signals were obtained in the Δhfq background (lane 5). To determine whether HilA overexpression could restore effector protein expression in the absence of Hfq, the wild-type and the Δhfq strains were transformed with plasmid pBAD-HilA (Lostroh et al., 2000), which carries a myc-tagged hilA gene under control of an arabinose-inducible PBAD promoter. Arabinose induction yielded comparable HilAmyc protein levels in both genetic backgrounds (Fig. 4A, right panel), and fully restored the intracellular levels of effector proteins in Δhfq cells to wild-type amounts (Fig. 4C, compare lanes 3 and 6). We next examined whether HilA overexpression could also restore effector protein secretion. Supernatants of the same cultures used for whole-protein determinations in Fig. 4C were examined for extracellular levels of the aforementioned effector proteins. In stark contrast to the full restoration of intracellular effector protein levels, HilA expression failed to significantly increase the extracellular amounts of these proteins in the Δhfq strain (lanes 11 and 12). HilA overexpression in the Δhfq background was therefore able to overcome the loss of expression of these effector proteins but not of their secretion.
One possible explanation for this secretion defect was that the hfq mutation does not permit assembly of a functional SPI1 secretion apparatus. The secreted PrgI protein, the main component of the needle of the SPI1-encoded TTSS, provides a testable marker for a functional secretion apparatus (Kimbrough and Miller, 2000; Kubori et al., 2000). We determined both the intra- and extracellular PrgI levels in all of the strains, and found that this protein was absent in the Δhfq mutant (Fig. 4C, lower panel, lanes 1 and 3 versus 5, lanes 7 and 9 versus 11). In contrast, HilA overexpression led to elevated intracellular and secreted PrgI levels in the wild-type but not hfq strains (lanes 4 and 6 versus 10 and 12). These results indicated that under aerobic growth conditions, Hfq affected SPI1 expression at multiple levels, and was required for the expression of the TTSS structural genes independent of HilA expression.
Effector protein secretion independent of hfq under SPI1-inducing conditions
As we had observed that the invasion defect of the Δhfq strain was less pronounced when grown under SPI1-inducing conditions, we considered whether this was the result of improved effector protein secretion. Indeed, supernatants of Δhfq cells cultured under SPI1-inducing conditions displayed a protein pattern close to the wild type (Fig. 5A, compare lanes 5 and 6), except for the flagellar protein, FliC. When these samples were probed on Western blots for the effectors SipC, SipD, SopB and SopE, a similar level of secretion as for the wild-type strain was evident for the hfq mutant (Fig. 5B). Furthermore, under these growth conditions, the Δhfq strain accumulated the needle protein, PrgI, to wild-type levels both intracellularly and in the supernatant, arguing that under this growth condition, Δhfq bacteria also possess a fully active SPI1 TTSS.
Impaired adhesion contributes to the non-invasive phenotype of Δhfq
Although the Δhfq strain appeared to show wild-type levels of expression in terms of SPI1 function when grown under SPI1-inducing conditions, it was puzzling that the mutant remained much less invasive. One important factor that contributes to Salmonella invasion of host cells in addition to SPI1 function is successful adhesion to epithelial cells, mediated by fimbrial adhesins. We therefore performed assays to compare the adhesion phenotypes of the wild-type and Δhfq strains. To better visualize bacteria, both strains were transformed with a low-copy plasmid that constitutively expresses green fluorescent protein (GFP). Transformants were grown under SPI1-inducing conditions, and used for infection of HeLa cells at a moi of 50. Following incubation at 37°C for 1 h, bacteria that had not attached to the HeLa cells were removed by extensive washing of the cells. The remaining bacteria and cells were fixed, and the number of bacteria per HeLa cell determined by fluorescence microscopy (Fig. S5A). For the wild-type strain, an average of ∼30 bacteria per HeLa cell were found to be adherent. In contrast, the average number observed with the Δhfq strain was significantly lower, i.e. ∼10 bacteria per HeLa cell. For both strains, we observed that a significant proportion of bacteria became internalized during the 1 h incubation step prior to counting. As the assay does not allow us to clearly distinguish extra- from intracellular bacteria, our calculation includes all bacteria associated with HeLa cells, based on the assumption that every internalization event was preceded by successful adhesion.
To better separate adhesion from invasion rates, bacterial adherence was also determined in HeLa cell infection assays without gentamicin treatment. To this end, serial dilutions of HeLa cells and adhered bacteria were plated on LB agar 30 min upon infection, and cfu determined (Fig. S5B). These experiments revealed a > twofold reduction in adhesion of the hfq deletion mutant as compared with wild-type Salmonella (25% adherence of wild-type compared with 11% of the hfq strain related to the input). In contrast, adherence of the two control strains, hfqHIS and hfq-C, did not significantly differ from the wild type (21% and 24% respectively). Collectively, the data suggest that a lower adhesion rate may contribute to the non-invasive phenotype of the hfq strain.
Δhfq is impaired in motility
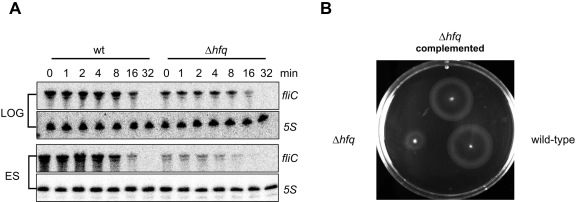
The strong Hfq dependence for expression of the phase 2 flagellin protein, FliC, suggested that Hfq would be required for Salmonella motility. To verify reduced FliC expression, we first analysed fliC mRNA levels in wild type and Δhfq Salmonella at different growth phases (Table 3 and Fig. 6A). Interestingly, loss of Hfq caused a mere 1.6-fold reduction of fliC mRNA levels in exponential phase, however, a sixfold reduction at early stationary phase (Table 3). We also compared fliC mRNA stability in wild-type strain and Δhfq cells, and found it largely unaffected by the hfq mutation at either growth phase (Fig. 6A). In contrast to fliC mRNA, we failed to detect fljB mRNA on any of these Northern blots (data not shown). Taken together, the reduced FliC expression of Δhfq is unlikely to result from phase variation of the invertible flagellar switch (fljB/fljA promoter), but rather from reduced fliC transcription.
Table 3.
Quantification of Hfq-dependent gene expression.
| Relative mRNA levelsa | Relative transcriptional/translational fusion activityb | ||
|---|---|---|---|
| Gene/OD600 | 0.3 | 2 | 2 |
| fliC | −1.6 | −6 | ND |
| ompC | 1.7 | 1.6 | 0.84/1.1 |
| ompD | 1.7 | 1.4 | 0.82/2.5 |
| PLtetO-gfp | ND | ND | 1.0 |
Fold change of mRNA levels in hfq strain as compared with SL1344 as determined by Northern hybridization.
Fold change of GFP reporter fusion activity in hfq strain as compared with SL1344.
Fig. 6.
The Δhfq strain is non-motile. A. Northern blot detection of fliC mRNA levels in wild-type and Δhfq cells at logarithmic and early stationary phase before and within 32 min after rifampicin treatment. Densitometry of the Northern blot signals showed that the fliC mRNA decays with the same half-life in both genetic backgrounds (∼9 min or ∼7 min in logarithmic or early stationary phase cultures respectively). 5S signals are shown as loading control. B. To measure motility, equal numbers of bacteria from each strain were inoculated onto a motility agar-plate. The image was obtained following 4 h of incubation at 37°C.
Next, we compared the motility of the wild-type and the Δhfq strains, harbouring either a control or complementation plasmid pStHfq-6H, on motility agar plates. Wild-type cells were motile and formed concentric motility rings around the point of inoculation (Fig. 6B). In contrast, the Δhfq mutant displayed impaired motility, as judged by the much smaller motility ring formed. The strongly reduced motility of Δhfq could also be seen by light microscopy of samples from liquid culture (data not shown). Complementation with plasmid pStHfq-6H fully restored motility. Two control strains, hfq-C and hfqHIS, were found to be as motile as the wild type (data not shown), further supporting that loss of motility was a direct consequence of the lack of Hfq.
Growth rate-dependent repression of OmpD
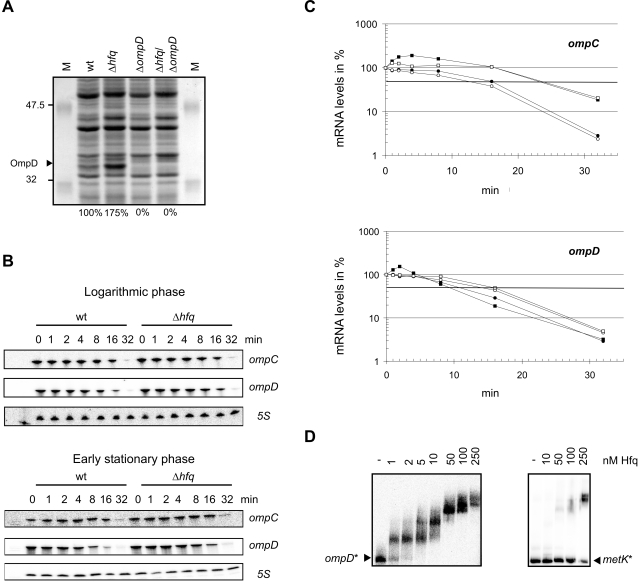
In addition to the positive regulation of secreted effector protein expression, the protein patterns obtained from different growth phases showed that Hfq was also involved in the repression of OmpD synthesis as cells progress into stationary phase (Fig. 3A). To confirm a negative regulatory role for Hfq in OmpD regulation, protein samples of wild-type, Δhfq, ΔompD and Δhfq/ΔompD strains grown to early stationary phase were compared (Fig. 7A). MALDI-TOF analysis of the 40 kDa protein band which showed higher levels of accumulation in the Δhfq strain unequivocally identified it as OmpD, consistent with the complete loss of this protein band in ΔompD and Δhfq/ΔompD cells. Using fluorescent dye staining, we also quantified the relative OmpD accumulation, and found approximately twofold elevated levels of this protein in whole cell lysates (Fig. 7A).
Fig. 7.
Hfq is essential for growth rate-dependent repression of OmpD. A. SDS-PAGE analysis of total protein prepared from wild-type, Δhfq, ΔompD and Δhfq/ΔompD bacteria grown to early stationary phase. OmpD protein levels as quantified by fluorescent staining (not shown) are given below each lane. B. Northern blot detection of ompC and ompD mRNA levels of wild-type and Δhfq bacteria grown to either logarithmic or early stationary phase prior to (0 min) and within 32 min of rifampicin treatment. 5S sRNA probing (loading control) is shown below each panel. C. Decay of ompC and ompD mRNA upon rifampicin treatment as derived from quantification of the Northern blot signals shown in (B). Logarithmic phase, wild-type (filled circles) or Δhfq (open circles); early stationary phase, wild-type (filled squares) or Δhfq (open squares). D. Hfq binds to ompD 5′ UTR RNA in vitro (gel mobility shift assay). Left panel: 1 nM of 32P-labelled ompD was incubated with increasing concentrations of Hfq protein (given above the lanes). Following a 15 min incubation at 37°C samples were run on a native 6% gel. Shown is an autoradiograph of the gel. A control gel shift assay with an Hfq-independent RNA derived from the metK 5′ UTR is shown in the right panel.
To learn more about the underlying mechanism of Hfq-dependent ompD regulation, we first determined the relative changes in ompC and ompD mRNA abundance at three different points during the growth phase (Table 3). We found that the Δhfq strain exhibited elevated ompC/D mRNA levels throughout growth. We also followed the decay of both mRNAs after rifampicin treatment (transcription block, Fig. 7B). Figure 7C shows that absence of Hfq slowed ompD mRNA decay twofold (half-lives: ∼9 min versus ∼16 min in wild-type and Δhfq strains), whereas ompC decay was not affected.
Next, we constructed transcriptional and translational reporter (GFP) plasmids for both mRNAs. Quantification of GFP reporter activity showed a slightly decreased ompD promoter activity (0.82-fold) at early stationary phase, whereas ompD translation was upregulated > 2.5-fold (Table 3). As the enhanced activity of the translational ompD fusion was consistent with elevated OmpD protein levels (Fig. 7A), we reasoned that Hfq may bind to the 5′ region of the ompD mRNA to interfere with its translation. To test this hypothesis, we synthesized a 5′ fragment of the ompD mRNA, encompassing its 5′ UTR and 118 nucleotides of the coding region, and performed in vitro mobility shift assays with purified Hfq protein. Figure 7D shows that Hfq binds this fragment with high affinity. Up to four different Hfq/ompD complexes are observed with increasing Hfq concentration, indicating that there are several Hfq binding sites in the ompD 5′ UTR. In contrast, no significant shift was observed with an Hfq-independent RNA (5′ UTR of metK) within a 250 nM range of Hfq (Fig. 7D). Taken together, these data suggests a direct role for Hfq in translational repression of the ompD mRNA.
Discussion
The RNA chaperone, Hfq, has recently been recognized as a major post-transcriptional regulator of bacterial gene expression which participates in numerous regulatory pathways (Valentin-Hansen et al., 2004). First identified as a host factor for replication of RNA phage Qβ in E. coli, Hfq has been shown to have a broad impact on physiology in several bacteria. The role of Hfq beyond phage replicative functions was first shown with an E. coli hfq::Ω mutation, which resulted in pleiotropic phenotypes related mainly to reduced survival of stress conditions (Tsui et al., 1994). Later, Hfq was found to be required in E. coli and Salmonella for efficient translation of rpoS mRNA, encoding the general stress sigma factor, σS (Brown and Elliott, 1996; Muffler et al., 1996). As RpoS is required for Salmonella proliferation in mice (Fang et al., 1992; Nickerson and Curtiss, 1997; Humphreys et al., 1999), it has been assumed that Hfq plays an important role in Salmonella virulence (e.g. Ding et al., 2004). However, the mechanisms by which Hfq affects the pathogenicity of Salmonella remained undefined. Previous work in E. coli established that Hfq also has regulatory functions independent of its effects on σS expression (Muffler et al., 1997). Likewise, B. abortus does not possess an RpoS-like σ factor (Roop et al., 2003), yet an hfq mutant of B. abortus has a pronounced virulence defect (Robertson and Roop, 1999). Similarly, the virulence defect of a V. cholerae hfq mutant was not accompanied by reduced σS levels (Ding et al., 2004).
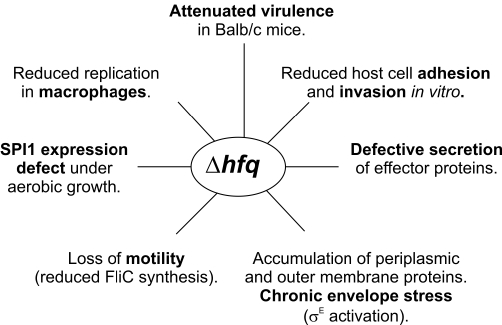
Peroral infection of the Salmonella hfq mutant revealed about the same degree of attenuation (Fig. 2A) as reported for a SalmonellaΔrpoS mutant, i.e. approximately a three-log difference in cfu recovered from the spleen 3 days post infection using a 10-fold higher infective dose (Nickerson and Curtiss, 1997). Generally, hfq mutants of several Salmonella strains exhibit four- to sevenfold reduced RpoS levels (Fig. S6; Brown and Elliott, 1996; Bang et al., 2005). This is about the degree of RpoS reduction observed in the mouse-avirulent strain, LT2, which has an altered rpoS start codon. At first glance, these observations appear to support a model in which reduced σS production would fully account for the attenuation of Δhfq. However, using a set of newly constructed SL1344 hfq mutant and control strains, we defined hfq phenotypes that relate to virulence and global gene expression (see Fig. 8 for a summary), and which are largely independent of σS.
Fig. 8.
Summary of phenotypes of the Salmonella hfq mutation determined in this study.
The most prominent virulence-associated phenotype we observed is the drastically reduced invasiveness of the Δhfq mutant (Table 1). The ability of Salmonellae to invade cultured non-phagocytic cells is dependent on the expression of SPI1-encoded genes (Lee et al., 1992), and is strongly dependent on growth rate and media. Two growth conditions showing maximal invasiveness have been defined: growth in LB with aeration to early stationary phase, and growth in low-oxygen, high-salt media (SPI1-inducing). We found that although the hfq mutant is defective for invasion under both conditions, the underlying mechanisms are different. When grown to early stationary phase, the Δhfq strain fails to activate the SPI1 transcription factor cascade, characterized by reduced HilA levels and the lack of SPI1 effector protein expression. Our observation that HilA overexpression resulted in the re-appearance of secreted protein expression indicated that the major target of Hfq regulation is HilA activation. This conclusion is also supported by the appearance of normal intracellular levels of SopB and SopE (Fig. 4C), both of which are encoded outside of SPI1 and whose expression requires the concerted function of InvF and SicA. The latter, SPI1-encoded genes are also highly dependent on HilA for expression (Darwin and Miller, 2000, and references therein).
The regulation of hilA promoter activity is complex, involving the coactivators HilC, HilD and RtsA, as well as other factors which act upstream of these proteins (Lostroh and Lee, 2001; Ellermeier et al., 2005; and references therein). A global transcriptome microarray analysis indicated that Δhfq cells have several-fold reduced levels of hilC/D and rtsA mRNAs (A. Sittka et al., unpubl. results), suggesting that Hfq affects signal transmission further upstream in the SPI1-activating cascade. Strikingly, complementation with the HilA plasmid restored intracellular levels of several effector proteins encoded within SPI1, yet not their secretion. The latter observation may result from a failure to assemble a functional SPI1 TTSS, because only traces of the needle protein, PrgI, were detected in supernatants of HilA-complemented Δhfq cells. The prgI gene is encoded within the SPI1 prgHIJKorgABC operon (Klein et al., 2000), and is directly controlled by HilA. These observations suggest that the role of Hfq as a novel factor of SPI1 gene activation may not be confined to promoting HilA expression. It remains possible that Hfq either is also involved in the mRNA stability of the prgHIJKorgABC operon transcript, or affects the translation of the encoded gene products. Further work is required to clarify the effects of Hfq on this subset of HilA-dependent genes.
In contrast to aerobic growth, under SPI1-inducing conditions the Δhfq mutant shows normal SPI1 gene expression, TTSS assembly (as judged by PrgI levels in the supernatant) and effector protein secretion (Fig. 5B). Under these growth conditions, the Δhfq mutant should have been capable of invasion of non-phagocytic cells, yet invasion was strongly reduced compared with the wild-type strain (Table 1). Our results from adhesion and motility assays as well as proteome analysis indicate several other factors may contribute to this impairment. The hfq mutant shows a significantly reduced ability to adhere to HeLa cells (Fig. S5), which is likely to affect the rate of invasion. The hfq mutant is non-motile (Fig. 6), due most likely to the loss of the flagellar subunit protein, FliC (Figs 3A–C and 6A). However, while flagella-mediated bacterial motility accelerates the invasion of Salmonella, motility per se is not required for invasion (van Asten et al., 2004). Finally, a preliminary proteome analysis (Table 2) showed differential regulation of numerous lipoproteins and OMPs, suggesting that Hfq is also involved in regulation of genes related to the bacterial envelope composition. Importantly, Δhfq cells exhibit strongly elevated levels of HtrA, also known as DegP. HtrA/DegP has recently been shown in Salmonella and E. coli to be part of the σE regulon that mediates the response to envelope stress (Rhodius et al., 2006; Skovierova et al., 2006), and activation of the σE pathway (by RpoE overexpression) results in a strong induction of htrA mRNA (Rhodius et al., 2006). Three additional proteins that promote OMP assembly, FkpA, YraP and YaeT, and whose genes are members of the σE core regulon (Rhodius et al., 2006; Skovierova et al., 2006), also showed elevated levels in the hfq mutant. In addition, two strictly σE-dependent small RNAs, MicA and RybB, showed promoter activation in the hfq mutant under the same conditions used in this study (Papenfort et al., 2006, and unpublished results). Interestingly, strong induction of the σE response was also observed in a V. cholerae hfq mutant (Ding et al., 2004). Based on the activation of multiple σE-dependent genes, the Δhfq strain appears to experience chronic envelope stress which would ultimately change outer membrane properties. In summary, we suggest that the multiple phenotypes of the hfq mutant on motility and adherence, and an apparent chronic cell envelope stress in Salmonella all contribute to the observed reduced invasiveness of the hfq mutant.
A comparison of the hfq phenotypes that relate to virulence of Salmonella and other previously studied pathogenic bacteria reveals interesting similarities yet also major differences. Hfq mutants of the rather closely related species, V. cholerae and P. aeruginosa, are severely attenuated for virulence in mice (Sonnleitner et al., 2003; Ding et al., 2004). In contrast, hfq mutants of L. monocytogenes and L. pneumophila show only mild virulence defects in Balb/c mice and an amoeba infection model respectively (Christiansen et al., 2004; McNealy et al., 2005). A mouse virulence defect was also described for the B. abortus hfq mutant, although Hfq did not appear to affect spleen colonization per se, but rather the survival and/or persistence in this organ (Robertson and Roop, 1999). Survival in macrophages was investigated for L. pneumophila, L. monocytogenes and B. abortus, and the effects of the respective hfq mutations were comparable to those described here for Salmonella, although the B. abortus hfq was affected in long-term macrophage survival (Robertson and Roop, 1999; Christiansen et al., 2004; McNealy et al., 2005). Thus far, L. monocytogenes is the only other species for which an hfq mutant has been studied with respect to non-phagocytic cell invasion, and unlike Salmonella, the L. monocytogenes hfq mutant was found to be fully invasive (Christiansen et al., 2004). Also in contrast to the Salmonella hfq mutant, the assembly of functional pili and secretion of cholera toxin was not affected in the hfq mutant of V. cholerae (Ding et al., 2004). In light of the variability and diversity of Hfq function(s) in virulence among these pathogens, the clear loss of SPI1 expression and the secretion phenotype shown here for Salmonella provide an excellent basis to dissect the mechanisms of Hfq functions in a well-characterized model pathogen.
Analyses of protein patterns on one- and two-dimensional gels showed that the expression of a large number of Salmonella genes is affected by Hfq. Classification of these genes according to the genome annotation of Salmonella LT2 (McClelland et al., 2001) shows that the encoded proteins belong to diverse functional categories (Table 2). The increase of GlpK and GlpQ in the hfq mutant is currently unexplained, but might indicate changes in glycerophospholipid metabolism (note that the glpK and glpQ genes are not linked). Other pronounced changes include OMPs such as OmpD, the flagellin FliC, and numerous periplasmic proteins. Given that Hfq has recently been in the spotlight as a small RNA-binding protein (Valentin-Hansen et al., 2004), the altered periplasm of Δhfq cells is of particular interest. Specifically, the ∼200 nt GcvB RNA of E. coli as well as its Yersinia pestis homologue was shown to negatively regulate the periplasmic proteins, OppA, DppA and GltI (Urbanowski et al., 2000; McArthur et al., 2006), which all accumulate to higher levels in the Δhfq strain (Fig. 3B). The molecular mechanism of GcvB action in these two species remains unknown, but OppA was found to strongly accumulate in an E. coliΔhfq mutant (Ziolkowska et al., 2006). Moreover, GcvB co-immunoprecipitates with E. coli Hfq (Zhang et al., 2003), suggesting that this protein mediates GcvB binding to trans-encoded target mRNAs. As the gcvB gene is conserved and expressed in Salmonella (Urbanowski et al., 2000; C.M. Sharma and J. Vogel, unpublished), it is tempting to speculate that the high levels of OppA, DppA and GltI observed here results from a loss of GcvB-mediated mRNA repression in the absence of Hfq.
Of the 71 proteins with altered levels in the hfq mutant (Table 2), five have no known homologues in E. coli (SipA, SipC, STM1254, STM1328 and STM2494). Of the remaining 66, seven overlap with previously published Hfq-associated E. coli mRNAs, i.e. CspD, Dps, LppA, LppB, OmpX, RplL and YfiA (Zhang et al., 2003). Notably, the majority of these are proteins whose expression was reduced, suggesting Hfq might function to stabilize their mRNAs, either directly or indirectly by promoting efficient translation.
One of the most drastic changes we observed in the absence of Hfq is the increase in OmpD levels (Fig. 7A). OmpD is a Salmonella-specific porin, and is the most abundant protein in the outer membrane under standard growth conditions. Together with the other major porins, OmpC and OmpF, it accounts for ∼1–2 × 105 porins per cell (Santiviago et al., 2003). Expression of this porin is regulated primarily at the level of transcription, is subject to catabolite repression, and the ompD promoter is repressed by low pH. However, post-transcriptional activation of OmpD expression under anaerobiosis has also been reported, and shown to depend on the global transcription regulator, FNR (Santiviago et al., 2003), whereas bile appears to repress ompD post-transcriptionally (Prouty et al., 2004). Despite its abundance, the physiological roles of OmpD remain unclear. Unlike the other two major porins, OmpC and OmpF, OmpD is not regulated by osmolarity (Santiviago et al., 2003). The only physiological role of OmpD elucidated thus far is its requirement for the efficient efflux of the toxic compound, methyl viologen (Santiviago et al., 2002). In contrast, possible contributions of OmpD to Salmonella pathogenicity remain a matter of debate. Two LD50 studies of Salmonella wild-type and ompD mutant strains in mice yielded inconsistent results (Dorman et al., 1989; Meyer et al., 1998). Other studies postulated a requirement of OmpD for adherence to human macrophages and intestinal epithelial cell lines (Negm and Pistole, 1998; Hara-Kaonga and Pistole, 2004). Intriguingly, the presence of ompD correlates with the ability of Salmonella serovars to grow in alternative, non-human hosts. Santiviago et al. (2003) identified ompD in all Salmonella serovars that have multiple mammalian hosts, e.g. S. typhimurium and Salmonella enteritidis, but its absence in Salmonella typhi, which is restricted to humans.
In any case, the conservation of ompD argues for an important function, and the data obtained here implicate Hfq as a novel factor of ompD mRNA regulation at the post-transcriptional level. Hfq binds with high affinity and presumably at multiple sites to the ompD 5′ UTR in vitro, and its absence stabilizes the ompD mRNA in vivo. Interestingly, both these observations bear striking similarity to the previously reported Hfq-dependent control of OmpA, the major OMP of E. coli, i.e. increased ompA mRNA stability in E. coli hfq mutants, and Hfq binding of this messenger (Vytvytska et al., 1998; Udekwu et al., 2005). Importantly, it has recently become clear that one role of Hfq in this regulation may be the promotion of MicA function, an Hfq-dependent sRNA that represses ompA mRNA translation in stationary phase (Rasmussen et al., 2005; Udekwu et al., 2005). There is ample evidence of fine tuning of E. coli OMP expression by Hfq-dependent sRNAs. In addition to MicA, six E. coli sRNAs, namely MicC, MicF, OmrA/B, RseX and RybB, were shown to mediate repression of single or multiple OMP-encoding mRNAs (reviewed in Guillier et al., 2006; Vogel and Papenfort, 2006). Similarly, unpublished results from our laboratory show that ompD mRNA is acted upon by the Salmonella homologues of the E. coli sRNAs, MicC and RybB. In addition, the SPI1-endoded 80 nt InvR RNA negatively regulates ompD expression. As all these sRNAs are Hfq-dependent, we hypothesize that the post-transcriptional effect of Hfq on ompD expression reported here is mediated by Hfq-dependent regulatory sRNAs.
In summary, this study implicates Hfq as a major post-transcriptional regulator of Salmonella gene expression. Unlike other abundant global regulatory proteins, e.g. Fis, IHF, H-NS and HU (Harrison et al., 1994; Wilson et al., 2001; Schechter et al., 2003; Mangan et al., 2006), Hfq is primarily known to act at the RNA level. Interestingly, similar to H-NS that recognizes AT-rich sequences in DNA, Hfq binds to AU-rich RNA species. It has recently been proposed that H-NS repression serves to silence newly acquired genomic loci with different GC-content, thus avoiding detrimental consequences from unregulated expression of these genes following their uptake by Salmonella (Lucchini et al., 2006; Navarre et al., 2006). Experiments are currently underway to determine if Hfq plays a similar role by specifically acting on AU-rich mRNAs of newly acquired genes. If so, Hfq may again turn out to be the ‘host factor’ as which it was originally described 40 years ago (Franze de Fernandez et al., 1968).
Experimental procedures
Oligonucleotides
The complete list of DNA oligonucleotides used for cloning and as probes in hybridization is provided as supplementary material (Table S2).
Bacterial strains, media and growth conditions
Growth in LB broth or on LB plates at 37°C was used throughout this study unless stated otherwise. SOC medium was used to recover transformants after heat shock or electroporation and prior to plating. Green plates for screening against lysogens in P22 transductions were prepared as described (Sternberg and Maurer, 1991). For SPI1 induction, cultures were inoculated in 5 ml LB containing 0.3 M NaCl in 15 ml Falcon tubes with a tightly closed lid. Cultures were incubated for 12 h at 37°C with shaking. To determine growth rates of strains, the inoculated culture was split in 12 aliquots and each aliquot was opened only once to measure OD600. Antibiotics (where appropriate) were applied at the following concentrations: 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 20 μg ml−1 chloramphenicol. For HilA expression from plasmid pCH112, cultures were grown to an OD600 of 1 and induced with l-arabinose in a final concentration of 0.05% until cells reached an OD600 of 2.
The bacterial strains used in this study are listed in Table 4. Chromosomal mutagenesis of Salmonella SL1344 followed the protocol described by Datsenko and Wanner (2000) with few modifications. Strain JVS-00008, which carries plasmid pKD46, was grown in LB at 28°C complemented with ampicillin and 0.2% l-arabinose to an OD600 of 0.5. Cells were collected by centrifugation (2 min, 11 000 g), washed three times with ice-cold H2O, and dissolved in 1/100 of the original culture volume. PCR products of marker genes (50 μl standard reactions) were DpnI-treated for 30 min at 37°C, and purified on Macherey-Nagel spin columns (NucleoSpin Extract II). One-fifth of the 25 μl column eluate (in water) was used for transformation. Forty microlitres of competent cells was mixed with the purified PCR product in a chilled cuvette (0.1 cm electrode gap) and electroporated (18 kV cm−1). Subsequently, 1 ml of pre-warmed SOC medium was added, and cells were recovered by incubation for 1 h at 37°C before selection on LB agar plates with the appropriate antibiotics. All mutations were moved to a fresh SL1344 background by phage P22 transduction.
Table 4.
Strains and plasmids used in this study.
| Strain | Relevant markers/genotype | Reference/source |
|---|---|---|
| S. typhimurium | ||
| SL1344 | StrRhisG rpsL xyl | Hoiseth and Stocker (1981), provided by D. Bumann, MPI-IB Berlin |
| JVS-00255 | SL1344 Δhfq::CmR | This study |
| JVS-00177 | SL1344hfq-6HIS-CmR | This study |
| JVS-00179 | SL1344hfq-CmR | This study |
| JVS-00756 | SL1344hilA-3xFLAG-KmR | This study |
| JVS-00405 | SL1344 Δspi1 (KmR cassette removed) | S. Pätzold, MPI-IB Berlin (unpublished) |
| JVS-00748 | SL1344 ΔrpoS::KmR | Kowarz et al. (1994) |
| JVS-00584 | SL1344 Δhfq (CmR cassette removed) | This study |
| JVS-00735 | SL1344 ΔompD::KmR | This study |
| JVS-00822 | SL1344 Δhfq::CmR/ΔompD::KmR | This study |
| E. coli | ||
| TOP10 | mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697galU galK rpsL endA1 nupG | Invitrogen |
| TOP10F′ | F′{lacIq Tn10 (TetR)} mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697galU galK rpsL endA1 nupG | Invitrogen |
| ER 2566 | F–λ–fhuA2 [lon]ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr) 114::IS10 R(mcr-73:: miniTn10)2 R(zgb-210::Tn10) (TetS ) endA1 [dcm] | New England Biolabs |
To construct the hfq deletion strain, the cat chloramphenicol-resistance gene was amplified from plasmid pKD3 with oligonucleotides JVO-0252 and JVO-0318. Strains hfq-C and hfqHIS were constructed in the same way, using primer pairs JVO-0252/JVO-0253 and JVO-0252/JVO-0319 respectively. Mutants were verified by colony PCR using primers JVO-0076/JVO-0077. For removal of the cat gene the Δhfq strain was transformed with the FLP helper plasmid pCP20 (for detailed procedure, see Datsenko and Wanner, 2000). The ompD deletion strain was constructed by replacing the gene with a kanamycin marker gene amplified from pKD4 with primers JVO-0817/JVO-0818. The deletion mutant was verified using oligonucleotides JVO-0818/0819. Chromosomal FLAG-tagging (3xFLAG) of hilA was carried out as described in Uzzau et al. (2001), using primers JVO-0837/0838 on template pSUB11. The chromosomal tagging was verified by PCR with oligonucleotides JVO-839/840, and sequencing of the PCR product.
Plasmids
Plasmids used, and details of their construction are described in Table 5. Maps of selected plasmids are provided in the supplementary material (Fig. S7). E. coli TOP10 and TOP10F′ strains were used for cloning. All plasmids were purified using the Machery-Nagel Plasmid QuickPure Kit. To transform Salmonella strains, these were rendered competent using the same protocol as described above, except that cells were cultured at 37°C without arabinose.
Table 5.
Plasmids used in this study.
| Name | Fragment | Comment | Origin/marker | Reference |
|---|---|---|---|---|
| pJV300 | ColE1 control plasmid, based on pZE12-luc, PLlacO promoter transcribes a ∼50 nt nonsense transcript (rrnB terminator) | ColE1/AmpR | This study | |
| pJV859-8 | PLtetO-gfp | GFP control plasmid (constitutive GFP expression) | pSC101*/CmR | Urban and Vogel (2006) |
| pJV968-1 | ‘lacZ’ | ColE control plasmid, carries 1.5 kb internal lacZ fragment | ColE1/AmpR | Vogel et al. (2004) |
| pVP003 | luc | Control plasmid; low-copy version of pZE12-luc | pSC101*/AmpR | This study |
| pVP004-1 | Hfq-6HIS | pStHfq-6H, expresses a HIS-tagged Hfq under control of its own promoter; includes 1014 bp upstream of hfq reading frame | pSC101*/AmpR | This study |
| pVP009 | Low-copy version of control plasmid pJV300 | pSC101*/AmpR | This study | |
| pVP012 | ‘lacZ’ | Low-copy version of control plasmid pJV968-1 | pSC101*/AmpR | This study |
| pVP019 | ompD::gfp | ompD translational GFP fusion plasmid | This study | |
| pVP020 | ompC::gfp | ompC translational GFP fusion plasmid | This study | |
| pAS009 | hfq | Overexpression plasmid of Salmonella hfq (cloned in N-terminal fusion vector pTYB 11) | M13/AmpR | This study |
| pAS0046 | gfp | Transcriptional fusions plasmid, based on pJV859-8 | pSC101*/CmR | This study |
| pAS0047-2 | PhilA -gfp | hilA transcriptional GFP fusion plasmid | pSC101*/CmR | This study |
| pAS0057-1 | PompC -gfp | ompC transcriptional GFP fusion plasmid | pSC101*/CmR | This study |
| pAS0058-1 | PompD -gfp | ompD transcriptional GFP fusion plasmid | pSC101*/CmR | This study |
| pJU004 | GFP control plasmid | pSC101*/CmR | Urban and Vogel (2006) | |
| pBAD/Myc-His A | pBAD control plasmid | pBR322/AmpR | Invitrogen | |
| pZS*24-MCS1 | luc | General expression vector | pSC101*/KmR | Lutz and Bujard (1997) |
| pBAD 18-Kn | pBAD control plasmid | pBR322/KmR | Guzman et al. (1995) | |
| pCH112 | PBAD-hilA-Myc-His | pHilA; hilA ORF in pBAD/Myc-His | pBR322/AmpR | Lostroh et al. (2000) |
| pKD3 | Template for mutant construction; carries chloramphenicol cassette | oriRγ/AmpR | Datsenko and Wanner (2000) | |
| pKD4 | Template for mutant construction; carries kanamycin cassette | oriRγ/AmpR | Datsenko and Wanner (2000) | |
| pKD46 | ParaB-γ-β-exo | Temperature sensitive red recombinase expression plasmid | oriR101/AmpR | Datsenko and Wanner (2000) |
| pCP20 | Temperature sensitive FLP recombinase expression plasmid | oriR101/AmpR, CmR | Datsenko and Wanner (2000) | |
| pSUB11 | Template for mutant construction; 3xFLAG linked to a KmR cassette | R6KoriV, AmpR | Uzzau et al. (2001) | |
| pZA31-luc | luc | General expression plasmid | p15A/CmR | Lutz and Bujard (1997) |
| pZE12-luc | luc | General expression plasmid | ColE1/AmpR | Lutz and Bujard (1997) |
| pTYB-11 | Protein overexpression plasmid (IMPACT-CN system) | M13/AmpR | NEB |
Control plasmids based on pZE12-luc were constructed as follows: to lower the copy number of plasmid pZE12-luc, the ColE1 origin was swapped to pSC101* by inserting the AvrII-SacI fragment of plasmid pZS*24-MCS1, resulting in pVP003. To obtain plasmid designated pVP012, a low-copy version of control plasmid pJV968-1, the 1.5 kb ‘lacZ’ XbaI/XhoI fragment of the latter was introduced into pVP003 by the same enzymes. Note that these plasmids lack the PLlacO promoter region of pZE12-luc, hence the insert is not transcribed.
To express Hfq-6HIS under control of its own promoter, low-copy vector pVP003 was digested with XhoI/XbaI and ligated to a PCR product obtained with the primer pair JVO-0370/0182 (JVO-0370 binds 1014 bp upstream of the hfq open reading frame (ORF) in miaA while JVO-182 adds a 6HIS-tag sequence followed by a stop codon to the last codon of hfq). For clarity, the obtained plasmid, pVP004-1, is designated in figures as pStHfq-6H.
Control plasmid pJV300 was obtained by ligation of a pZE12-luc derived PCR product. The −1 site of promoter PLlacO is fused to the second position of the XbaI site (which is destroyed upon cloning). Transcription from the PLlacO promoter now yields a ∼50 nt nonsense transcript derived from the rrnB terminator on pJV300. To obtain a low-copy version of this plasmid, the origin was changed to pSC101* as described above, yielding pVP009.
To clone transcriptional GFP fusions, a PCR fragment was amplified from plasmid pJV859-8 (GFP expression plasmid) using oligonucleotides JVO-0888/pZE-XbaI. JVO-0888 introduces stop codons after a XhoI and NheI site in all three ORFs, a ribosome binding site, a 7 bp spacer, and the sequence of the first six amino acids (aa) of the GFP coding region with a silent mutation at position 6 (T(r)C) to destroy the GFP internal NheI site. Plasmid pJV859-8 was cut XhoI (removing the promoter region, the ribosome binding site and the sequence for the first 142 aa of GFP), gel-purified, and the vector backbone ligated to the PCR fragment digested with the same enzyme. Due to the internal XhoI site in the GFP coding region (cuts in the sequence after aa 142) this leads to a promoterless transcriptional fusion plasmid (used as a negative control plasmid in transcriptional fusion experiments). The resulting plasmid was designated pAS0046. For construction of the ompC-gfp transcriptional fusion plasmid pAS0057-1 and the ompD-gfp transcriptional fusion plasmid pAS0058-1, pAS0046 was digested with AatII/NheI and ligated to PCR products amplified with primer pairs JVO-0801/0805 and JVO-0806/0807 respectively, cut with the same enzymes.
For translational ompD::gfp and ompC::gfp fusions, PCR fragments of oligonucleotides JVO-0726/0802 and JVO-0717/0801 respectively, were inserted into plasmid pJV859-8 by AatII/NheI cloning, yielding plasmids pVP019 (GFP fusion to 15th aa of OmpD) and pVP020 (GFP fusion to 12th aa of OmpC) respectively.
To overexpress and purify Salmonella Hfq protein, the hfq coding region was amplified with primer pair JVO-0078/0084. The PCR product was SapI digested and ligated to the N-terminal fusion vector pTYB11 cut with enzymes SapI/SmaI, yielding plasmid pAS009.
P22 transduction
P22 lysates were prepared from soft agar plate lysates of donor strains using P22 phage HT/105-1 by standard procedures. Transductions were performed as described by Sternberg and Maurer (1991) using P22 phage HT/105-1 and further purified on Green plates. For unknown reasons, we were not able to prepare lysates of the hfq deletion mutant, hence Δhfq/P22 lysates were prepared from this strain upon complementation with plasmid pVP004. Transformants were verified by PCR.
Gentamicin protection (invasion) assays
The invasion assay was performed as described in Isberg and Falkow (1985). HeLa cells (ATCC CCL2) were seeded in RPMI medium (Gibco), supplemented with 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, and containing 10 μg ml−1 penicillin and streptomycin in 12 well plates with a density of 1 × 105 per well the day before or 0.5 × 105 per well 2 days before infection respectively. At the day of infection HeLa cells reached a density of 1–2 × 105. When seeded 2 days before infection medium was changed the day before the assay was performed. One hour prior to infection medium was changed to RPMI containing no antibiotics.
Bacterial cultures were inoculated 1/100 from overnight cultures into fresh medium. For experiments with cultures in early stationary phase cultures were grown in LB (with 50 μg ml−1 ampicillin if indicated) at 37°C, 220 rpm, with normal aeration. For experiments with SPI1-induced bacteria, cultures were grown for 12 h in 15 ml Falcon tubes containing 5 ml LB/0.3 M NaCl (with 50 μg ml−1 ampicillin if indicated) at 37°C, 220 rpm, under limited oxygen conditions.
HeLa cells were infected with a moi of 10 with 100 μl of bacterial suspension in RPMI medium. The suspension was plated in serial dilutions on LB plates and incubated o/n at 37°C for determination of the input.
Bacterial cells were centrifuged (37°C, 250 g, 10 min) onto the HeLa cell monolayer, followed by a 50 min incubation step at 37°C in an atmosphere containing 5% CO2. One hour after infection medium was changed to RPMI (containing 50 μg ml−1 gentamicin) to kill non-invasive bacterial cells. Incubation was carried on for additional 60 min. After 2 h of infection medium was changed for the 6 h time point to RPMI containing 10 μg ml−1 gentamicin and incubation carried on for additional 4 h. For the 2 h time point cells were washed two times in PBS buffer and collected by scraping HeLa cells from the bottom of each well in PBS/0.1% Triton X-100. Dilutions in PBS were plated on LB plates and incubation carried out o/n at 37°C. Six hours after incubation samples for the second time point are treated the same way. Rate of invasion was calculated according to recovered bacterial cells related to the input. Experiments were carried out in duplicates.
Macrophage survival assay
Infection of macrophage cell lines was performed as described in Thompson et al. (2006). The macrophage cell line used was RawB, a derivative of Raw 264.7 (ATCC TIB-71). Macrophages were seeded in 12 well plates 1 day prior to infection at 1 × 105 cells per well. Next day bacteria were harvested for infection at early stationary growth phase (OD600∼2–3). Macrophages were infected with a moi of 1. Bacterial cells were centrifuged (37°C, 250 g, 10 min) onto the macrophages, followed by a 20 min incubation step at 37°C in an atmosphere containing 5% CO2. Thirty minutes in total after infection medium was changed to RPMI (containing 50 μg ml−1 gentamicin) to kill non-invasive bacterial cells. Incubation was carried on for additional 30 min. Medium was changed for the 4 and 24 h time points to RPMI containing 10 μg ml−1 gentamicin and incubation carried on for additional 3 or 23 h respectively. The number of intracellular bacteria was determined 1, 4 and 24 h after infection and given in per cent related to the input. Experiments were carried out in triplicates and data are representative of two independent experiments.
HeLa cell adhesion assay
The adhesion assay was performed as described in Hara-Kaonga and Pistole (2004). In brief, bacteria were grown for 12 h under SPI1-inducing conditions. One hundred microlitres of HeLa cells (5 × 105 per ml in RPMI medium) was incubated with 100 μl of bacterial suspension in RPMI medium for 60 min with a moi of 50 at 37°C in 96 well plates. Infections were carried out in triplicates. Non-adherent bacteria were removed by washing cells 4× with 200 μl PBS at 400 g. Each sample was resuspended in 50 μl PBS/4% formaldehyde. Each well was sampled three times, and 10 HeLa cells were analysed per sampling. Cells were counted with 1000× magnification using an Eclipse 50i microscope (Nikon).
In a further adhesion assay similar to the macrophage assay by Buchmeier and Heffron (1989), 1 × 105 HeLa cells per well ml−1 were infected with a moi of 10 for 30 min with bacteria grown to early stationary phase (bacteria were spun for 10 min on the HeLa cell monolayer followed by 20 min incubation at 37°C). Each well was washed three times with 1 ml PBS and cells were collected by scraping HeLa cells from the bottom of each well in PBS/0.1% Triton X-100. Dilutions in PBS were plated on LB agar and incubation carried out o/n at 37°C. Rate of adhesion and invasion (determined in parallel as above) was calculated according to recovered bacterial cells related to the input. Experiments were carried out in triplicates.
Animal infections
Bacterial cultures for mice infections were grown in l-broth to early stationary phase (OD600 of 2–3), harvested by centrifugation, and diluted to the appropriate cfu ml−1 in sterile PBS for infections. For peroral infections, strains were resuspended at 109 cfu ml−1, and 0.1 ml of the resuspensions (∼108 bacteria) used to infect groups of five Balb/c mice per strain. The total infective dose was determined in parallel by plating dilutions to agar plates with or without selection, where appropriate. After 72 h, the mice were sacrificed by euthanization in a CO2 chamber, and spleens were removed for determination of organ bacterial loads. Isolated spleens were washed once in 70% ethanol, once in PBS and homogenized in 1 ml of PBS. Cell resuspensions were lysed by addition of 1 ml of 0.2% Triton X-100 in deionized, distilled water and incubation at room temperature for 15 min. Dilutions of the cell lysates were plated to agar plates with or without antibiotic selection where appropriate for enumeration of total intracellular bacteria. Intraperitoneal infections were performed by injection of 0.1 ml of a 1:1 mixture of bacterial suspensions of 2 × 106 cfu ml−1 of wild-type and mutant strains into the peritoneal space, yielding a final infective dose of approximately 105 cfu ml−1 for each strain per animal. Forty-eight hours after the infections, mice were sacrificed and spleens isolated and processed as above. The CI was calculated from the ratios of total input and recovered wild-type and chloramphenicol-resistant Δhfq cfu as previously described (Shea et al., 1996).
Motility assay
Cultures were diluted 1/100 into fresh media and incubated at 37°C/220 rpm to an OD600 of 2. One microlitre of culture was inoculated in motility agar plates (LB/0.3% agarose), followed by incubation for 4 h at 37°C.
Whole cell protein fractions
Culture samples were taken according to 1 OD600. Samples were spun 2 min at 16 100 g at 4°C. The cell pellet was resuspended in 1× sample loading buffer (1× SLB; Fermentas) to a final concentration of 0.01 OD µl−1. Samples were heated 5 min at 95°C. For small and large SDS-PAGE 0.1 OD and 0.2 OD, respectively, were loaded per lane.
Secreted protein fractions
The protocol for extraction of secreted protein fractions was modified from the protocol described in Kaniga et al. (1995). Culture samples were taken either from regular LB cultures at OD 2 or after 12 h of growth or after 12 h of growth in SPI1-induction media, and spun 20 min at 16 100 g at 4°C. Proteins from the supernatant were precipitated by adding 25% TCA to a final concentration of 5% followed by 20 min centrifugation at 16 100 g, 4°C. The pellet was washed 2× in ice-cold acetone and air dried. The pellet was resuspended in 1× SLB to a final concentration of 1 OD/10 μl. Samples were heated 5 min at 95°C. For small and large SDS-PAGE 1 OD and 2 OD, respectively, were loaded per sample.
Periplasmic protein fractions
Periplasmic proteins were extracted following the cold osmotic shock procedure described by Neu and Heppel (1965). Overnight cultures were inoculated 1/100 in fresh media and grown to an OD600 of 2. Cells were harvested (30 min, 4000 g, 4°C) and the pellet was resuspended at room temperature in ‘shock buffer’ (30 mM Tris-HCl, pH 8.0, 20% sucrose). EDTA, pH 8.0 was added at a final concentration of 1 mM. Cells were incubated for 10 min at room temperature with occasional shaking. Cells were collected by centrifugation (30 min, 4000 g, 4°C) and the pellet resuspended in 10 ml ice-cold 5 mM MgSO4. After incubation for 10 min with occasional shaking in an ice-water bath, the suspension was centrifuged as mentioned above. The supernatant is the cold osmotic shock-fluid.
Membrane fractions
The total membrane protein fraction was extracted essentially as described (Matsuyama et al., 1984). Culture samples were taken at OD600 of 2 (4 OD total) and spun 20 min at 16 100 g at 4°C. Pellets were washed 1× in 2 ml 10 mM phosphate buffer (pH 7.2). Pellets were resuspended in 0.5 ml of the same buffer. Cells were disrupted by sonication on ice (cycle duty 80%, tip limit 9, four cycles of 30 s with 1 min break on ice). The supernatant was cleared of unbroken cells by centrifugation for 10 min at 1400 g, 4°C. Cell envelopes were recovered by centrifugation of the supernatant for 30 min at 16 100 g, 4°C. After resuspending the pellet in 2 ml phosphate buffer containing 2% Triton X-100 the samples were incubated for 30 min at 37°C. The insoluble fraction was recovered by 30 min centrifugation at 16 100 g at room temperature. After one wash in 2 ml phosphate buffer followed by 5 min centrifugation at 16 100 g the pellet was resuspended in 50 μl phosphate buffer (results in approximately 100 μg in 50 μl). Five microlitres per sample was separated on 10% SDS-PAGE.
Western blot
Commercially available antibodies and antisera used in this study are listed in Table S3. 0.01 or 0.02 OD and 0.1 or 0.2 OD whole cell and secreted protein fractions, respectively, were separated via SDS-PAGE. Proteins were blotted for 60 min at 100 V at 4°C in a cable tank blotter (Peqlab) onto PVDF (Perkin Elmer) membrane in transfer buffer (25 mM Tris base, 190 mM Glycin, 20% Methanol). Blots were rinsed 1× in TBST20 buffer (20 mM Tris base, 150 mM NaCl, 0.1% Tween 20). Membranes were blocked for 1 h in 10% dry milk in TBST20. Hybridization as follows: appropriate antisera or antibodies (in 3% BSA, TBST20; see Table S3 for dilutions) for 1 h at room temperature, 5 × 6 min wash in TBST20,α-Rabbit-HRP or α-mouse-HRP (1:5000 in 3% BSA in TBST20) for 1 h at room temperature, 6 × 10 min wash in TBST20. Blots were developed using Western Lightning (Perkin Elmer) in a Fuji LAS-3000.
Two-dimensional gel analysis and protein identification
Sample preparation from Salmonella cultures at the growth phases given in the respective figure legends, analysis by high-resolution two-dimensional electrophoresis, protein staining, and peptide mass fingerprinting, were performed at the MPI-IB protein analysis core facility (http://info.mpiib-berlin.mpg.de/jungblut/) according to previously published standard protocols (Jungblut and Seifert, 1990; Klose and Kobalz, 1995; Doherty et al., 1998; Jungblut et al., 2000).
Protein quantification by fluorescent stain
Cultures of the wild-type, the hfq mutant, the ompD mutant, and the hfq/ompD double mutant strain were grown with aeration at 37°C, 220 rpm to OD 2. Total protein samples corresponding to 0.1 OD culture were separated on SDS-PAGE (15% gel). Gels were stained with Sypro Ruby (Bio-Rad) following the manufacture's protocol. Protein levels were analysed using the fluorescence mode of a phosphorimager (Phosphorimager, FLA-3000 Series, Fuji) using a 473 nm laser and filter O58. Band intensities were quantified with AIDA software (Raytest, Germany).
Protein overexpression and purification
Overexpression and purification of Salmonella Hfq was carried out as published for E. coli Hfq (Møller et al., 2002) using the IMPACT (Intein Mediated Purification with Affinity Chitin-binding Tag)-CN system (New England Biolabs) according to the manufacture's protocol. Strain ER 2566 carrying plasmid pAS009 was grown to OD of 0.5, and Hfq expression was induced by addition of IPTG (final concentration of 0.5 mM). Following growth for 15 h at 15°C, cells were disrupted using a French press (three passages, 1000 PSI). On-column cleavage of the Hfq moiety was carried out for 24 h at room temperature. The Hfq protein eluate was dialysed against a buffer containing 125 mM NaCl, 12 mM Tris/HCl pH 7.6, 0.5 mM EDTA and concentrated in Vivaspin columns.
Stability experiments, RNA isolation and Northern detection
Overnight cultures were diluted 1/100 in fresh medium and grown to exponential (OD 0.3) and early stationary phase (OD 2). Rifampicin was added to a final concentration of 500 μg ml−1. Incubation was continued at 37°C, 220 rpm, and aliquots (5 ml for OD 0.3; 1.7 ml for stationary phase) were withdrawn prior to or 1, 2, 4, 8, 16 and 32 min after rifampicin addition, mixed with 0.2 vol. of stop solution (5% water-saturated phenol, 95% ethanol), and snap-frozen in liquid nitrogen. After thawing on ice, bacteria were pelleted by centrifugation (2 min, 16 100 g, 4°C), and RNA was isolated using the Promega SV total RNA purification kit as described (Kelly et al., 2004). The purified RNA was quantified on a Nanodrop machine (NanoDrop Technologies).
RNA samples (∼5 μg) were denatured for 5 min at 95°C in loading buffer containing 95% formamide, separated on 8.3 M urea – 5% polyacrylamide gels (PAGE), and transferred to Hybond-XL membranes (GE Healthcare) by electro-blotting (1 h, 50 V, 4°C) in a tank blotter (Peqlab). Membranes were hybridized at 42°C with gene-specific [32P] end-labelled oligodeoxyribonucleotides, random-labelled PCR fragments, or at 70°C with riboprobes, in Rapid-hyb Buffer (GE Healthcare).
ompC transcripts were detected with a random-labelled ([32P] dCTP; Rediprime II labelling kit, GE Healthcare) PCR fragment generated with primer pair JVO-0717/0719. To detect the ompD and hilA mRNAs, PCR fragments generated with primer pairs JVO-0751/0934 and JVO-1298/1299, respectively, were in vitro transcribed from the T7 promoter (added by primers JVO-0934 and JVO-1299) in the presence of [32P]-α-UTP using Ambion's T7 polymerase Maxiscript kit. Riboprobes were purified over a G50 column. fliC and fljB transcripts were probed using end-labelled oligodeoxyribonucleotides JVO-1592 and JVO-1595. For normalization of RNA amounts 5S signals were detected using end-labelled oligodeoxyribonucleotide JVO-0322. Following hybridization for 2 h, membranes hybridized with riboprobes were washed at 65°C in three subsequent 15 min steps in SSC (2×, 1× or 0.5×)/0.1% SDS solutions, after rinsing the membrane first in 2× SSC/0.1% SDS. Membranes hybridized with PCR fragments were rinsed in 2× SSC/0.1% SDS, followed by 15 min washes in 2× (65°C), 1× and 0.5× (42°C) SSC/0.1% SDS. For end-labelled oligodeoxyribonucleotides hybridization membranes were rinsed in 5× SSC followed by three wash steps at 42°C in SSC (5×, 1× and 0.5× respectively). Signals were visualized on a phosphorimager (Phosphorimager, FLA-3000 Series, Fuji), and band intensities quantified with AIDA software (Raytest, Germany).
Gel mobility shift assay
The ompD DNA template for in vitro transcription with T7 RNA polymerase was generated with the primers JVO-1186/-1058. It starts with a T7 promoter fused to the +1 transcriptional start site of OmpD (mapped with 5′RACE; V. Pfeiffer et al., in preparation) at position −69 relative to the ompD AUG start codon, and ends with the 39th codon of the ompD coding sequence. In vitro transcription was performed using the Megascript kit (Ambion, #1333), followed by DNase I digestion (1 unit, 15 min, 37°C). Following extraction with phenol : chloroform : isopropanol (25:24:1 v/v), the RNA was precipitated overnight at −20°C with 1 vol. of isopropanol. RNA integrity was checked on a denaturing polyacrylamide gel. 20 pmol RNA was dephosphorylated with 10 units of calf intestine alkaline phosphatase (New England Biolabs) in a 20 μl reaction at 37°C for 1 h. Following phenol extraction, the RNA was precipitated overnight with ethanol/sodium acetate and 20 μg glycogen. The dephosphorylated RNA was 5′ end-labelled with 32P-γATP (20 μCi), using 1 unit of polynucleotide kinase (New England Biolabs) for 30 min at 37°C in a 20 μl reaction. Unincorporated nucleotides were removed using MicrospinTM G-50 Colums (GE Healthcare), followed by purification of the labelled RNA on a denaturing polyacrylamide gel (6%/7 M urea). Upon visualization of the labelled RNA by exposure on a phosphorimager, the RNA was cut from the gel and eluted with RNA elution buffer (0.1 M sodium acetate, 0.1% SDS, 10 mM EDTA) at 4°C overnight, followed by phenol extraction and precipitation as before.
Binding assays were performed in 1× structure buffer (100 mM Tris pH 7, 1 M KCl, 100 mM MgCl2, provided along with RNase T1 from Ambion #2283) as follows: 5′-labelled RNA (0.01 pmol of ompD mRNA; final concentration in binding reaction: ∼1 nM) and 1 μg of yeast RNA (final concentration: 4.3 μM) were incubated with increasing concentrations of Hfq in 10 μl reactions at 37°C for 15 min. The Hfq dilutions (1, 2, 3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, 500 or 1000 nM; calculated for the Hfq hexamer) were prepared in 1× dilution buffer (1× structure buffer with 1% glycerol, 0.1% Triton X-100). Prior to gel run, the binding reactions were mixed with 3 μl of loading buffer (50% glycerol, 0.5× TBE, 0.2% bromphenolblue), and electrophoresed on native 6% polyacrylamide gels in 0.5× TBE buffer at 300 V at 4°C for 3 h. Gels were dried, and analysed using a phosphorimager (see above).
To synthesize the Hfq-independent metK control RNA, a DNA template for T7 RNA polymerase in vitro transcription was amplified with primers JVO-1701/1702. The resulting RNA spans the entire 5′ UTR (129 nt) according to the +1 transcriptional start site mapped in Wei and Newman (2002) and 80 bp of the metK coding region. In vitro transcription and the labelling reaction were performed as described for ompD RNA.
Fluorescence measurements
Strains carrying the GFP fusion plasmids were inoculated from single colonies in 20 ml LB medium supplemented with 20 μg ml−1 chloramphenicol and incubated with aeration at 37°C/220 rpm. At the indicated cell density, 3 × 100 μl culture were transferred to a 96 well plate, and fluorescence was measured at 37°C using a VICTORTM3 machine (1420 Multilable Counter, Perkin Elmer). All experiments were done in triplicates. Plasmid pJV859-8, which expresses GFP from a constitutive PLtetO promoter, served as a control. In transcriptional fusion studies, strains carrying plasmid pAS0046 served as background control, while plasmid pJU004 was used in translational fusion studies. A detailed protocol of fluorescence measurement will be described elsewhere (Urban and Vogel, 2006).
Acknowledgments
We thank Monica Schmid, Ursula Zimny-Arndt and Peter Jungblut (MPI-IB Berlin protein analysis core facility) for 2D gel analysis and peptide identification, and Robert Hurwitz and Ralf Winter (MPI-IB Berlin protein purification core facility) for help with Hfq purification. We are indebted to Michael Kolbe and Vivien Wolter (MPI-IB Berlin), WD Hardt (ETH Zurich) and Regine Hengge (Free University Berlin), for their generous gifts of antisera. We thank Peter Schwerk for help with animal experiments. K.T. was supported by operating funds from the Institut für Mikrobiologie und Tierseuchen, Freie Universität Berlin. Work in the Vogel lab is supported by the Max Planck Society, Germany.
Supplementary material
The following supplementary material is available for this article online:
Summary of Hfq-dependent changes of protein expression.
Oligonucleotides used in this study.
Commercially available antibodies and antisera used in this study.
Growth characteristics of Salmonella strains under SPI1-inducing conditions.
The Dhfq mutant is defective for invasion and intracellular replication.
The Dhfq strain shows an invasion and intracellular growth defect in intestinal epithelial cells and J774A murine macrophage.
The hfq mutation leads to various differences in protein levels.
The hfq mutant shows reduced adhesion.
RpoS expression is Hfq-dependent in SL1344.
Physical maps of plasmids.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- van Asten FJ, Hendriks HG, Koninkx JF, van Dijk JE. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int J Med Microbiol. 2004;294:395–399. doi: 10.1016/j.ijmm.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- Bang IS, Frye JG, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol. 2005;56:811–823. doi: 10.1111/j.1365-2958.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- Brown L, Elliott T. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier NA, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael GG, Weber K, Niveleau A, Wahba AJ. The host factor required for RNA phage Qbeta RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J Biol Chem. 1975;250:3607–3612. [PubMed] [Google Scholar]
- Chaudhuri RR, Khan AM, Pallen MJ. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 2004;32:D296–D299. doi: 10.1093/nar/gkh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- Collazo CM, Galan JE. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Miller VL. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Miller VL. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol Microbiol. 2000;35:949–960. doi: 10.1046/j.1365-2958.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Doherty NS, Littman BH, Reilly K, Swindell AC, Buss JM, Anderson NL. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:355–363. doi: 10.1002/elps.1150190234. [DOI] [PubMed] [Google Scholar]
- Dorman CJ, Chatfield S, Higgins CF, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989;57:2136–2140. doi: 10.1128/iai.57.7.2136-2140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int J Med Microbiol. 2002;291:479–485. doi: 10.1078/1438-4221-00156. [DOI] [PubMed] [Google Scholar]
- Eichelberg K, Galan JE. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Bono H, Ogata H, Fujibuchi W, Nishioka T, Sato K, Kanehisa M. Organizing and computing metabolic pathway data in terms of binary relations. Pac Symp Biocomput. 1997;00:175–186. [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Regnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Kaonga B, Pistole TG. OmpD but not OmpC is involved in adherence of Salmonella enterica serovar typhimurium to human cells. Can J Microbiol. 2004;50:719–727. doi: 10.1139/w04-056. [DOI] [PubMed] [Google Scholar]
- Harrison JA, Pickard D, Higgins CF, Khan A, Chatfield SN, Ali T, et al. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol Microbiol. 1994;13:133–140. doi: 10.1111/j.1365-2958.1994.tb00408.x. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- Jones AM, Goodwill A, Elliott T. Limited role for the DsrA and RprA regulatory RNAs in rpoS regulation in Salmonella enterica. J Bacteriol. 2006;188:5077–5088. doi: 10.1128/JB.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut PR, Seifert R. Analysis by high-resolution two-dimensional electrophoresis of differentiation-dependent alterations in cytosolic protein pattern of HL-60 leukemic cells. J Biochem Biophys Methods. 1990;21:47–58. doi: 10.1016/0165-022x(90)90044-d. [DOI] [PubMed] [Google Scholar]
- Jungblut PR, Bumann D, Haas G, Zimny-Arndt U, Holland P, Lamer S, et al. Comparative proteome analysis of Helicobacter pylori. Mol Microbiol. 2000;36:710–725. doi: 10.1046/j.1365-2958.2000.01896.x. [DOI] [PubMed] [Google Scholar]
- Kaminski PA, Desnoues N, Elmerich C. The expression of nifA in Azorhizobium caulinodans requires a gene product homologous to Escherichia coli HF-I, an RNA-binding protein involved in the replication of phage Q beta RNA. Proc Natl Acad Sci USA. 1994;91:4663–4667. doi: 10.1073/pnas.91.11.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Trollinger D, Galan JE. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JC, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JR, Fahlen TF, Jones BD. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun. 2000;68:3368–3376. doi: 10.1128/iai.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose J, Kobalz U. Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis. 1995;16:1034–1059. doi: 10.1002/elps.11501601175. [DOI] [PubMed] [Google Scholar]
- Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa SI. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol. 1999;34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Jones BD, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- Lostroh CP, Bajaj V, Lee CA. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol Microbiol. 2000;37:300–315. doi: 10.1046/j.1365-2958.2000.01991.x. [DOI] [PubMed] [Google Scholar]
- Lucas RL, Lee CA. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2001;183:2733–2745. doi: 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:746–752. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur SD, Pulvermacher SC, Stauffer GV. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 2006;6:52. doi: 10.1186/1471-2180-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- McNealy TL, Forsbach-Birk V, Shi C, Marre R. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol. 2005;187:1527–1532. doi: 10.1128/JB.187.4.1527-1532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Mangan MW, Lucchini S, Danino V, Croinin TO, Hinton JC, Dorman CJ. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Inokuchi K, Mizushima S. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coli K-12. J Bacteriol. 1984;158:1041–1047. doi: 10.1128/jb.158.3.1041-1047.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer PN, Wilmes-Riesenberg MR, Stathopoulos C, Curtiss R., 3rd Virulence of a Salmonella typhimurium OmpD mutant. Infect Immun. 1998;66:387–390. doi: 10.1128/iai.66.1.387-390.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DM, Bajaj V, Lee CA. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao H, Watanabe H, Nakayama S, Takeda T. yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq) Mol Microbiol. 1995;18:859–865. doi: 10.1111/j.1365-2958.1995.18050859.x. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Negm RS, Pistole TG. Macrophages recognize and adhere to an OmpD-like protein of Salmonella typhimurium. FEMS Immunol Med Microbiol. 1998;20:191–199. doi: 10.1111/j.1574-695X.1998.tb01127.x. [DOI] [PubMed] [Google Scholar]
- Neu HC, Heppel LA. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- Nickerson CA, Curtiss R., 3rd Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. σE-dependent sRNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol Med Microbiol. 2004;41:177–185. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe-Saule V, Coynault C, Norel F. The live oral typhoid vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol Lett. 1995;126:171–176. doi: 10.1111/j.1574-6968.1995.tb07412.x. [DOI] [PubMed] [Google Scholar]
- Robertson GT, Roop RM., Jr The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Roop RM, Gee JM, Robertson GT, Richardson JM, Ng WL, Winkler ME. Brucella stationary-phase gene expression and virulence. Annu Rev Microbiol. 2003;57:57–76. doi: 10.1146/annurev.micro.57.030502.090803. [DOI] [PubMed] [Google Scholar]
- Santiviago CA, Fuentes JA, Bueno SM, Trombert AN, Hildago AA, Socias LT, et al. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol Microbiol. 2002;46:687–698. doi: 10.1046/j.1365-2958.2002.03204.x. [DOI] [PubMed] [Google Scholar]
- Santiviago CA, Toro CS, Hidalgo AA, Youderian P, Mora GC. Global regulation of the Salmonella enterica serovar typhimurium major porin, OmpD. J Bacteriol. 2003;185:5901–5905. doi: 10.1128/JB.185.19.5901-5905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- Schechter LM, Jain S, Akbar S, Lee CA. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2003;71:5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovierova H, Rowley G, Rezuchova B, Homerova D, Lewis C, Roberts M, Kormanec J. Identification of the sigmaE regulon of Salmonella enterica serovar Typhimurium. Microbiology. 2006;152:1347–1359. doi: 10.1099/mic.0.28744-0. [DOI] [PubMed] [Google Scholar]
- Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004;279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sternberg NL, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- Thompson A, Rolfe MD, Lucchini S, Schwerk P, Hinton JC, Tedin K. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J Biol Chem. 2006;281:30112–30121. doi: 10.1074/jbc.M605616200. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Winkler ME. Transcriptional patterns of the mutL-miaA superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie. 1994;76:1168–1177. doi: 10.1016/0300-9084(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl1040. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Vytvytska O, Jakobsen JS, Balcunaite G, Andersen JS, Baccarini M, von Gabain A. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc Natl Acad Sci USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Newman EB. Studies on the role of the metK gene product of Escherichia coli K-12. Mol Microbiol. 2002;43:1651–1656. doi: 10.1046/j.1365-2958.2002.02856.x. [DOI] [PubMed] [Google Scholar]
- Wilmes-Riesenberg MR, Foster JW, Curtiss R., 3rd An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Libby SJ, Freet AM, Boddicker JD, Fahlen TF, Jones BD. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol Microbiol. 2001;39:79–88. doi: 10.1046/j.1365-2958.2001.02192.x. [DOI] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Ziolkowska K, Derreumaux P, Folichon M, Pellegrini O, Regnier P, Boni IV, Hajnsdorf E. Hfq variant with altered RNA binding functions. Nucleic Acids Res. 2006;34:709–720. doi: 10.1093/nar/gkj464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of Hfq-dependent changes of protein expression.
Oligonucleotides used in this study.
Commercially available antibodies and antisera used in this study.
Growth characteristics of Salmonella strains under SPI1-inducing conditions.
The Dhfq mutant is defective for invasion and intracellular replication.
The Dhfq strain shows an invasion and intracellular growth defect in intestinal epithelial cells and J774A murine macrophage.
The hfq mutation leads to various differences in protein levels.
The hfq mutant shows reduced adhesion.
RpoS expression is Hfq-dependent in SL1344.
Physical maps of plasmids.