Abstract
Tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK), a member of the TNF family, is a multi-functional cytokine that regulates cellular proliferation, angiogenesis, inflammation and apoptosis. In this study, we investigated TWEAK expression in periodontally diseased tissues and the effect of TWEAK on human gingival fibroblasts (HGF). Reverse transcription–polymerase chain reaction (RT–PCR) analysis and immunohistochemistry revealed that TWEAK and the TWEAK receptor, fibroblast growth factor-inducible 14 (Fn14), mRNA and protein were expressed in periodontally diseased tissues. HGF expressed Fn14 and produced interleukin (IL)-8 and vascular endothelial growth factor (VEGF) production upon TWEAK stimulation in a dose-dependent manner. The IL-8 and VEGF production induced by TWEAK was augmented synergistically by simultaneous stimulation with transforming growth factor (TGF)-β1 or IL-1β. IL-1β and TGF-β1 enhanced Fn14 expression in a dose-dependent manner. Moreover, TWEAK induced intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) expression on HGF in a dose-dependent manner. The ICAM-1 expression induced by TWEAK was augmented by TGF-β1. On the other hand, the TWEAK-induced VCAM-1 expression was inhibited by TGF-β1. Phosphatidylinositol 3-kinase (PI3K) and nuclear factor-kappaB (NF-κB) inhibitor inhibit both ICAM-1 and VCAM-1 expression induced by TWEAK. However, mitogen-activated protein kinase (MEK) and c-Jun NH2-terminal kinase (JNK) inhibitor enhanced only VCAM-1 expression on HGF. These results suggest that TWEAK may be involved in the pathophysiology of periodontal disease. Moreover, in combination with IL-1β or TGF-β1, TWEAK may be related to the exacerbation of periodontal disease to induce proinflammatory cytokines and adherent molecules by HGF.
Keywords: Fn14, gingival fibroblast, periodontal disease, TWEAK
Introduction
The tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) was first identified as a new member of the TNF superfamily that induced cell death in some tumour cell lines [1–3]. The human TWEAK gene is expressed in different cell types and encodes a ∼30 kDa type 2 transmembrane protein that can be cleaved to generate a ∼18 kDa soluble factor with biological activity. Recently, Wiley et al. identified fibroblast growth factor-inducible 14 (Fn14) as a TWEAK receptor with a physiological affinity [4]. Fn14 is a type 1 transmembrane protein that is composed of only one cysteine-rich domain in the extracellular region and a short cytoplasmic region containing a TNF receptor-associated factor (TRAF)- binding motif. Saitoh et al. reported recently that TWEAK stimulates the nuclear factor κB (NF-κB) signalling pathway via TRAF molecules [5].
TWEAK mRNA is expressed in a variety of tissues and cell types, with relatively high levels found in most major organs, including the heart, brain, skeletal muscle and pancreas. Tissues related to the immune system also show the presence of TWEAK mRNA including spleen, lymph nodes and thymus [1,6]. TWEAK mRNA expression has also been detected in various human tumour cell lines [1,6,7], human peripheral blood lymphocytes [1,6,7], mouse peritoneal macrophages [8] and WI-38 human fibroblasts [9].
Several groups have reported that TWEAK is able to increase the expression of various molecules involved in the inflammatory response. Indeed, TWEAK stimulation induces interleukin (IL)-8 secretion in human tumour cell lines [1,10], WI-38 fibroblasts [1] and astrocytes [11]. TWEAK also increases IL-6 secretion and intercellular adhesion molecule-1 (ICAM-1) expression when added to astrocytes cell cultures, and it has been proposed that this cytokine may play a role in brain inflammation [11]. In recent studies, Chicheportiche et al. [12] demonstrated that TWEAK treatment of human dermal fibroblasts increased the production of proteolytic enzyme matrix metalloproteinase (MMP)-1 and the proinflammatory molecules prostaglandin E2, IL-6, IL-8, regulated upon activation normal T cell expressed and secreted (RANTES) and interferon-inducible protein 10 (IP-10). A similar TWEAK response was observed using human synoviocytes isolated from patients with rheumatoid arthritis or advanced osteoarthritis. These investigators concluded that TWEAK could be involved in the pathogenesis of chronic inflammatory disease.
Periodontal disease is characterized as chronic inflammation associated with Gram-negative bacteria in the oral cavity [13,14], resulting in soft tissue destruction and periodontal bone resorption. The host-immune response to these bacteria has been suggested to be associated with the alteration or even progress of this disease, and inflammatory cells, such as T cells, B cells and macrophages, are related to the exacerbation of periodontal disease [15]. In particular, proinflammatory cytokines such as IL-1β and TNF-α are involved in periodontal tissue destruction [16]. However, the TWEAK expression and the proinflammatory role of TWEAK in periodontal disease are uncertain.
The aim of this study was to examine TWEAK and Fn14 expression in periodontally diseased tissues and to reveal the effects of TWEAK on chemokine, vascular endothelial growth factor (VEGF) and adhesion molecule expression by human gingival fibroblasts (HGF).
Materials and methods
Gingival tissue biopsies and cell culture
Tissue biopsies were sampled from the inflamed gingiva of patients at surgery who were diagnosed with chronic periodontitis, or from the gingivae of clinically healthy subjects. All gingival biopsy sites in the chronic periodontitis group exhibited radiographic evidence of bone destruction, as well as having clinical probing depths greater than 4 mm, with sulcular bleeding on probing; otherwise, the patients were systemically healthy. Samples of gingival tissues were obtained from 17 chronic periodontitis patients (four males and 13 females, aged 49–82 years) and seven healthy control subjects (seven females, aged 23–40 years). We used four clinically healthy gingival samples and nine chronic periodontitis samples for reverse transcription–polymerase chain reaction (RT–PCR), and three clinically healthy gingival samples and eight chronic periodontitis samples for immunohistochemical staining. We used HGF isolated from three clinically healthy gingiva during routine distal wedge surgical procedures. Gingival specimens were cut into small pieces and transferred to culture dishes. The HGF that grew from the gingivae were cultured primarily on 100 mm2 uncoated plastic dishes in Dulbecco’s modified Eagle medium (DMEM; Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and antibiotics (penicillin G; 100 units/ml, streptomycin; 100 mg/ml) at 37°C in humidified air with 5% CO2. Confluent cells were transferred and cultured for use in the present study. After three to four subcultures by trypsinization, the cultures contained homogeneous, slim and spindle-shaped cells growing in characteristic swirls. The cells were used for experiments after five passages. In selected experiments, HGF were cultured for 1 h in the presence or in the absence of SB203580 (20 µM; Santa Cruz Biotechnology, Santa Cruz, CA, USA), PD98059 (20 µM; Calbiochem, La Jolla, CA, USA), SP600125 (20 µM; Sigma), LY294002 (20 µM; Calbiochem), rapamycin (50 nM; Santa Cruz Biotechnology) or MG-132 (50 µM; Calbiochem), prior to incubation with the various stimuli. Informed consent was obtained from all subjects participating in this study. The study was performed with the approval and compliance of the University of Tokushima Ethical Committee.
RNA extraction and RT–PCR analysis
Total RNA was prepared from gingival biopsies or HGF using the Rneasy total RNA isolation Kit (Qiagen, Hilden, Germany). Single-strand cDNA for a PCR template was synthesized from 48 ng of total RNA using a primer, oligo(dT)12−18 (Invitrogen, Carlsbad, CA, USA) and superscript III reverse transcriptase (Invitrogen) under the conditions indicated by the manufacturer. Specific primers were designed from cDNA sequence for TWEAK, Fn14 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH). Each cDNA was amplified by PCR using Hot star Taq DNA polymerase (Qiagen). The sequences of the primers were as follows: TWEAK-F (5′-CCCTGCGCTGCCTGGAGGAA-3′), TWEAK-R (5′-AGACCAGGGCCCCTCAGTGA-3′), Fn14-F (5′-CCAAGCTCCTCCAACCACAA-3′), Fn14-R (5′-TGGGGCCTAGTGTCAAGTCT-3′), GAPDH-F (5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′), and GAPDH-R (5′-CATGTGGGCCATGAGGTCCACCAC-3′). The conditions for PCR were 1 × 95°C, 15 min; 35 × 94°C, 1 min, 59°C, 1 min, 72°C, 1 min; and 1 × 72°C, 10 min. The products were analysed on a 1·5% agarose gel containing ethidium bromide. The expected sizes of the PCR products for TWEAK, Fn14 and GAPDH were 200 base pairs (bp), 260 bp and 985 bp, respectively. We could not detect any bands when we performed PCR without adding the cDNA template in this study.
Immunohistochemistry
Gingival biopsies were immediately embedded in the optical cutting temperature (OCT) compound (Miles Laboratories Inc., Elkhart, IN, USA) and quenched and stored in liquid nitrogen. The specimens were cut in 6 µm sections using a cryostat (SFS; Bright Instrumental Company, Huntingdon, UK) and collected on poly l-lysine-coated slides. TWEAK expression was analysed with specific antibodies; mouse anti-human TWEAK antibody (clone: CARL-1; Biolegend, San Diego, CA, USA, 5 µg/ml), mouse anti-human Fn14 antibody (clone: ITEM-1, Biolegend, 5 µg/ml); we used an isotype-matched control antibody as the negative control. The sections were reacted with specific antibodies overnight at 4°C. After washing with phosphate-buffered saline (PBS), the sections were incubated with biotinylated anti-mouse and rabbit immunoglobulins (Universal Antibody; Dako, Kyoto, Japan) for 20 min at room temperature and washed with PBS to remove any unreacted antibodies. The sections were then treated with peroxidase-conjugated streptavidin (Dako) for 10 min, and washed and reacted with DAB (3,3-diamino-benzidine tetrahydrochloride; Dako) in the presence of 3% H2O2 to develop the colour. The sections were counterstained with haematoxylin and mounted with glycerol. We did not detect any staining when we used the isotype-matched control antibody.
Cytokine production by HGF
HGF were stimulated with TWEAK (Peprotech, Rocky Hill, NJ, USA), TNF-α (Peprotech), IL-1β (Peprotech) and transforming growth factor (TGF)-β1 (Peprotech) for 24 h. The endotoxin levels in the cytokines we purchased from Peprotech were less than 0·1 ng per µg. The supernatants from HGF were collected and IL-8 and VEGF concentrations of the culture supernatants were measured in triplicate with enzyme-linked immunosorbent assay (ELISA). Duoset (R&D systems, Minneapolis, MI, USA) was used for the IL-8 determinations and a human VEGF ELISA development Kit (Peprotech) for VEGF. Detection ranges for the IL-8 and VEGF ELISAs were 20–2000 and 32–4000 pg/ml, respectively. All assays were performed according to the manufacturer’s instructions, and cytokine levels were determined using a standard curve prepared for each assay.
Flow cytometric analyses
Following the required time in culture, the cells were washed twice with ice-cold PBS. HGF were harvested by incubation with PBS-4 mmol/l ethylenediamine tetraacetic acid (EDTA). Most of the cells were rounded-up following this treatment and could be removed by gentle agitation. Any cells that failed to detach were removed with gentle scraping. The cells were washed twice with ice-cold PBS and incubated (20 min on ice) in PBS−1% bovine serum albumin (BSA). The cells were incubated with mouse anti-human ICAM-1 antibody (clone 8·4A6, Sigma, 5 µg/ml), mouse anti-human vascular cell adhesion molecule-1 (VCAM-1) antibody (clone 1.G11B1, Cymbus Biotechnology Ltd, Hants, UK; 5 µg/ml), mouse anti-human Fn14 antibody (5 µg/ml; Biolegend) or an isotype control antibody on ice for 30 min. After washing three times with PBS−1% BSA (Sigma), the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse F(ab′) 2 fragment (Dako) for 30 min on ice. After washing three times with PBS−1% BSA, the cells were analysed immediately with flow cytometry (Epics XL-MCL; Coulter, Hialeah, FL, USA).
Statistical analysis
Statistical significance was analysed using Student’s t-test. P-values < 0·05 were considered significant.
Results
TWEAK and Fn14 expression in periodontal tissue
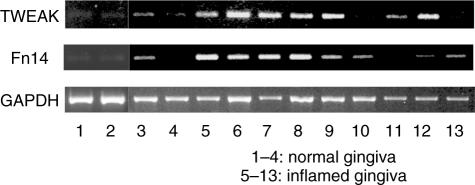
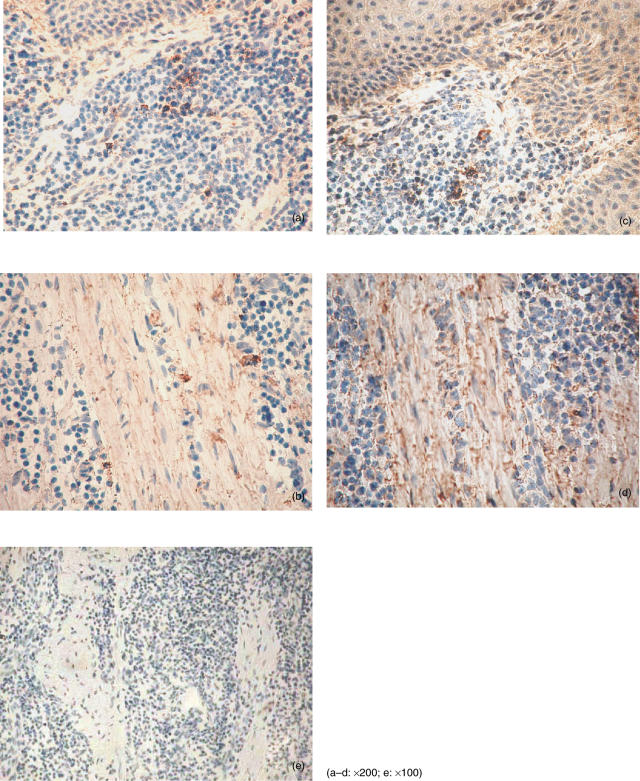
We first examined the TWEAK and Fn14 mRNA expression in whole gingival tissue. Both TWEAK mRNA and Fn14 mRNA were detected in one of four samples of clinically normal gingiva. However, the expression was very weak. TWEAK and Fn14 mRNA were detected in seven of nine samples and eight of nine samples derived from periodontal diseased tissues, respectively (Fig. 1). We carried out immunohistochemical staining to investigate the expression of TWEAK and Fn14 in periodontally diseased tissues (Fig. 2). Morphologically, mononuclear cells (Fig. 2a) and fibroblasts (Fig. 2b) expressed TWEAK in periodontally diseased tissue. Fn14 was expressed by mononuclear cells (Fig. 2c) or fibroblasts (Fig. 2d) in periodontally diseased tissue.
Fig. 1.
Reverse transcription–polymerase chain reaction (RT–PCR) analysis of tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) and fibroblast growth factor-inducible 14 (Fn14) mRNA expression in periodontally diseased tissue. Total RNA was prepared from four clinically healthy gingival samples (pocket depth 2 mm) and nine periodontally diseased gingival samples (pocket depth 4–10 mm). The expressions of TWEAK, Fn14 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) mRNAs in periodontal tissues were analysed by RT–PCR, as described in the Methods.
Fig. 2.
Tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) and fibroblast growth factor-inducible 14 (Fn14) immunostainings in periodontally diseased tissue. Immunohistochemical staining of human periodontally diseased tissues with anti-TWEAK antibody (a, b), anti-Fn14 antibody (c, d) and control mouse IgG (e). Original magnification for each photograph was ×200 (a–d) or ×100 (e).
HGF express Fn14
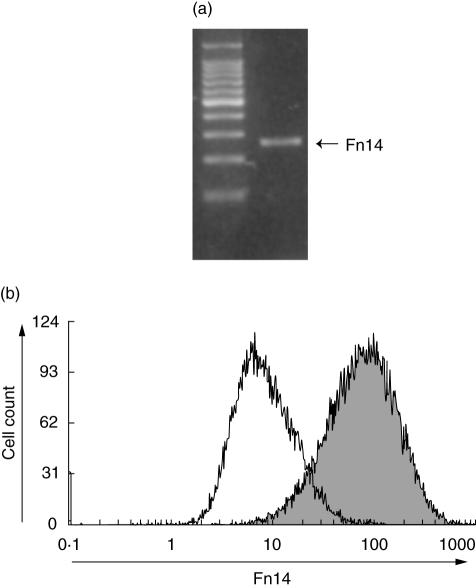
To examine whether TWEAK may act on HGF, we first examined the expression of Fn14 by HGF. RT–PCR analysis showed that non-stimulated HGF express Fn14 mRNA (Fig. 3a). Flow cytometric analysis showed a higher fluorescence from cells incubated with anti-Fn14 monoclonal antibody (MoAb) compared to those incubated with control antibody, indicating that Fn14 was expressed significantly on the cell surface (Fig. 3b).
Fig. 3.
Fibroblast growth factor-inducible 14 (Fn14) expression by HGF. (a) Total RNA was prepared from non-stimulated human gingival fibroblasts (HGF). The expression of Fn14 mRNA in non-stimulated HGF was analysed by reverse transcription–polymerase chain reaction (RT–PCR), as described in the Methods. (b) Flow cytometric analysis of Fn14 expression by non-stimulated HGF. The filled area represents Fn14-specific fluorescence and the open area represents the background level of fluorescence caused by secondary antibody.
TWEAK induces IL-8 production by human gingival fibroblasts
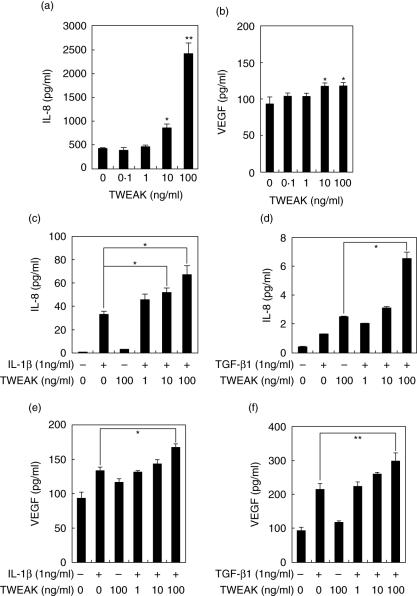
Because cultured HGF express Fn14, we then investigated whether TWEAK could stimulate HGF. We examined IL-8 production, because previous studies have shown that IL-8 was involved in the pathogenesis of periodontal disease. As shown in Fig. 4a, TWEAK induced IL-8 production in a concentration-dependent manner. It has been reported that IL-1β and TGF-β1 induced IL-8 production by HGF. We were thus interested in examining the effects of TWEAK on IL-1β (Fig. 4c) or TGF-β1 (Fig. 4d)-induced IL-8 production. Notably, TWEAK increased IL-1β or TGF-β1 induced IL-8 production by HGF in a concentration-dependent manner.
Fig. 4.
Tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK)-induced interleukin (IL)-8 and vascular endothelial growth factor (VEGF) by human gingival fibroblasts (HGF). HGF were treated with TWEAK (0·1, 1, 10 or 100 ng/ml), and the supernatants were collected after 24 h. The expression levels of IL-8 (a) and VEGF (b) in the supernatants were measured using enzyme-linked immunosorbent assay (ELISA). HGF were treated with TWEAK (0·1, 1, 10, 100 ng/ml) with or without IL-1β (1 ng/ml), and the supernatants were collected after 24 h. The expression levels of IL-8 (c) and VEGF (e) in the supernatants were measured with ELISA. HGF were treated with TWEAK (0·1, 1, 10, 100 ng/ml) with or without transforming growth factor (TGF)-β1 (1 ng/ml), and the supernatants were collected after 24 h. The expression levels of IL-8 (d) and VEGF (f) in the supernatants were measured with ELISA. Data are representative of three different HGF samples from three different donors. The results were calculated as the mean ± s.d. of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the medium or between two results.
TWEAK induces VEGF production by HGF
We next examined the effects of TWEAK on VEGF production by HGF because it has been reported that TWEAK was involved in angiogenesis. As shown Fig. 4c, TWEAK slightly induced VEGF production. It has been reported that IL-1β and TGF-β1 induced VEGF production by HGF. We were thus interested in investigating the effects of TWEAK on IL-1β (Fig. 4e) or TGF-β1-induced VEGF (Fig. 4f) production. Notably, TWEAK showed a synergistic effect on TGF-β1-induced VEGF production by HGF in a concentration-dependent manner. These results indicated that TWEAK and IL-1β/TGF-β1 acted synergistically for VEGF in HGF.
IL-1β and TGF-β1-enhanced Fn14 expression on HGF
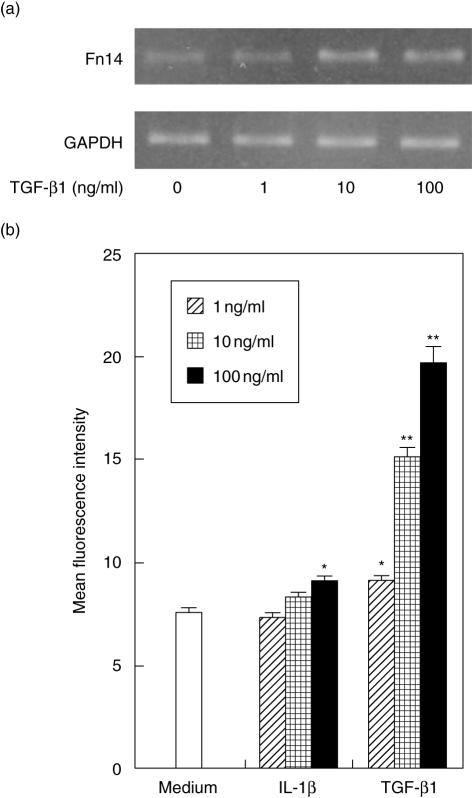
Because IL-1β and TGF-β1 up-regulated the IL-8 and VEGF production induced by TWEAK, we examined the effect of IL-1β or TGF-β1 on Fn14 expression by HGF. TGF-β1 stimulation induced Fn14 mRNA expression in a dose-dependent fashion (Fig. 5a). IL-1β treatment slightly induced Fn14 mRNA expression in HGF (data not shown). Flow cytometry data shows that both IL-1β and TGF-β1 up-regulated the Fn 14 expression on HGF in a dose-dependent manner (Fig. 5b).
Fig. 5.
Interleukin (IL)-1β and transforming growth factor (TGF)-β1 induced fibroblast growth factor-inducible 14 (Fn14) by human gingival fibroblasts (HGF). (a) HGF were treated with TGF-β1 (0·1, 1 or 10 ng/ml) and total RNA was prepared after 4 h stimulation. The expression of Fn14 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) mRNA in HGF was analysed by reverse transcription–polymerase chain reaction (RT–PCR), as described in the Methods. (b) HGF were treated with IL-1β (0·1, 1 or 10 ng/ml) or TGF-β1 (0·1, 1 or 10 ng/ml) and the cells were collected after 24 h. The expression levels of Fn14 on the surface of HGF were measured using flow cytometry. The data are representative of three different HGF samples from three different donors. The results were calculated as mean ± s.d. of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the medium.
TGF-β1 modulated TWEAK-induced ICAM-1 and VCAM-1 expression by HGF
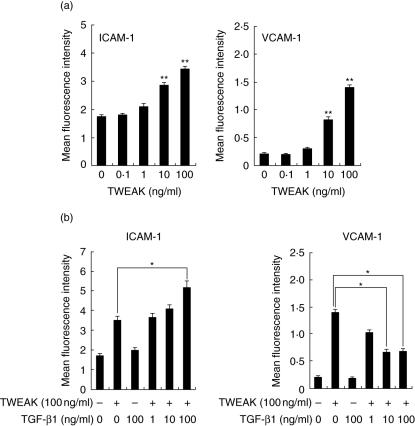
We next examined the effects of TWEAK on ICAM-1 and VCAM-1 expression by HGF because it has been reported that TWEAK was related to inflammation. As shown Fig. 6a, TWEAK enhanced both ICAM-1 and VCAM-1 expression by HGF in a concentration-dependent manner, although VCAM-1 expression on HGF was marginal. It has been reported that TGF-β1 modulated the adherent molecule expression by HGF. We were thus interested in investigating the effects of TGF-β1 on TWEAK-induced ICAM-1 and VCAM-1 expression by HGF (Fig. 6b). TGF-β1 showed a synergistic effect on the TWEAK-induced ICAM-1 expression by HGF in a concentration-dependent manner. On the other hand, TGF-β1 showed inhibitory effects on theTWEAK-induced VCAM-1 expression by HGF. These results indicate that TGF-β1 modulated differentially the TWEAK-induced ICAM-1 and VCAM-1 expression by HGF.
Fig. 6.
Tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) and transforming growth factor (TGF)-β1 controlled intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) expression on human gingival fibroblasts (HGF). (a) HGF was treated with TWEAK (0·1, 1, 10 or 100 ng/ml) and the cells were collected after 24 h. The expression levels of ICAM-1 or VCAM-1 on the surface of HGF were measured using flow cytometry. (b) HGF were treated with TWEAK (100 ng/ml) and TGF-β1 (1, 10 or 100 ng/ml) and the cells were collected after 24 h. The expression levels of ICAM-1 and VCAM-1 on the surface of HGF were measured using flow cytometry. The data are representative of three different HGF samples from three different donors. The results were calculated as mean ± s.d. of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the medium.
Signal transduction inhibitors modulate differentially ICAM-1 and VCAM-1 expression on HGF induced by TWEAK
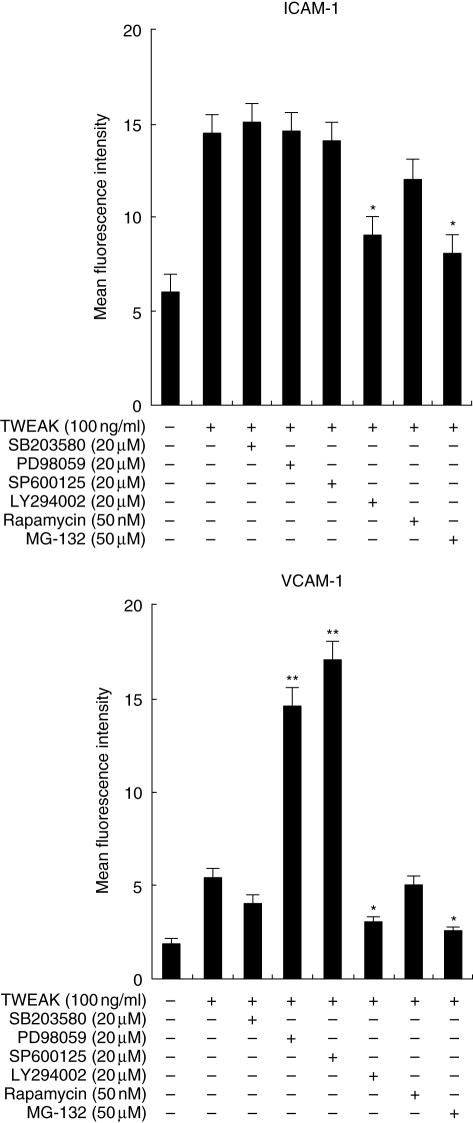
To determine whether p38 mitogen-activated protein kinase (MAPK), mitogen-activated protein kinase kinase (MEK), c-Jun NH2-terminal kinase (JNK), phosphatidylinositol 3-kinase (PI3K), mammalian target of rapamycin (mTOR) or NF-κB are required for ICAM-1 or VCAM-1 expression on HGF in response to TWEAK, the effects of several inhibitors on ICAM-1 and VCAM-1 expression by HGF were examined using flow cytometric analysis (Fig. 7). LY294002 (PI3K inhibitor) and MG-132 (NF-κB inhibitor) inhibit both the ICAM-1 and VCAM-1 expression induced by TWEAK. SB203580 (p38MAPK inhibitor) and rapamycin (mTOR inhibitor) did not modulate the expression of either molecule. On the other hand, PD98059 (MEK inhibitor) and SP600125 (JNK inhibitor) enhanced only the VCAM-1 expression induced by TWEAK.
Fig. 7.
Effects of mitogen-activated protein kinase (MAPK), PI3K inhibitor, mammalian target of rapamycin (mTOR) inhibitor and nuclear factor-kappaB (NF-κB) inhibitor on tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK)-stimulated intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) expression on human gingival fibroblasts (HGF). Cells were preincubated with SB203580 (20 µM), PD98059 (20 µM), SP600125 (20 µM), LY294002 (20 µM), rapamycin (50 nM) or MG-132 (50 µM) for 1 h and then incubated with TWEAK (100 ng/ml). After 24 h incubation, the cells were collected and the expression levels of ICAM-1 and VCAM-1 on the surface of the HGF were measured using flow cytometry. The data are representative of three different HGF samples from three different donors. The results were calculated as the mean ± s.d. of one representative experiment performed in triplicate. Dimethylsulphoxide did not modulate the ICAM-1 and VCAM-1 expression induced by TWEAK on HGF. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the TWEAK stimulation without inhibitors.
Discussion
In this study, we demonstrated that TWEAK and Fn14 were expressed in periodontally diseased tissues. TWEAK induced IL-8 and VEGF production by HGF through Fn14 and, moreover, enhanced ICAM-1 and VCAM-1 expression on HGF. TWEAK and TGF-β1/IL-1β synergize to induce IL-8 or VEGF production and ICAM-1 expression by HGF. These results suggest that TWEAK may be involved in the pathogenesis of periodontal disease.
TWEAK induced IL-8, VEGF, ICAM-1 and VCAM-1 by HGF. It has been shown that TWEAK induced ICAM-1, IL-8 and MCP-1 by human umbilical vein endothelial cells [17], IL-8 by certain tumour cell lines [1], IL-8, IP-10 and RANTES from human dermal fibroblasts and synoviocytes [12]. Thus, TWEAK-induced molecules were related to inflammation by various kinds of cells, and may be involved in inflammatory responses throughout the human body.
Because TGF-β1 and IL-1β have been implicated in the regulation of inflammation and are expressed in periodontally diseased tissues [18–20], we were interested in examining the effect of TGF-β1 or IL-1β on TWEAK-induced IL-8, VEGF, ICAM-1 and VCAM-1 expression by HGF. It has been reported that TGF-β1 augmented the TWEAK-induced RANTES production by human keratinocytes [21]. Additionally, we revealed that TGF-β1 and IL-1β enhanced IL-8, VEGF and ICAM-1 induced by TWEAK from HGF.
It is uncertain as to how Fn14 expression is controlled. Jin et al. reported that TGF-β1 and IFN-γ did not affect Fn14 expression by keratinocytes [21]. However, we revealed that TGF-β1 and IL-1β enhanced Fn14 expression by HGF and this phenomenon may be related to synergistic effects on TGF-β1 or IL-1β-induced IL-8, VEGF and ICAM-1 expression. Thus, it seems that the pattern of Fn14 expression induced by cytokines varies depending on the cell types.
It has been reported that TWEAK activated NF-κB and MAPK cascades in RAW cells [22]; consequently, we used NF-κB and MAPK inhibitors in this experiment. It is uncertain as to whether TWEAK activates the PI3K cascade. However, it is known that the TNF-α family activates PI3K [23,24], therefore we used the PI3K inhibitor. In this study, we revealed that TWEAK-induced ICAM-1 expression was inhibited by a PI3K inhibitor and NF-κB inhibitor, but not by MAPK inhibitors. It has been reported by Chen et al. that TNF-α has been shown to induce ICAM-1 expression mediated through the activation of NF-κB, but not p38 MAPK, ERK and JNK in A549 epithelial cells [25]; our reports agree. We showed that the TWEAK-induced VCAM-1 expression was enhanced by a MEK inhibitor and JNK inhibitor, and inhibited by a PI3K inhibitor and NF-κB inhibitor. It has been reported that activation of p38 MAPK induced VCAM-1 expression, and activation of JNK inhibited VCAM-1 expression induced by TNF-α in chondrosarcoma cells [26]. It has also been described that p38 MAPK inhibitor did not affect TNF-α-induced VCAM-1 expression in Sertoli cells [27]. Wang et al. have reported that the activation of ERK, p38 MAPK, JNK and NF-κB pathways is essential for IL-1β-induced VCAM-1 expression in human tracheal smooth muscle cells [28]. The discrepancies in these previous reports imply that there are divergent pathways leading to VCAM-1 expression, depending on the nature of the stimuli and cell types.
We showed that TGF-β1 enhanced TWEAK-induced ICAM-1 expression and suppressed TWEAK-induced VCAM-1 expression. It has been reported that TGF-β1 induced increases in JNK, p38 MAPK, ERK phosphorylation and activity by human lung fibroblasts [29,30]. It is known that PI3K inhibitors suppress the function of TGF-β1 by fibroblasts, such as collagen gene expression and ADAM12 expression [31]. It has also been reported that the DNA binding activities of NF-κB of synovial fibroblasts are increased by TGF-β1 [32]. These reports mean that TGF-β1 is involved in the p38 MAPK, ERK, JNK, PI3K and NF-κB activation of fibroblasts. The balance of these signal transduction pathway activations might be important for ICAM-1 and VCAM-1 expression by HGF. It is probable that ERK and JNK activation may be related to the inhibition of VCAM-1 expression by TGF-β1; further investigation will be necessary.
An immunohistochemical study has revealed that mononuclear cells in periodontally diseased tissues expressed TWEAK and Fn14. It has been reported that T cells, B cells and macrophages are infiltrated mainly in periodontally diseased tissues [15]. Moreover, TWEAK has been detected by human peripheral blood lymphocytes [1] and mouse peritoneal macrophages [8], and macrophages expressed Fn14 on the cell surface [33]. Our results and previous reports might explain how TWEAK produced by lymphocytes and macrophages might stimulate both HGF and macrophages in periodontally diseased tissues; further investigation will be necessary to elucidate this hypothesis.
It is known that Fn14 is a major TWEAK receptor. Recently, it has been reported that a second TWEAK receptor exists on RAW264·7 cells [22]. However, which molecule is the second TWEAK receptor is still unknown. HGF may express not only Fn14, but also the second TWEAK receptor. Research on the second TWEAK receptor should be continued.
In summary, we have shown a novel biological activity of TWEAK on human gingival fibroblasts. Because IL-8, VEGF, ICAM-1 and VCAM-1 are involved in the inflammatory response in periodontally diseased tissues, induction of these molecules by TWEAK in HGF may be related to the pathophysiology of periodontal disease. Furthermore, we show that cytokines such as IL-1β and TGF-β1 enhanced the effects of TWEAK on HGF. This means that cytokine balances in periodontally diseased tissues will be important for the progression of inflammation. Because many kinds of cytokines exist in periodontally diseased tissues, further investigation of cytokine networks in periodontal tissues will be important.
Acknowledgments
This study was supported by a grant-in-aid from the Ministry of Education, Science and Culture of Japan.
References
- 1.Chicheportiche Y, Bourdon PR, Xu H, et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem. 1997;272:32401–10. doi: 10.1074/jbc.272.51.32401. [DOI] [PubMed] [Google Scholar]
- 2.Schneider P, Schwenzer R, Haas E, et al. TWEAK can induce cell death via endogenous TNF and TNF receptor 1. Eur J Immunol. 1999;29:1785–92. doi: 10.1002/(SICI)1521-4141(199906)29:06<1785::AID-IMMU1785>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama M, Kayagaki N, Yamaguchi N, Okumura K, Yagita H. Involvement of TWEAK in interferon gamma-stimulated monocyte cytotoxicity. J Exp Med. 2000;192:1373–80. doi: 10.1084/jem.192.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiley SR, Cassiano L, Lofton T, et al. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–46. doi: 10.1016/s1074-7613(01)00232-1. [DOI] [PubMed] [Google Scholar]
- 5.Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–12. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 6.Marsters SA, Sheridan JP, Pitti RM, Brush J, Goddard A, Ashkenazi A. Identification of a ligand for the death-domain-containing receptor Apo3. Curr Biol. 1998;8:525–8. doi: 10.1016/s0960-9822(98)70204-0. [DOI] [PubMed] [Google Scholar]
- 7.Pratdet-Balade B, Medema JP, Lopez-Fraga M, et al. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK–APRIL fusion protein. EMBO J. 2002;21:5711–20. doi: 10.1093/emboj/cdf565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicheportiche Y, Fossati-Jimack L, Moll S, Ibnou-Zekri N, Izui S. Down-regulated expression TWEAK mRNA in acute and chronic inflammatory pathologies. Biochem Biophys Res Commun. 2000;279:162–5. doi: 10.1006/bbrc.2000.3913. [DOI] [PubMed] [Google Scholar]
- 9.Semov A, Semova N, Lacelle C, et al. Alternation in TNF- and IL-related gene expression in space-flow WI-38 human fibroblasts. FASEB J. 2002;279:899–901. doi: 10.1096/fj.01-1002fje. [DOI] [PubMed] [Google Scholar]
- 10.Kaptein A, Jansen M, Dilaver G, et al. Studies on the interaction between TWEAK and the death receptor WSL-1/TRAMP (DR3) FEBS Lett. 2000;485:135–41. doi: 10.1016/s0014-5793(00)02219-5. [DOI] [PubMed] [Google Scholar]
- 11.Saas P, Boucraut J, Walker PR, et al. TWEAK stimulation of astrocytes and the proinflammatory consequences. Glia. 2000;32:102–7. [PubMed] [Google Scholar]
- 12.Chicheportiche Y, Chicheportiche R, Sizing I, et al. Proinflammatory activity of TWEAK on human dermal fibroblasts and synoviocytes: blocking and enhancing effects of anti-TWEAK monoclonal antibodies. Arthritis Res. 2002;4:126–33. doi: 10.1186/ar388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzink JL, Taner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;1:648–59. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 14.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 15.Taubman MA, Eastcott JW, Shimauchi H, Takeichi O, Smith DJ. Modulatory role of T lymphocytes in periodontal inflammation. In: Genco RJ, Hamada S, Megenhagen SE, et al., editors. Molecular pathogenesis of periodontal disease. Washington, DC: American Society for Microbiology; 1994. p. 14757. [Google Scholar]
- 16.Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003;74:391–401. doi: 10.1902/jop.2003.74.3.391. [DOI] [PubMed] [Google Scholar]
- 17.Harada N, Nakayama M, Nakano H, Fukuchi Y, Yagita H, Okumura K. Pro-inflammatory effect of TWEAK/Fn14 interaction on human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2002;299:488–93. doi: 10.1016/s0006-291x(02)02670-0. [DOI] [PubMed] [Google Scholar]
- 18.Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodont Res. 1991;26:230–42. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Zee E, Everts V, Beertsen W. Cytokines modulate routes of collagen breakdown. Review with special emphasis on mechanisms of collagen degradation in the periodontium and the burst hypothesis of periodontal disease progression. J Clin Periodontol. 1997;24:297–305. doi: 10.1111/j.1600-051x.1997.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 20.Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–66. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- 21.Jin L, Nakao A, Nakayama M, et al. Induction of RANTES by TWEAK/Fn14 interaction in human keratinocytes. J Invest Dermatol. 2004;122:1175–9. doi: 10.1111/j.0022-202X.2004.22419.x. [DOI] [PubMed] [Google Scholar]
- 22.Polek TC, Talpaz M, Darnay BG, Spivak-Kroizman T. TWEAK mediates signal transduction and differentiation of RAW264.7 cells in the absence of Fn14/TweakR. J Biol Chem. 2003;278:32317–23. doi: 10.1074/jbc.M302518200. [DOI] [PubMed] [Google Scholar]
- 23.Morel J, Audo R, Hahme M, Combe B. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces rheumatoid arthritis synovial fibroblast proliferation through mitogen-activated protein kinases and phosphatidylinositol 3-kinase/Akt. J Biol Chem. 2005;280:15709–18. doi: 10.1074/jbc.M414469200. [DOI] [PubMed] [Google Scholar]
- 24.Gronning LM, Cederberg A, Miura N, Enerback S, Tasken K. Insulin and TNF alpha induce expression of the forkhead transcription factor gene Foxc2 in 3T3-L1 adipocytes via PI3K and ERK 1/2-dependent pathways. Mol Endocrinol. 2002;16:873–83. doi: 10.1210/mend.16.4.0803. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Chou C, Sun Y, Huang W. Tumor necrosis factor alpha-induced activation of downstream NF-kappaB site of the promoter mediates epithelial ICAM-1 expression and monocyte adhesion. Involvement of PKCalpha, tyrosine kinase, and IKK2, but not MAPKs, pathway. Cell Signal. 2001;13:543–53. doi: 10.1016/s0898-6568(01)00171-1. [DOI] [PubMed] [Google Scholar]
- 26.Ju JW, Kim SJ, Jun CD, Chun JS. p38 kinase and c-Jun N-terminal kinase oppositely regulates tumor necrosis factor alpha-induced vascular cell adhesion molecule-1 expression and cell adhesion in chondrosarcoma cells. IUBMB Life. 2002;54:293–9. doi: 10.1080/15216540215674. [DOI] [PubMed] [Google Scholar]
- 27.De Cesaris P, Starace D, Riccioli A, Padula F, Filippini A, Ziparo E. Tumor necrosis factor-alpha induces interleukin-6 production and integrin ligand expression by distinct transduction pathways. J Biol Chem. 1998;273:7566–71. doi: 10.1074/jbc.273.13.7566. [DOI] [PubMed] [Google Scholar]
- 28.Wang CC, Lin WN, Lee CW, et al. Involvement of p42/p44 MAPK, p38 MAPK, JNK, and NF-kappaB in IL-1beta-induced VCAM-1 expression in human tracheal smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L227–37. doi: 10.1152/ajplung.00224.2004. [DOI] [PubMed] [Google Scholar]
- 29.Khalil N, Xu YD, O’Connor R, Duronio V. Proliferation of pulmonary interstitial fibroblasts is mediated by TGF-beta 1 induced release of extracellular FGF-2 and phosphorylation of p38 MAPK and JNK. J Biol Chem. 2005;280:43000–9. doi: 10.1074/jbc.M510441200. [DOI] [PubMed] [Google Scholar]
- 30.Finlay GA, Thannickal VJ, Fanburg BL, Paulson KE. Transforming growth factor-beta 1-induced activation of the ERK pathway/activator protein-1 in human lung fibroblasts requires the autocrine induction of basic fibroblast growth factor. J Biol Chem. 2000;275:27650–6. doi: 10.1074/jbc.M000893200. [DOI] [PubMed] [Google Scholar]
- 31.Le Pabic H, Bonnier D, Wewer UM, et al. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056–66. doi: 10.1053/jhep.2003.50205. [DOI] [PubMed] [Google Scholar]
- 32.Cheon H, Yu SJ, Yoo DH, Chae IJ, Song GG, Sohn J. Increased expression of proinflammatory cytokines and metalloproteinase-1 by TGF-beta 1 in synovial fibroblasts from rheumatoid arthritis and normal individuals. Clin Exp Immunol. 2002;127:547–52. doi: 10.1046/j.1365-2249.2002.01785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S-H, Kang Y-J, Kim W-J, et al. TWEAK can induce pro-inflammatory cytokines and matrix metalloproteinase-9 in macrophages. Circ J. 2004;68:396–9. doi: 10.1253/circj.68.396. [DOI] [PubMed] [Google Scholar]