Abstract
The objective of this study was to investigate the prevalence and clinical significance of a spectrum of autoantibodies in systemic lupus erythematosus and incomplete lupus syndromes using a proteome microarray bearing 70 autoantigens. Microarrays containing candidate autoantigens or control proteins were printed on 16-section slides. These arrays were used to profile 93 serum samples from patients with systemic lupus erythematosus (SLE (n = 33), incomplete LE (ILE; n = 23), first-degree relatives (FDRs) of SLE patients (n = 20) and non-autoimmune controls (NC; n = 17). Data were analysed using the significance analysis of microarray (SAM) and clustering algorithms. Correlations with disease features were determined. Serum from ILE and SLE patients contained high levels of IgG autoantibodies to 50 autoantigens and IgM autoantibodies to 12 autoantigens. Elevated levels of at least one IgG autoantibody were detected in 26% of SLE and 19% of ILE samples; elevated IgM autoantibodies were present in 13% of SLE and 17% of ILE samples. IgG autoantibodies segregated into seven clusters including two specific for DNA and RNA autoantigens that were correlated with the number of lupus criteria. Three IgG autoantibody clusters specific for collagens, DNA and histones, were correlated with renal involvement. Of the four IgM autoantibody clusters, two were correlated negatively with the number of lupus criteria; none were correlated with renal disease. The IgG : IgM autoantibody ratios generally showed a stepwise increase in the groups following disease burden from NC to SLE. Insights derived from the expanded autoantibody profiling made possible with the antigen array suggest differences in autoreactivity in ILE and SLE. Determining whether the IgM aurotreactivity that predominates in ILE represents an early stage prior to IgG switching or is persistent and relatively protective will require further longitudinal studies.
Keywords: autoantibodies, autoantigens, autoimmunity, incomplete lupus, proteome microarray, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is a multi-system autoimmune disease that can be associated with organ failure and premature mortality. In the US population the overall prevalence is estimated at 0·05%, but in some segments of the population, such as minority females, the prevalence may be at least 10-fold greater [1]. Establishing a diagnosis of SLE is complex and relies on a list of criteria that were developed originally over 20 years ago [2]. A major component of the diagnostic approach is detection of anti-nuclear antibodies (ANAs). Although an ANA is generally required for a diagnosis of SLE, the test is non-specific and has a high rate of false positive results [3]. Recently published findings suggest that autoantibodies can be detected in the serum prior to the onset of clinical disease, with the number and complexity of these antibodies increasing up to the point of diagnosis [4]. This result raises the possibility that risk profiles for lupus could be detected prior to onset of clinical symptoms, which would in turn make possible early institution of definitive therapies.
In other studies, we have shown that autoantibody complexity is increased in patients with incomplete lupus (ILE) syndromes defined as having at least one but less than four of the criteria for SLE [5]. These findings are consistent with observations suggesting that 10–50% of patients with ILE progress to SLE within 5 years [6,7]. We have considered that focusing on ILE patients and first-degree relatives of lupus patients may permit development of feasible approaches to early identification of those who have the greatest probability of disease progression.
The use of protein arrays to characterize a wide spectrum of autoantibodies has been applied to characterize patients with rheumatoid arthritis [8]. We have reported previously using a similar approach with a ‘glomerular proteome array’ to identify the autoantibody clusters that best predict lupus disease activity [9]. These previous studies demonstrated that the protein array has greater sensitivity and a broader dynamic range than enzyme-linked immunosorbent assay (ELISA). In the present study, we expanded the number of autoantigens on the array to 70, covering most of specificities reported to be present in patients with SLE, rheumatoid arthritis (RA), multiple sclerosis (MS), Sjögren's syndrome, inflammatory muscle diseases and other autoimmune disorders. These arrays were used to screen the autoantibody profiles in 93 serum samples from individuals with SLE or ILE, first-degree relatives (FDRs) of SLE patients and non-autoimmune control subjects. The goal of this study was to compare autoantibody profiles in ILE and FDRs, who are also at enhanced risk for SLE [10], to patients with SLE. An advantage of focusing on samples from individuals in the ILE category is that the autoantibody profile might be more closely related to inciting or primary events and less obscured by tissue damage and other long-term disease-related changes. Unaffected FDRs provide the additional advantage of carrying out analyses in a group with elevated risk with few, if any, drug effects.
Materials and methods
Patient recruitment and serum samples
Rheumatology clinic patients were recruited at UT South-western Medical Center at Dallas, including subjects from clinics at Parkland Memorial Hospital, a large publicly funded facility, and from the Aston Ambulatory Care Center. Some individuals from a local community-based practice were also included. Studies were carried out on a total of 76 subjects in three categories related to lupus: (1) 33 patients with systemic lupus erythematosus (SLE) who satisfied four or more of the American College of Rheumatology (ACR) classification criteria [2,11]; (2) 23 patients with incomplete lupus erythematosus (ILE), defined as having at least one but less than four of the criteria for SLE; and (3) 20 unaffected FDRs, who were related to individuals in either of the other two groups (Table 1). The ILE group included some individuals who were also classified as having either anti-phospholipid syndrome or Sjögren's syndrome. The mean number of lupus criteria was greater in SLE (5·0 ± 0·3) than in ILE (2·1 ± 0·2; P < 0·0001). A non-autoimmune control (NC) group (n = 17) of healthy individuals or patients with non-autoimmune conditions such as osteoarthritis was also included. ANA and anti-DNA antibodies were measured in serum samples from these patients using ELISA (Inova Diagnostics, San Diego, CA, USA). ANA values were significantly higher in SLE and ILE than in the FDR or NC groups (P < 0·01) and were not significantly different between the FDR and NC groups (P > 0·1).
Table 1.
Demographic and clinical characteristics of the four study groups: systemic lupus erythematosus (SLE), incomplete lupus (ILE), first-degree relatives (FDR), non-autoimmune controls (NC).
| SLE | ILE | FDR | NC | |
|---|---|---|---|---|
| Number | 33 | 23 | 20 | 17 |
| % Female | 90 | 94 | 84 | 91 |
| % AA | 43 | 16 | 16 | 27 |
| % Hispanic | 24 | 19 | 11 | 18 |
| Age (years) | 38·6 ± 2·9* | 46·9 ± 3·2 | 46·6 ± 4·4 | 38·9 ± 5·3 |
| SLE criteria | 5·0 ± 0·3 | 2·1 ± 0·2 | – | – |
| ANA (EU)** | 118 ± 11 | 103 ± 18 | 37 ± 11 | 16 ± 4 |
| DNA (EU) | 317 ± 99 | 156 ± 44 | 132 ± 46 | n.d. |
Values for age, criteria and antibodies are expressed as mean ± s.e.m.
Anti-nuclear antibodies (ANA) and DNA values are expressed as enzyme-linked immunosorbent assay (ELISA) units (EU). n.d. indicates that testing for anti-DNA was not done.
No other criteria were required for enrolment. Subjects could have other comorbid conditions, be on any form of treatment and have any disease duration. All subjects were sampled between November 2003 and December 2004. The population was predominantly female (90%), consistent with the gender bias of human autoimmune diseases. Forty-two per cent of the subjects were under-represented minorities, either African American (24%) or Hispanic (18%; Table 1). Although the SLE group had a higher percentage of African Americans (43%) than any of the other three groups, this was not statistically significant when all four groups were compared using a one-way analysis of variance (anova) (P = 0·7) or when the SLE group was compared to the ILE group only using Fisher's exact test (P = 0·08).
The subjects gave consent for entry into the Dallas Regional Autoimmune Disease Registry. All research carried out under the auspices of this registry has been approved by the UT Southwestern Institutional Review Board. Medical records were reviewed to determine numbers of SLE criteria present at any time during the disease course, using the ACR diagnostic criteria. SLE patients were categorized as having renal involvement based on a biopsy-proven diagnosis at any time during the disease course.
Study participants had blood drawn for isolation of serum, which was aliquoted and stored at −70°C. Serum samples were pretreated with DNAse-I (50 U/ml) for 30 min at room temperature in buffer containing 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, pH 8·3.
Autoantigens
Forty new autoantigens were added to the previously described glomerular proteome array, so the revised autoantigen array included 74 proteins (70 autoantigens and four control proteins). The added specificities were selected based on a survey of the literature to identify autoantigens implicated in various human autoimmune diseases, including SLE, rheumatoid arthritis, multiple sclerosis and Sjögren's syndrome. Most of the added autoantigens were purchased from Diarect Ag (Freiburg, Germany) except C1q, which was from Alpha Diagnostic (San Antonio, TX, USA), β2-microglobulin from RDI Division of Fitzgerald Industries (Concord, MA, USA) and Jo-1 and topoisomerase from Sigma Chemical Co. (St Louis, MO, USA). The optimal printing concentration for all antigens (Ags) was determined to be 1 mg/ml except for cardiolipin, for which the optimal concentration was determined to be 0·1 mg/ml. A complete listing of the antigens used and their sources is available from the authors on request.
Manufacture of autoantigen microarrays
Antigens were diluted to the optimal printing concentration in phosphate-buffered saline (PBS) (137 mM NaCl, 9 mM KOH, 11·3 mM NaH2PO4, pH 7) and transferred to 384-well plates. Rabbit anti-human IgG and IgM were diluted to 0·1 mg/ml and used as internal controls and also for normalization of the autoantibody data. A MicroGrid II microarrayer was used to spot the antigens and antibodies onto Nitrocellulose-coated 16-pad FAST™ slides (Whatman Schleicher & Schleicher BioScience, Keene, NH, USA). The 16-pad FAST slides were selected because this format allowed printing of 16 arrays on one slide, which is ideal for high throughput analysis, and required far fewer materials. The spotter was programmed so that each pad was printed with the exactly the same pattern of proteins. The antigens were printed in duplicate but distributed randomly on the slides. After printing, the slides were kept in a chamber with 70% humidity for 4 h at room temperature. The spotted protein arrays can be stored at 4°C for up to 6 months without loss of activity.
Hybridization and scanning of autoantigen microarrays
On the day of hybridization, the autoantigen slides were brought to room temperature for 30 min. A 16-section frame was applied to each slide to separate the individual arrays. Blocking buffer [1% bovine serum albumin (BSA) in PBS with Tween (PBST)] was added (50 µl) and the slides were agitated slowly for 30 min at room temperature. The pretreated serum samples were diluted 1 : 100 in blocking buffer, a dilution found in preliminary experiments to optimize signal-to-noise ratio for the majority of targets, and 50 µl of this dilution was added to each array, followed by incubation with agitation for 60 min. Each array was then washed with 100 µl of washing buffer (PBS with 0·05% v/v Tween 20, pH 7·4) three times for 5 min each with agitation. Cy3-labelled anti-human IgG and Cy5-labelled anti-human IgM (Jackson ImmunoResearch, West Grove, PA, USA) at 1 : 500 dilution were next applied to each array followed by 60 min of incubation at room temperature. The arrays were then washed with buffer as before, the frame was removed, the slide was immersed in PBS for 1 min and then spun dry. A Genepix 4000B scanner with laser wavelengths 532 nm (for cy3) and 635 nm (for cy5) was used to generate Tif images for analysis.
Array results for four autoantibodies, SSA, SSB, chromatin and dsDNA, were compared to values obtained in a commercial ELISA (Inova Diagnostics, San Diego, CA, USA). The results were highly correlated for each antibody, with correlation coefficients (r2 values) ranging from 0·69 to 0·86, with P < 0·001 for each.
Data analysis
The image of each array was analysed using Genepix Pro6·0 software to generate a Gene Pix Results (GPR) file. The net fluorescence intensities (nfi) were normalized using anti-human IgG/IgM spotted onto each array. To derive the nfi, the absolute fluorescence intensity (spot fluorescence intensity minus background intensity) for any given antigen was divided by the absolute fluorescence intensity of the Ig control spots, and the resulting ratio was multiplied by a factor of 1000 for each antigen. Tests of significance between groups were carried out using Student's t-test or a one-way anova for multiple groups. Correlations between continuous variables were determined using Pearson's r and dichotomized variables were compared using Fisher's exact test. A P-value < 0·05 was considered significant.
Data obtained from duplicate spots were averaged. For intergroup comparisons, a one-way anova was used (GraphPad Prism 4·0). Diagrams with row-wise and column-wise clustering were generated using Cluster and Treeview software (versions 2·1 and 1·6, respectively; http://rana.lbl.gov/EisenSoftware.htm). Fluorescence intensities that were higher than the row mean were coloured red, those that fell below the row mean were coloured green, and cells with signals close to the mean were left black. Missing data were denoted with grey.
Results
The 16-pad nitrocellulose coated FAST™ slides were used as the carrier of 74 proteins for higher throughput detection of autoantibodies. Of the 70 autoantigens tested, 13 (including entactin, elastin, dermatan sulphate, aggrecan, alpha-actinin, amyloid, phosphatidylinositol, cytochrome p450, topoisomerase, hen egg lysozyme, BSA, ovalbumin) were not targeted by the IgG autoantibodies (nfi < 50) in any of the experimental samples, and 20 of them (aggrecan, alpha-actinin, C1q, chondroitin, dermatan sulphate, elastin, gliadin, Goodpasture antigen, cytochrome p450, proteoglycan, heparan sulphate, hyaluronic acid, entactin, phosphatidylinositol, ssDNA, topoisomerase, vimentin, HEL and ovalbumin) were not targeted by the IgM autoantibodies (nfi < 25) in any samples. For the remaining 57 IgG autoantibody specificities and 50 IgM autoantibody specificities, the penetrance of each individual autoantibody in each experimental group was calculated using the mean of the NC group plus 3 standard deviations (s.d.) as the cut-off. For the IgG autoantibodies, the average penetrance for the NC, FDR, ILE and SLE groups was 1·4%, 3·6%, 19·1% and 26%, respectively. Both the ILE and SLE groups had significantly higher penetrance of IgG autoantibodies than the NC and FDR groups (P < 0·001). For the IgM autoantibodies, the average penetrance of NC, FDR, ILE and SLE was 0·25%, 3·2%, 17·2% and 13·2%, respectively. The ILE patients had significantly higher penetrance of IgM autoantibodies than SLE (P < 0·05) (Table 2).
Table 2.
Penetrance of IgG and IgM autoantibodies in subject groups.a
| IgG autoantibodies (%) | IgM autoantibodies (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antigensb | IgG cut-off valuec | NC (n = 17) | FDR (n = 20) | ILE (n = 23) | SLE (n = 33) | IgM cut-off valuec | NC (n = 17) | FDR (n = 20) | ILE (n = 23) | SLE (n = 33) |
| B2-glycoprotein I | 99·1 | 0·0 | 5·0 | 33·3 | 35·3 | 45·5 | 0·0 | 5·3 | 17·4 | 6·1 |
| C1q | 68·7 | 0·0 | 5·0 | 19·0 | 35·3 | 11·4 | 0·0 | 5·3 | 8·7 | 9·1 |
| Cardolipin | 750·3 | 0·0 | 5·0 | 9·5 | 8·8 | 76·3 | 0·0 | 5·3 | 8·7 | 6·1 |
| CENP-A | 911·7 | 0·0 | 5·0 | 14·3 | 14·7 | 52·7 | 0·0 | 5·3 | 21·7 | 3·0 |
| CENP-B | 1852·3 | 0·0 | 5·0 | 9·5 | 2·9 | 117·1 | 0·0 | 5·3 | 21·7 | 12·1 |
| Chromatin | 151·4 | 5·9 | 0·0 | 33·3 | 55·9 | 60·9 | 5·9 | 0·0 | 13·0 | 6·1 |
| Collagen I | 169·6 | 0·0 | 0·0 | 28·6 | 26·5 | 27·5 | 0·0 | 5·3 | 8·7 | 6·1 |
| Collagen II | 129·7 | 0·0 | 0·0 | 23·8 | 29·4 | 37·1 | 0·0 | 5·3 | 13·0 | 9·1 |
| Collagen III | 139·5 | 5·9 | 0·0 | 14·3 | 14·7 | 18·9 | 0·0 | 0·0 | 8·7 | 9·1 |
| Collagen IV | 53·0 | 0·0 | 0·0 | 23·8 | 38·2 | 9·8 | 0·0 | 0·0 | 8·7 | 18·2 |
| Cytochrome C | 416·9 | 0·0 | 5·0 | 14·3 | 5·9 | 98·4 | 0·0 | 5·3 | 21·7 | 6·1 |
| dsDNA | 191·6 | 5·9 | 0·0 | 14·3 | 32·4 | 54·9 | 0·0 | 0·0 | 17·4 | 15·2 |
| Fibrinogen IV | 1520·7 | 0·0 | 5·0 | 9·5 | 11·8 | 102·0 | 0·0 | 0·0 | 17·4 | 9·1 |
| GBM | 144·0 | 0·0 | 0·0 | 19·0 | 26·5 | 53·6 | 0·0 | 5·3 | 17·4 | 6·1 |
| GBM (dissociated) | 94·4 | 5·9 | 0·0 | 9·5 | 26·5 | 34·9 | 0·0 | 5·3 | 13·0 | 6·1 |
| Glomerular sonicate | 107·9 | 5·9 | 0·0 | 19·0 | 20·6 | 19·5 | 0·0 | 5·3 | 13·0 | 3·0 |
| Goodpasture antigen | 127·4 | 5·9 | 5·0 | 4·8 | 2·9 | 19·6 | 0·0 | 5·3 | 13·0 | 15·2 |
| Hemocyanin | 678·4 | 0·0 | 5·0 | 9·5 | 2·9 | 98·4 | 0·0 | 0·0 | 30·4 | 9·1 |
| Heparin | 804·7 | 0·0 | 5·0 | 4·8 | 2·9 | 90·8 | 0·0 | 10·5 | 8·7 | 3·0 |
| Histone H1 | 83·1 | 5·9 | 5·0 | 23·8 | 41·2 | 9·4 | 0·0 | 5·3 | 30·4 | 18·2 |
| Histone H2A | 111·7 | 0·0 | 5·0 | 19·0 | 20·6 | 19·7 | 0·0 | 5·3 | 8·7 | 12·1 |
| Histone H2B | 170·0 | 0·0 | 10·0 | 28·6 | 44·1 | 25·9 | 0·0 | 0·0 | 21·7 | 21·2 |
| Histone H3 | 375·6 | 0·0 | 5·0 | 9·5 | 11·8 | 162·0 | 0·0 | 5·3 | 13·0 | 6·1 |
| Histone H4 | 404·3 | 0·0 | 5·0 | 23·8 | 26·5 | 135·4 | 0·0 | 0·0 | 8·7 | 6·1 |
| HSPG | 2962·5 | 0·0 | 5·0 | 14·3 | 0·0 | 102·5 | 0·0 | 5·3 | 17·4 | 9·1 |
| Intrinsic factor | 873·3 | 0·0 | 5·0 | 0·0 | 2·9 | 102·0 | 0·0 | 0·0 | 13·0 | 9·1 |
| JO-1 | 354·4 | 0·0 | 5·0 | 9·5 | 29·4 | 170·3 | 0·0 | 5·3 | 17·4 | 6·1 |
| Ku (p70/p80) | 238·1 | 5·9 | 0·0 | 28·6 | 35·3 | 45·8 | 0·0 | 0·0 | 17·4 | 33·3 |
| La/SS-B | 474·3 | 5·9 | 5·0 | 42·9 | 17·6 | 36·9 | 0·0 | 0·0 | 39·1 | 21·2 |
| Laminin | 2385·5 | 0·0 | 5·0 | 14·3 | 0·0 | 71·0 | 0·0 | 0·0 | 13·0 | 6·1 |
| Myosin | 548·7 | 0·0 | 0·0 | 0·0 | 8·8 | 72·7 | 0·0 | 0·0 | 13·0 | 6·1 |
| PCNA | 282·1 | 0·0 | 5·0 | 19·0 | 29·4 | 138·8 | 0·0 | 5·3 | 21·7 | 15·2 |
| Ribosomal phosphoprotein | 86·3 | 0·0 | 5·0 | 52·4 | 58·8 | 68·8 | 0·0 | 5·3 | 21·7 | 9·1 |
| Ro/SS-A (52KDs) | 766·4 | 0·0 | 5·0 | 33·3 | 35·3 | 441·6 | 0·0 | 5·3 | 21·7 | 6·1 |
| Ro/SS-A (60KDa) | 272·9 | 5·9 | 5·0 | 38·1 | 41·2 | 15·6 | 0·0 | 5·3 | 39·1 | 33·3 |
| Scl-100 | 868·6 | 5·9 | 0·0 | 9·5 | 8·8 | 108·7 | 0·0 | 0·0 | 13·0 | 6·1 |
| Scl-70 | 433·4 | 0·0 | 5·0 | 14·3 | 11·8 | 36·4 | 0·0 | 0·0 | 21·7 | 18·2 |
| ssDNA | 51·1 | 0·0 | 0·0 | 23·8 | 44·1 | 5·1 | 0·0 | 0·0 | 0·0 | 3·0 |
| Thyroid peroxidase | 1303·0 | 0·0 | 5·0 | 4·8 | 8·8 | 51·1 | 0·0 | 5·3 | 8·7 | 6·1 |
| Tissue transglutaminase | 244·8 | 0·0 | 5·0 | 9·5 | 26·5 | 52·0 | 0·0 | 0·0 | 8·7 | 12·1 |
| Total histone | 176·5 | 0·0 | 5·0 | 14·3 | 26·5 | 26·2 | 0·0 | 10·5 | 17·4 | 12·1 |
| U1-snRNP-68 | 374·7 | 0·0 | 5·0 | 4·8 | 52·9 | 61·6 | 0·0 | 5·3 | 17·4 | 27·3 |
| U1-snRNP-A | 45·5 | 0·0 | 5·0 | 23·8 | 73·5 | 3·6 | 5·9 | 5·3 | 39·1 | 54·5 |
| U1-snRNP-BB’ | 89·3 | 0·0 | 0·0 | 52·4 | 70·6 | 14·1 | 0·0 | 0·0 | 30·4 | 45·5 |
| U1-snRNP-C | 61·5 | 0·0 | 5·0 | 28·6 | 61·8 | 7·9 | 0·0 | 0·0 | 17·4 | 39·4 |
| Vimentin | 99·6 | 0·0 | 10·0 | 9·5 | 17·6 | 8·7 | 0·0 | 0·0 | 21·7 | 15·2 |
| Vitronectin | 155·2 | 0·0 | 0·0 | 33·3 | 20·6 | 59·3 | 0·0 | 5·3 | 13·0 | 6·1 |
A total of 93 samples were tested; non-autoimmune controls (NC) = 17, first-degree relatives (FDR) = 20, incomplete lupus (ILE) = 23, systemic lupus erythematosus (SLE) = 33.
GBM, glomerular basement membrane; HSPG, heparan sulphate proteoglycan; PCNA, proliferating cell nuclear antigen.
Cut-off was set as mean value plus 3 s.d. of all samples in NC group for each antigen. Values represent % above cut-off value.
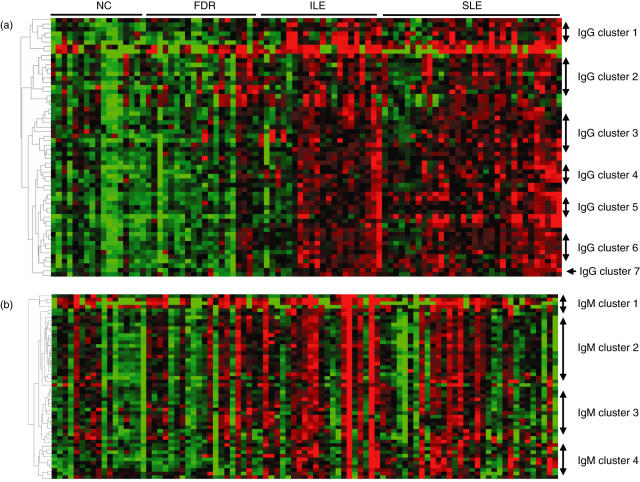
The 57 IgG autoantibody specificities across all serum samples were then analysed using hierarchical clustering. The IgG autoantibody heat map shows that most of the samples in the ILE and SLE groups have elevated antibody levels for a majority of the autoantigens (Fig. 1a). Most clusters included antibodies which showed relatedness to each other, making this a useful approach for examination of correlations with disease features. Seven clusters can be distinguished from the IgG autoantibody heat map. Cluster 1 is comprised of seven antigens including SSA(Ro), SSB(La), Jo-1, microglobulin and tRNA synthetases. Cluster 2 consists of 12 antibodies including fibrinogens, haemocyanin, thyroglobulin, matrigel, laminin, heparin, hyaluronic acid, Goodpasture antigen, heparin, haemocyanin, gliadin and proteoglycan. Cluster 3 contains 12 specificities including collagens I, II and III, cytochrome C, myosin, Scl100, CENP-B, CENP-A, proliferating cell nuclear antigen, vitronectin, tissue transglutaminase and intrinsic factor. Cluster 4 contains seven antigens including chromatin, dsDNA, ssDNA, glomerular sonicate, Ku and glomerular basement membrane (GBM) disassociated. Cluster 5 has eight antigens, including U1sn-RNPs (A, BB, C, 68KDa), liver cytosolic protein type 1 (LC1), GBM, β2-glycoprotein I and ribosomal phosphoprotein P0. Cluster 6 includes nine antigens including all the histones (total, H1, H2A, H2B, H3 and H4), Scl70 and vimentin. Cardiolipin and chondroitin sulphate were separated from all of the other clusters and were designated as cluster 7.
Fig. 1.
(a) Heat map of the 58 IgG autoantibody reactivities in serum samples. The average signal intensity of each antigen in each sample was normalized to the average intensity of total IgG which was printed in six replicates on the arrays as internal control. The normalized fluorescent intensity (nfi) data were used to generate the heat map. For each antigen (Ag), the reactivity intensities are depicted on a relative scale, where reactivities above the mean of all samples are coloured red, reactivities below are coloured green and reactivities close to the mean are black. The left margin dendrogram indicates seven distinct clusters of Ags, with reactivities that clustered together in the tested samples. (b) The heat map of the 50 IgM autoantibody reactivities in four groups of sera, normalized and clustered as described in (a). The left margin dendrogram indicates four distinct clusters of Ags.
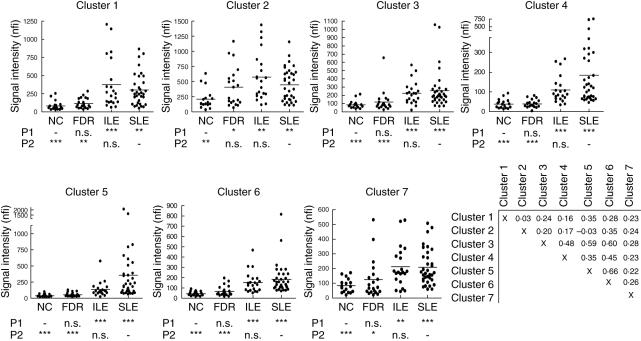
IgG autoantibody reactivities in each cluster were compared between the different sample groups. For all autoantibody clusters, no statistically significant differences were found between ILE and SLE, and only cluster 2 showed a difference between FDR and NC (P < 0·05). However, all autoantibody clusters showed significantly increased reactivity level in ILE and SLE compared with the two unaffected groups (P < 0·01). Most of the IgG autoantibody clusters demonstrated a stepwise increase in antibody reactivities following disease burden from NC, FDR, ILE and SLE, with the SLE patients exhibiting the highest level of IgG autoantibody in most cases (Fig. 2).
Fig. 2.
Average normalized fluorescent intensity (nfi) of the autoantibodies for the four subject groups are plotted. Reactivities of the seven IgG autoantibody clusters, as defined in Fig. 1a. Cluster 1 contains eight antibodies (β2-microglobulin, Ro/SSA 50KDa, Ro/SSA 62KDa, La/SSB, Jo-1, threonyl-tRNA synthetase and alanyl-tRNA synthetase); cluster 2 contains 12 antibodies (hyaluronic acid, Goodpasture antigen, heparin, haemocyanin, gliadin, fibrinogen S, fibrinogen IV, thyroid peroxidase, thyroglobulin, matrigel, laminin and heparin sulphate proteoglycan); cluster 3 contains 12 antibodies (collagen I, collagen II, collagen III, cytochrome C, myosin, PM/Scl100, CENP-B, CENP-A, proliferating cell nuclear antigen, vitronectin, tissue transglutaminase and intrinsic factor); cluster 4 contains seven DNA-related antibodies (chromatin, dsDNA, ssDNA, glomerular sonicate, Ku, GBM disassociated, collagen IV); cluster 5 contains eight antibodies (U1-snRNP-A, U1-snRNP-BB′, U1-snRNP-C, U1-snRNP-68, LC1, GBM, β2-glycoprotein I and ribosomal phosphoprotein P0); cluster 6 contains nine antibodies (total histone, histones H1, H2A, H2B, H3, H4, C1q, Scl70 and vimentin); cluster 8 contains two antibodies (cardolipin and chondroitin sulphate). A one-way analysis of variance (anova) non-parametric test was performed to compare all groups with non-autoimmune controls (NC) or with systemic lupus erythematosus (SLE), and the results are listed under each plot. P1 represents the comparison of each group to the NC group; P2 compares each group to SLE. *P < 0·05; **P < 0·01; ***P < 0·001, n.s.: not significant. Correlations between the seven IgG autoantibodies are shown in the final panel; values represent Pearson's r.
Relatively high correlations were observed between four of the IgG autoantibody clusters, including clusters 5 and 6 (U1snRNPs and histones, r = 0·66), clusters 3 and 6 (collagens and histones, r = 0·6), clusters 3 and 5 (collagens and U1snRNPs, r = 0·59), clusters 3 and 4 (collagens and DNA-related, r = 0·48) and clusters 4 and 6 (DNA-related and histones, r = 0·45) (Fig. 2).
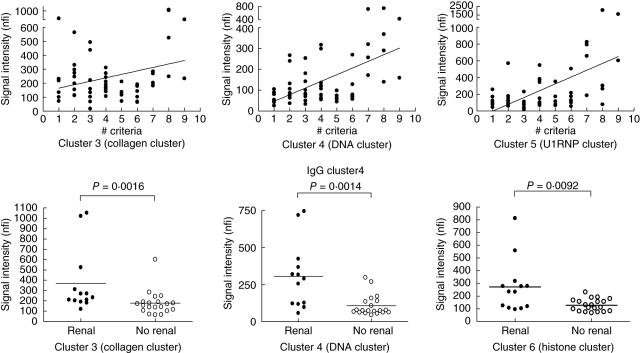
Correlation analyses were carried out in the ILE and SLE groups to compare the IgG autoantibody reactivity level within each cluster with the number of ACR diagnostic criteria fulfilled. Three IgG autoantibody clusters were positively correlated with the number of criteria. Clusters 4 (the DNA cluster) and 5 (U1-snRNP cluster) showed higher correlations with the number of ACR diagnostic criteria for lupus (r-values of 0·50 and 0·52, respectively; P < 0·001; Fig. 3), while cluster 3 (collagens) was weakly correlated with disease criteria (r = 0·28, P = 0·03). The SLE samples were grouped further according to the presence or absence of renal involvement. Samples from SLE patients with renal disease had significantly higher IgG reactivities than those without renal involvement in three of the autoantibody clusters, those related to collagen (cluster 3, P = 0·0016), DNA (cluster 4, P = 0·0014) and histones (cluster 6, P = 0·0092) (Fig. 3).
Fig. 3.
Correlation of IgG autoantibody reactivities with American College of Rheumatology (ACR) diagnostic criteria for systemic lupus erythematosus (SLE) (top panels) and with the presence of renal disease (bottom panels). The autoantibody reactivities in clusters 3, 4 and 5 showed positive correlation with ACR diagnostic criteria. IgG reactivities in clusters 3 (collagen cluster), 4 (DNA cluster) and cluster 6 (histone cluster) were correlated with the presence of renal involvement.
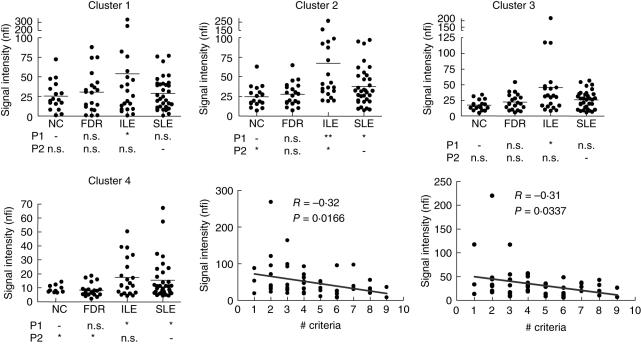
Four IgM autoantibody clusters were identified based on the heat map (Fig. 1b). Generally, the lupus sample groups (ILE and SLE) had higher IgM autoantibody reactivity than the non-lupus groups (NC and FDR) in all clusters (Fig. 4). Interestingly, the ILE group showed significantly higher IgM reactivity than the SLE group for most of the autoantigens. Levels of IgM reactivity in clusters 2 (including DNAs, u1snRNP68, histone H3, H4, collagen I, fibrinogen IV, GBM, Jo-1, Ro/SSA, La/SSB, Scl100, ribosomal phosphoprotein P0, CENP-B) and 3 (collagen II, III and IV, glomerular sonicate, fibrinogen S, matrigel, Scl70, Ku, myosin, total histone and laminin) were correlated negatively with the number of SLE criteria (r-values −0·32 and −0·31, P < 0·05; Fig. 4). No significant differences in IgM reactivities were observed between the SLE groups with and without renal disease (data not shown).
Fig. 4.
IgM autoantibody reactivities in the four subject groups plotted as average normalized fluorescent intensity (nfi). Clusters were defined as in Fig. 1b. IgM cluster 1 contains four antibodies including amyloid, β2-microglobulin, PL12 and PL7; cluster 2 contains 22 autoantibodies including dsDNA, chromatin, RNP-68, histone H3, H4, collagen I, fibrinogen IV, haemocyanin, intrinsic factor, cytochrome C, β2-glycoprotein I, glomerular basement membrane (GBM), GBM disassociate, Jo-1, vitronectin, Ro/SSA (52KDa), La/SSB, PM/Scl100, ribosomal phosphoprotein P0, CENP-B and heparin; cluster 3 contains 16 autoantibodies including collagen II, III and IV, glomerular sonicate, fibrinogen S, matrigel, Scl70, Ku, myosin, tissue transglutaminase (TTG), proliferating cell nuclear antigen (PCNA), liver cytosolic protein type 1 (LC1), TTG, total histone, heparin sulphate proteoglycan (HSPG) and laminin; cluster 4 contains 10 antibodies including RNP-A, RNP-BB′, RNP-C, Ro/SSA (60KDa), histones 1, 2 A and 2B, CENP-A, cardolipin and thyroglobulin. Correlation of IgM autoantibodies with the American College of Rheumatology (ACR) diagnostic criteria in the systemic lupus erythematosus (SLE) and incomplete LE (ILE) samples are shown. The autoantibody reactivities in clusters 2 and 3 were correlated negatively with the ACR diagnostic criteria. A one-way analysis of variance (anova) non-parametric test was performed to compare all groups as described for Fig. 2.
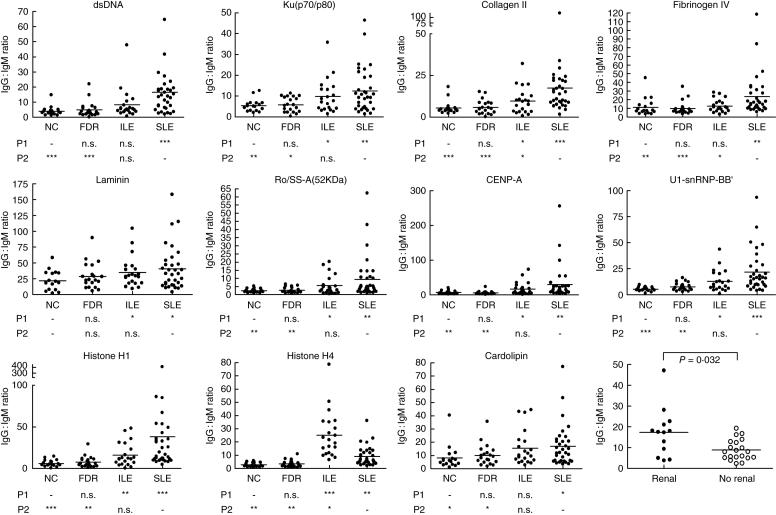
The IgG : IgM ratio calculated for each autoantigen showed a stepwise increase from NC, FDR, ILE to SLE (Fig. 5). The exception was anti-histone H4, which was higher in ILE than in SLE (P < 0·05). The IgG : IgM ratios for autoantigens in IgG cluster 4 (including dsDNA, ssDNA, chromatin, GBM, glomerular sonicate and Ku) were significantly higher in the SLE subset with renal involvement than in the subset without renal involvement (P = 0·032; Fig. 5).
Fig. 5.
Calculated IgG : IgM ratios for 11 autoantigens shown for four subject groups. A one-way analysis of variance (anova) non-parametric test was used to compare the sample groups and results are listed under each plot. P1, all groups compared to non-autoimmune controls (NC); P2, all groups compared to systemic lupus erythematosus (SLE). *P < 0·05; **P < 0·01; ***P < 0·001, n.s., not significant. The IgG : IgM ratio also was compared between the SLE subgroups with and without renal disease for the antigens in IgG cluster 4 (the DNA cluster) using Student's t-test (final panel).
Discussion
The association of a wide range of autoantibodies with lupus has been recognized for many years [12]. Given the clinical heterogeneity of SLE, patterns of autoantibody expression have been very useful in defining subsets of the disease, such as the well-documented sensitivity of antibodies to double-stranded DNA to detect the activity of lupus nephritis [13]. However, until recently the spectrum of autoantibodies that could be reasonably measured was limited by the availability of reagents and by technical obstacles to large-scale assays. Newer approaches that permit measurement of up to a dozen autoantibodies simultaneously using small volumes of serum have entered clinical laboratories [14]. Clustering algorithms using data from these types of assays can be a useful tool for clinical evaluation and prognosis [15]. Yet even the newer multiplex assays include only a small fraction of the autoantibodies that are present in these patients. One recent review suggests that over 100 autoantibodies may be present in SLE patients [16]. This raises the possibility that the relatively small spectrum of antibodies that are routinely measured cannot assess adequately this multi-faceted and heterogeneous disease. Measurements that include a broader range of specificities have potential to provide significant novel insights into diagnosis, pathogenesis and prognosis in patients with SLE and other autoimmune diseases [17,18]. The method used in the present study detected antibodies to 70 different proteins; the slide capacity could accommodate readily more than 100 additional specificities.
Previously, we have reported that the autoantigen array can uncover novel patterns of antibody reactivity, some of which correlate with the presence of glomerulonephritis or with overall disease activity measurements in SLE [9]. These previous studies both confirmed known associations and at the same time suggested novel hypotheses regarding how antibodies might be pathogenic in lupus nephritis. The present findings add further validity to the slide-based approach. For example, the cluster of IgG antibodies to DNA-related antigens was correlated with disease severity and was elevated in subjects with renal involvement, consistent with what is known about these types of antibodies in lupus nephritis [19]. Several novel clusters of IgG autoantibodies were identified to have correlation with either disease criteria or renal involvement. The autoantibodies to the histone cluster (including histones, Scl70 and vimentin) were found to be significantly elevated in SLE sera with renal involvement compared to those without renal disease, which is in agreement with other results indicating that both anti-histone and anti-DNA antibodies are associated with renal disease in SLE [20]. Another cluster that correlated with renal disease includes collagens, consistent with the proposed involvement of antibodies to collagen in the pathogenesis of lupus nephritis [21]. In contrast, antibodies to RNA-related antigens correlated with the number of lupus criteria, but not with renal involvement, consistent with the observation that patients with these autoantibodies tend to have milder renal disease [22]. In addition, the autoantibodies in SLE were largely of the IgG isotype, in agreement with other data indicating that high-affinity IgG antibodies are pathogenic in lupus patients, while IgM antibodies are relatively protective [23].
One unexpected observation in the present study was that IgM autoantibodies were relatively higher in ILE patients than in SLE patients (P < 0·05). The two major clusters of IgM autoantibodies (clusters 3 and 4, including anti-DNA, anti-RNA, anti-histone, anti-collagen and anti-GBM) showed negative correlations with disease criteria, and the IgG : IgM ratio of autoantibodies to DNA-related autoantigens was elevated significantly in SLE samples with renal disease compared to SLE samples without renal involvement. These results suggest that IgM autoantibodies are not pathogenic, and may even be protective [24,25]. In fact, treatment of (NZB × NZW)F1 lupus-prone mice with IgM anti-dsDNA results in a delayed onset of proteinuria and a reduced degree of renal pathology [23].
Two explanations for the increased presence of IgM autoantibodies in ILE might be hypothesized. The first is that because the ILE patients most probably include a subgroup which is on a pathway towards developing SLE, the autoantibody response in these individuals may not yet have matured or switched from IgM to IgG. The second possibility is that ILE patients may represent or include a stable group in which persistence of IgM autoantibodies has a protective effect, blocking some of the more devastating disease manifestations of SLE such as nephritis. This scenario would be consistent with other studies suggesting that ILE represents a good prognosis subgroup [26]. These two hypotheses cannot be evaluated adequately in a cross-sectional sample, but longitudinal analyses that are in progress in our patients should provide further insights.
One conspicuous exception to the predominant IgM profile in ILE was histone H4, which showed a markedly increased IgG : IgM ratio in ILE (Fig. 4). Previous studies have demonstrated that both IgG and IgM antibodies to H1, H2A/H2B and H3 histone proteins were most prevalent among lupus patients, while many of the relatives of the SLE patients had IgM anti-H4 antibodies [27,28]. In addition, IgG and IgA anti-H4 antibodies are uncommon in lupus patient sera compared with antibodies to other individual histones (H1, H2A, H2B, H3) [29]. Our data suggest that the IgG : IgM ratio of anti-histone H4 might be a relatively specific marker for identifying the ILE subset.
The other group of interest in the present study is the FDRs, who are clinically normal yet who show some increases in autoreactivity. In the IgG analysis, for example, cluster 2 (containing hyaluronate, heparin, fibrinogen, matrigel, laminin, thyroid antigens and others) showed significantly greater levels in the FDRs than in the NC group (P < 0·05). Furthermore, in cluster 6 (including antibodies to histones and vimentin), the FDRs included five individuals with levels greater than 100, while none of the NC were reactive at this level (P = 0·0356, Fisher's exact test). A similar analysis of cluster 7 containing cardiolipin and chondroitin sulphate antibodies yielded a borderline level of statistical significance (P = 0·073). These findings with autoreactive antibodies are consistent with other results using gene expression analysis and support the hypothesis that FDRs have a liability for development of autoimmune disease [30].
In summary, the present findings validate the utility of an antigen-based array for detection of autoreactive antibodies in autoimmune patients and their family members. The facility of the assay, its relatively inexpensive cost and the large amount of data generated make it especially attractive for screening individuals in large-scale studies [31,32]. Longitudinal analyses are needed to determine the prognostic value of autoantibody burden as a predictor of poor outcome, especially in the subset of ILE patients who progress to severe disease. Confirmation of the specific autoantibodies of interest will still require additional verification by standard techniques, and the array is not yet an appropriate replacement for the assays currently in clinical use. However, the value of the autoantigen array as a screening tool remains unmatched by other currently available approaches.
Acknowledgments
The authors are grateful for the assistance of Drs Sukumar Narasimhulu and Azza Mutwally Badr for the collection and processing of the blood samples and to Yun Lian for help with data analysis.
References
- 1.Hochberg MC, Rus V. Systemic lupus erythematosus. In: Silman AJHM, editor. Epidemiology of the rheumatic diseases. Oxford: Oxford University Press; 2001. pp. 123–40. [Google Scholar]
- 2.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 3.Egner W. The use of laboratory tests in the diagnosis of SLE. J Clin Pathol. 2000;53:424–32. doi: 10.1136/jcp.53.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 5.Wandstrat AE, Carr-Johnson F, Branch V, et al. Autoantibody profiling to identify individuals at risk for systemic lupus erythematosus. J Autoimmun. 27:153–60. doi: 10.1016/j.jaut.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Vila LM, Mayor AM, Valentin AH, Garcia-Soberal M, Vila S. Clinical outcome and predictors of disease evolution in patients with incomplete lupus erythematosus. Lupus. 2000;9:110–15. doi: 10.1191/096120300678828073. [DOI] [PubMed] [Google Scholar]
- 7.Stahl HC, Nived O, Sturfelt G. Outcome of incomplete systemic lupus erythematosus after 10 years. Lupus. 2004;13:85–8. doi: 10.1191/0961203304lu477oa. [DOI] [PubMed] [Google Scholar]
- 8.Hueber W, Kidd BA, Tomooka BH, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–55. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 9.Li QZ, Xie C, Wu T, et al. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. J Clin Invest. 2005;115:3428–39. doi: 10.1172/JCI23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Linden MW, Westendorp RG, Zidane M, Meheus L, Huizinga TW. Autoantibodies within families of patients with systemic lupus erythematosus are not directed against the same nuclear antigens. J Rheumatol. 2001;28:284–7. [PubMed] [Google Scholar]
- 11.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 12.Reichlin M. ANAs and antibodies to DNA. their use in clinical diagnosis. Bull Rheum Dis. 1993;42:3–5. [PubMed] [Google Scholar]
- 13.Balow JE. Clinical presentation and monitoring of lupus nephritis. Lupus. 2005;14:25–30. doi: 10.1191/0961203305lu2055oa. [DOI] [PubMed] [Google Scholar]
- 14.Shovman O, Gilburd B, Barzilai O, et al. Evaluation of the BioPlex 2200 ANA screen: analysis of 510 healthy subjects: incidence of natural/predictive autoantibodies. Ann NY Acad Sci. 2005;1050:380–8. doi: 10.1196/annals.1313.120. [DOI] [PubMed] [Google Scholar]
- 15.To CH, Petri M. Is antibody clustering predictive of clinical subsets and damage in systemic lupus erythematosus? Arthritis Rheum. 2005;52:4003–10. doi: 10.1002/art.21414. [DOI] [PubMed] [Google Scholar]
- 16.Sherer Y, Gorstein A, Fritzler MJ, Shoenfeld Y. Autoantibody explosion in systemic lupus erythematosus: more than 100 different antibodies found in SLE patients. Semin Arthritis Rheum. 2004;34:501–37. doi: 10.1016/j.semarthrit.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Hueber W, Utz PJ, Steinman L, Robinson WH. Autoantibody profiling for the study and treatment of autoimmune disease. Arthritis Res. 2002;4:290–5. doi: 10.1186/ar426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson WH, DiGennaro C, Hueber W, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 19.Cervera R, Vinas O, Ramos-Casals M, et al. Anti-chromatin antibodies in systemic lupus erythematosus: a useful marker for lupus nephropathy. Ann Rheum Dis. 2003;62:431–4. doi: 10.1136/ard.62.5.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes-Hernandez J, Ordi-Ros J, Labrador M, et al. Antihistone and anti-double-stranded deoxyribonucleic acid antibodies are associated with renal disease in systemic lupus erythematosus. Am J Med. 2004;116:165–73. doi: 10.1016/j.amjmed.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 21.Lefkowith JB, Kiehl M, Rubenstein J, et al. Heterogeneity and clinical significance of glomerular-binding antibodies in systemic lupus erythematosus. J Clin Invest. 1996;98:1373–80. doi: 10.1172/JCI118924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Migliorini P, Baldini C, Rocchi V, Bombardieri S. Anti-Sm and anti-RNP antibodies. Autoimmunity. 2005;38:47–54. doi: 10.1080/08916930400022715. [DOI] [PubMed] [Google Scholar]
- 23.Werwitzke S, Trick D, Kamino K, et al. Inhibition of lupus disease by anti-double-stranded DNA antibodies of the IgM isotype in the (NZB × NZW) F1 mouse. Arthritis Rheum. 2005;52:3629–38. doi: 10.1002/art.21379. [DOI] [PubMed] [Google Scholar]
- 24.Okamura M, Kanayama Y, Amastu K, et al. Significance of enzyme linked immunosorbent assay (ELISA) for antibodies to double stranded and single stranded DNA in patients with lupus nephritis: correlation with severity of renal histology. Ann Rheum Dis. 1993;52:14–20. doi: 10.1136/ard.52.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krippner H, Merle S, Jorgens K, Pirlet K. Antibodies to dsDNA and ssDNA in the immunoglobulin classes IgG and IgM: prognostic value in the course of SLE. Z Rheumatol. 1984;43:265–71. [PubMed] [Google Scholar]
- 26.Swaak AJBH, Smeenk RJ, et al. Incomplete lupus erythematosus: results of a multicentre study under the supervision of the EULAR Standing Committee on International Clinical Studies Including Therapeutic Trials (ESCISIT) Rheumatology (Oxf) 2001;40:89–94. doi: 10.1093/rheumatology/40.1.89. [DOI] [PubMed] [Google Scholar]
- 27.Shoenfeld Y, Segol O. Anti-histone antibodies in SLE and other autoimmune diseases. Clin Exp Rheumatol. 1989;7:265–71. [PubMed] [Google Scholar]
- 28.Shoenfeld Y, Segol G, Segol O, et al. Detection of antibodies to total histones and their subfractions in systemic lupus erythematosus patients and their asymptomatic relatives. Arthritis Rheum. 1987;30:169–75. doi: 10.1002/art.1780300207. [DOI] [PubMed] [Google Scholar]
- 29.Cohen MG, Pollard KM, Webb J. Antibodies to histones in systemic lupus erythematosus: prevalence, specificity, and relationship to clinical and laboratory features. Ann Rheum Dis. 1992;51:61–6. doi: 10.1136/ard.51.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maas K, Chen H, Shyr Y, Olsen NJ, Aune T. Shared gene expression profiles in individuals with autoimmune disease and unaffected first-degree relatives of individuals with autoimmune disease. Hum Mol Genet. 2005;14:1305–14. doi: 10.1093/hmg/ddi141. [DOI] [PubMed] [Google Scholar]
- 31.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–47. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 32.Laustrup H, Heegaard NH, Voss A, Green A, Lillevang ST, Junker P. Autoantibodies and self-reported health complaints in relatives of systemic lupus erythematosus patients: a community based approach. Lupus. 2004;13:792–9. doi: 10.1191/0961203304lu2015oa. [DOI] [PubMed] [Google Scholar]